Abstract
Despite stringent biosecurity measures, infections by bacterial food pathogens such as Campylobacter are a recurrent problem in industrial poultry houses. As the main transmission route remains unclear, persistence of these infections has been linked to bacterial survival and possibly multiplication within protozoan vectors. To date, however, virtually no information is available on the diversity and occurrence of free-living protozoa in these environments. Using a combination of microscopic analyses of enrichment cultures and molecular methods (denaturing gradient gel electrophoresis [DGGE]) on natural samples, we show that, despite strict hygiene management, free-living protozoa are common and widespread throughout a 6-week rearing period in both water and dry samples from commercial poultry houses. Protozoan communities were highly diverse (over 90 morphotaxa and 22 unique phylotypes from sequenced bands) and included several facultative pathogens and known bacterial vectors. Water samples were consistently more diverse than dry ones and harbored different communities, mainly dominated by flagellates. The morphology-based and molecular methods yielded markedly different results: amoebic and, to a lesser degree, ciliate diversity was seriously underestimated in the DGGE analyses, while some flagellate groups were not found in the microscopic analyses. Some recommendations for improving biosecurity measures in commercial poultry houses are suggested.
The microaerobic bacterial genus Campylobacter (Campylobacteraceae) is recognized as the primary cause of food-borne diseases in industrialized countries, with poultry products as one of the main infection sources (3). To prevent contamination of the flocks, stringent biosecurity and control measures are recommended in industrial broiler houses in Europe (26). These include the application of a down period of at least 2 weeks between rearing cycles, during which the pathogen hosts (chickens) are absent and the broiler houses are cleansed and disinfected (23). However, despite these measures, recurrent contamination of flocks is common. Numerous studies have been carried out to identify the main infection sources and transmission routes during the rearing process, but to date these remain unclear (32). Campylobacter strains are not detected in the empty broiler houses after the cleansing and disinfection process (47). Epidemiological investigations have shown that most flocks become infected only 2 to 3 weeks after the introduction of new chicks, ruling out vertical transmission via contaminated eggs as the primary infection source (32). It has also been shown that the presence of animal vectors (e.g., rodents, insects, birds, pets, and livestock) plays only a minor role in the transmission of the pathogens (32). It thus seems that other, as yet not identified, vehicles may be present that allow the bacteria to persist in the environment and/or rapidly colonize the chickens.
There is increasing evidence, mainly derived from in vitro cocultivation studies, that bacteria, including Campylobacter (5, 30, 41), can survive and probably even multiply within protozoa (reviewed, e.g., in reference 43). The bacteria are able to survive protozoan grazing using pre- and postingestion adaptations (31). These interactions may protect the bacteria from unfavorable conditions such as desiccation and disinfection and in this way can have a major impact on their epidemiology, survival, and transmission (30). Some pathogens, such as Legionella pneumophila and Mycobacterium, have been shown to become more virulent after coculture with protozoa (see references 8 and 9).
The present study forms part of a larger investigation aimed at evaluating the role of protozoa in the transmission of Campylobacter jejuni and Campylobacter coli in industrial broiler houses and reports on the results of a comprehensive study on the occurrence and diversity of protozoa in various habitats inside commercial poultry farms. Since to date very little information is available on this topic and on the occurrence of protozoa in anthropogenic environments in general, the main aims of our study were the following: (i) to assess the biodiversity of the protozoan communities present in commercial broiler houses throughout a rearing period; (ii) to evaluate to what degree the spatial and temporal dynamics of these communities can be related to farm-specific environmental conditions and management strategies (including cleansing and disinfection) and within-farm microhabitat; and (iii) to translate these findings into a number of management and biosecurity recommendations. Protozoa are difficult to survey in any locality as many of them are present in a cryptic state; i.e., they are too rare to detect or are present as resting cysts (12). In order to get an inventory of their true diversity, we combined an incubation approach (aimed at enriching the pool of cryptic taxa) and microscopical observations with molecular-genetic fingerprints of the original samples.
MATERIALS AND METHODS
Sampling strategy and collection.
Three farms with different characteristics and management strategies, located in the northern part of Belgium, were selected for investigation (Table 1). Farms A and B had two and farm C had four broiler houses. One-day-old chicks were delivered to the farms and placed on litter composed of commercial packed wood shavings (farms A and B) or straw directly from the field (farm C). Pasteurized feed was supplied by an automatic auger system with feed trays, and water was provided through nipple drinker systems (farm A) or drinking cups (farms B and C). The water at farms A and B came directly from a local well (60-m depth), while farm C used (chlorinated) mains water. All broiler houses had a hygiene barrier with an anteroom and walk-over benches. All three farms took precautions so that possible vectors of bacterial pathogens (such as rodents, wild birds, and other farm animals) could not enter the broiler houses. At farm A, an all in-all out system of breeding was carried out. In the first week of the down period between rotating flocks, litter and remaining feed were removed from the houses. Before the broiler houses were cleansed, all demountable devices (drinking cups and nipples, feeding trays, water pipes, etc.) were removed and cleansed. Cleansing and disinfection procedures are listed in Table 1. One or 2 days before the introduction of the chickens, litter, water, and feed were provided, and the appropriate temperature (around 35°C) was set in the broiler houses.
TABLE 1.
General information and management characteristics of the three farms
| Condition or parameter | Description | Farma
|
||
|---|---|---|---|---|
| A | B | C | ||
| General information | ||||
| Broiler breed | Cob | X | ||
| Ross | X | X | ||
| pH of water tank/pipeline anteroom | Neutral (pH 7.24-7.99) | X | ||
| Alkaline (pH 8.00-8.68) | X | X | ||
| Presence of other animals (industrial animals or pets) | X | X | ||
| Campylobacter statusb | C. jejuni | X | ||
| C. coli | X | X | ||
| Salmonella statusb | Salmonella spp. | X | ||
| Cleansing and disinfection | ||||
| Mechanical cleansing | X | X | X | |
| Demountable devices | Alkali metals (sodium hydroxide) | X | ||
| Sodium hydroxide, nonionic surface-active compounds | X | |||
| Alkali metals | X | |||
| Cleansing water pipes | Hydrogen peroxide, colloidal silver | X | ||
| Hydrogen peroxide | X | |||
| Peracetic acid, hydrogen peroxide | X | |||
| High-pressure and disinfection | Formaldehyde, glutaraldehyde | X | X | |
| Quaternary ammonium, glutaraldehyde, isopropanol, formaldehyde | X | |||
| Disinfection broiler houses | Formaldehyde solution | X | ||
| Formalin atomization | X | X | ||
| Presence of disinfection bath | Quaternary ammonium, glutaraldehyde, isopropanol, formaldehyde; alternatively, peracetic acid, hydrogen peroxide | X | ||
X indicates the presence of the condition or parameter.
Campylobacter and Salmonella status was determined during the third, final sampling phase (Fig. 1).
Sampling took place between August and October 2006. At each farm, samples were collected from the different broiler houses during three phases of a single rearing period (Fig. 1). The first set of samples was taken during the down period 1 to 3 days before the chickens arrived but after decontamination and disinfection, when the temperature was equal to the outside temperature (17.7°C to 20.5°C) (phase 1). The second set was taken 3 to 5 days after the introduction of the chickens, with temperatures ranging between 30.2 and 34.4°C (phase 2). On the last sampling occasion (days 40 to 41) temperatures ranged between 21.1 and 25.5°C (phase 3). Samples were collected from nine different microhabitats within the broiler houses, as follows: (i) 5-liter water samples from the well (only farms A and B); (ii) 2- to 4-liter water samples collected from the pipelines located in the anteroom; (iii) 250-ml water samples from the pipelines located inside the broiler houses; (iv) 50-ml water samples from the drinking nipples or cups; (v) litter; (vi) food; (vii) various samples from dry and moist areas within the broiler houses, including dust, films on the exterior surface of the water pipelines, damp areas on walls, and litter; (viii) cecal droppings/feces (only phases 2 and 3); and (ix) condensation water within the broiler houses. Where possible, 2 to 9 replicates were taken per microhabitat per sampling phase. All samples from microhabitats 1 to 4 are collectively referred to as water samples/habitat while samples from microhabitats 5 to 9 are termed dry samples/habitat. In total, 388 samples (including replicates) were collected. All samples were taken with sterile equipment and collected in sterile flasks. pH was measured from water collected from the pipelines located in the anteroom (Table 1). Aliquots of the samples from dry microhabitats were diluted in 10 ml of sterile demineralized water. Cecal droppings collected on the last sampling phase were investigated for the presence of Campylobacter and Salmonella according to the method of Rasschaert et al. (37). DNA from water samples and diluted solid samples were collected on 0.22-μm-pore-size white GSWP filters (Millipore) and stored at −20°C.
FIG. 1.
Sampling strategy. Arrows indicate the three sampling phases. The bold line corresponds to the period when decontamination and disinfection methods were applied.
Morphology-based diversity assessment of enrichment cultures.
Aliquots (600 to 750 μl) of all samples were put in petri dishes and diluted by adding 30 to 40 ml of water from the pipelines in the anteroom (farms B and C) or from the well (farm A). The water was filtered through 0.22-μm-pore-size filters, and the suspension was then enriched by the addition of a sterile, uncooked rice grain as a carbon source to stimulate bacterial growth (7). The cultures were stored at constant room temperature and shielded from direct sunlight (36). Protozoan diversity and occurrence (presence/absence) was determined 1 week after enrichment using upward and inverted microscopy (Olympus CX41 and CKX41, respectively, both bright-field and phase-contrast microscopy). Live protozoa were identified on the basis of morphology and locomotion using standard identification methods (14, 16, 17, 18, 19, 20, 33, 35, 40). Fast-moving free-living ciliates and flagellates were slowed down by using petroleum jelly (Vaseline) (15). If necessary for identification, organisms were fixed with lugol-formalin saturated with sodium tetraborate-sodium thiosulfate (39). Taxa were identified down to genus or species levels where possible. All taxa were classified according to the recent eukaryote classification of Adl et al. (1). Organisms that could not be assigned to a known species or genus were assigned to a morphogroup (ciliate, flagellate, or amoeboid).
Molecular community profiling using DGGE.
As preliminary analyses of a selection of samples negative for the microscopic analyses also proved to be negative for DNA (Table 2), no further molecular analyses were performed on other samples that were negative for microscopy. Pilot denaturing gradient gel electrophoresis (DGGE) analysis generally yielded similar profiles (data not shown) for replicate samples from the same microhabitat (taken within the same broiler house on the same sampling occasion). We therefore selected only a few replicates per microhabitat (total number of samples, 97) and pooled PCR products from the same microhabitats (from the same broiler house on the same sampling occasion). This finally resulted in 30 (pooled) PCR products that were analyzed using DGGE. Seven of these still turned out to be negative.
TABLE 2.
Occurrence of protozoa in commercial broiler houses ordered by farm and habitat for both the microscopic (morphology) and PCR-based analyses
| Habitat and feature | Prevalence of protozoa by farm and method of analysis (no. of positive samples/total no. of samples [%])a
|
|||||
|---|---|---|---|---|---|---|
| Farm A
|
Farm B
|
Farm C
|
||||
| Morphology | PCR | Morphology | PCR | Morphology | PCR | |
| Water | ||||||
| Well | 1/1 | 1/1 | 2/2 | 0/1 | * | * |
| Pipelines in the anteroom | 7/7 | 1/3 | 4/4 | 0/1 | 10/12 | 1/5 |
| Pipelines inside the broiler houses | * | * | 6/6 | 2/2 | 17/17 | 6/6 |
| Drinking nipples or cups | 30/30 | 2/10 | 36/36 | 5/5 | 102/103 | 24/24 |
| Total for water samples | 38/38 (100) | 4/14 (28.6) | 48/48 (100) | 7/9 (77.8) | 129/132 (97.7) | 31/35 (88.6) |
| Dry | ||||||
| Litter | 0/5 | 0/1 | 1/5 | — | 7/12 | — |
| Food | 0/11 | 0/1 | 2/8 | — | 5/24 | — |
| Dry and moist areas | 6/13 | 5/8 | 8/19 | 6/10 | 25/31 | 10/11 |
| Cecal droppings/feces | 0/8 | — | 0/4 | — | 1/8 | — |
| Condensation water within the broiler houses | 0/17 | 0/4 | * | * | 5/5 | 2/4 |
| Total for dry samples | 6/54 (11.1) | 5/14 (35.7) | 11/36 (30.6) | 6/10 (60.0) | 43/80 (53.8) | 12/15 (80.0) |
| Total for all samples | 44/92 (47.8) | 9/28 (32.1) | 59/84 (70.2) | 13/19 (68.4) | 172/212 (81.1) | 43/50 (86.0) |
*, no samples taken; —, no samples analyzed (see Materials and Methods).
DNA was extracted using the bead-beating method with phenol extraction and ethanol precipitation (52). Extracted DNA was purified on a Wizard column (Promega) according to the manufacturer's recommendations. PCR amplification was carried out using the general eukaryotic primers and cycling program described by van Hannen et al. (48), except that 30 instead of 25 amplification cycles were used. PCR amplification procedures were performed with a Techne Genius temperature cycler (Techne Incorporated, Cambridge, United Kingdom). Each 50-μl mixture contained 4 μl of template DNA, primers at a concentration of 25 pmol, each deoxynucleoside triphosphate at a concentration of 0.3 mM, 1.3 mM MgCl2, 800 ng of bovine serum albumin (Roche Diagnostics GmbH, Germany), 5 μl of 10× PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl) (Invitrogen, Paisley, United Kingdom), and 4U of Taq DNA polymerase (Invitrogen); the mixture was adjusted to a final volume of 50 μl with sterile water (Sigma). The presence of PCR products was determined by analyzing 8 μl of product on 1% agarose gels, staining with ethidium bromide, and comparison with a molecular weight marker (TrackIt 100 bp DNA ladder; Invitrogen).
DGGE was performed with the D-Code system from Bio-Rad Laboratories (Hercules, CA) as described by van Hannen et al. (48). Equal amounts of PCR products (650 ng/μl) were applied to the gels. Electrophoresis was performed for 16 h at 150 V; temperature was set at 60°C. DGGE gels were stained with ethidium bromide and photographed on a UV transillumination table with a Kodak Easy Share P880 camera. For DGGE standards, DNA was extracted from protozoan isolates with an AquaPure Genomic DNA kit (Bio-Rad Laboratories). PCR-DGGE analyses (see above) were used to assess the position of all isolates in the gels, after which a selection of eight bands, covering the entire gel gradient, was pooled to be used as a standard in the DGGE analyses. Three standard lanes were included per gel. The positions of the standards were used to align the digitized DGGE images using a semiautomatic procedure present in the software package Bionumerics, version 5.1 (Applied Maths BVBA). Sequence information of the bands (see below) was used to check the grouping of bands into band classes. All data were combined in a binary (presence/absence) matrix.
Nucleotide sequences of DGGE bands of interest were obtained by direct sequencing of DNA amplicons from excised bands. Sequencing was performed with an ABI Prism kit (PE Biosystems) using the primers 1427F (no GC-clamp) and 1616R (48) and an Applied Biosystems ABI 3130XL genetic analyser. After forward and reverse sequences were combined, bands were identified by screening the partial 18S rRNA gene sequences against GenBank and EMBL sequences using BLAST (4).
Data analysis.
Variation in the diversity and community composition of eukaryotic microorganisms was analyzed using univariate and multivariate statistical techniques, respectively. Because eukaryote-specific primers were used for the DGGE analyses, nonprotozoan protists (such as fungi and some microalgae) were amplified as well. While all DGGE bands were included in the multivariate data analyses, only bands that were positively identified as protozoa were used for Fig. 2B and 3B, so comparisons could be made to the morphology-based diversity data (see Fig. 2A and 3A). The number of data, however, was too low to allow (univariate) statistical analysis of the variation patterns in molecular protozoan diversity.
FIG. 2.
Pie chart showing the relative proportion of hetero- and mixotrophic eukaryotic taxonomic groups as determined by morphology (A) and sequencing of excised DGGE bands (only unique sequences were taken into account) (B).
FIG. 3.
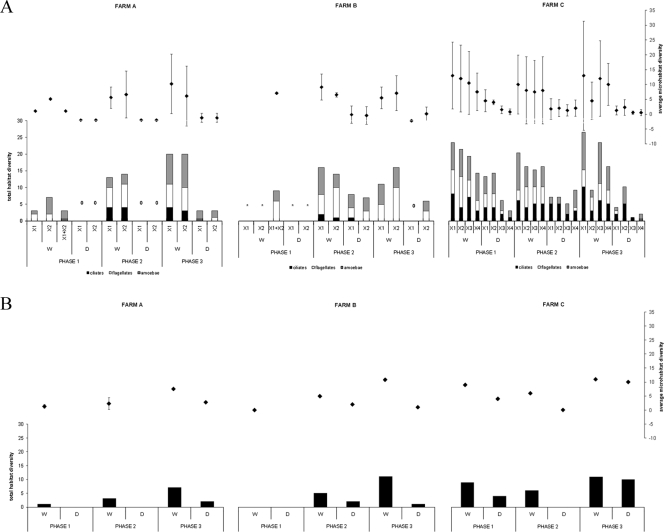
Variability in species richness within a farm during the three phases determined by average microhabitat and total habitat protozoan diversity. Diversity was assessed on the basis of morphology per broiler house (shown for the three main protozoan morphogroups: ciliates, flagellates, and amoebae) (A) and genetic profiling (B). *, no samples could be taken; 0, no protozoa were detected. The different broiler houses at any one farm are designated X1 to X4, and X1+X2 stands for samples from sources common to the indicated broiler houses (e.g., water from the well and litter and food from the supply). Error bars indicate the standard deviations.
Univariate statistical analysis.
Due to the complexity of the data set and missing values, various separate analyses had to be performed in order to test the statistical significance of the observed variation in morphology-based diversity between farms, broiler houses within farms, habitat (water versus dry), and sampling phases (see Fig. 3A). All analyses were performed with SPSS, version 15.0, for Windows; all data were log transformed to satisfy assumptions of normality and equality of variances. We calculated average microhabitat diversity per habitat (i.e., the average taxon richness of the different microhabitats present in the water and the respective dry habitat, calculated for each broiler house and sampling occasion; note that before averaging data for the microhabitats, total diversity was pooled for the different replicates per microhabitat) and total habitat diversity (i.e., cumulative taxon richness present in the water and the respective dry habitat for each broiler house and sampling occasion) (see Fig. 3A). Because no significant differences in microhabitat diversity were observed between the different broiler houses of a farm (see two-way analysis of variance below [ANOVA]), broiler houses (per farm and per sampling phase) were used as replicates in order to investigate differences in average microhabitat and total habitat taxon richness between farms, habitats, and sampling phases (repeated measures analysis of variance and paired t tests below).
First, to test for differences in average microhabitat diversity between broiler houses and habitat within each farm, two-way ANOVA was performed for each farm separately, with broiler house and habitat as fixed factors. No distinction was made between the sampling phases. For these analyses, data were averaged over the three sampling phases, and the microhabitats were considered as replicates. Second, repeated measures (RM) ANOVA was performed to assess whether significant differences in average microhabitat and total habitat diversity existed between farms, sampling phases, and habitat. Farm and habitat were used as fixed factors; broiler houses were considered as replicates (see above). Because of missing data (sampling phase 1 from farm B), only data from sampling phases 2 and 3 were analyzed. Finally, to detect whether significant changes in average microhabitat and total habitat taxon richness occurred throughout the whole rearing period (phases 1 to 3 for farms A and C; phases 2 to 3 for farm B), paired t tests were performed.
Multivariate statistical analysis.
Cluster analysis (unweighted pair group method with arithmetic mean [UPGMA] with the Dice similarity coefficient) (Bionumerics, version 5.1) and ordination techniques (detrended correspondence analysis [DCA]) (CANOCO for Windows, version 4.5) (44) were used to analyze variation patterns in community composition (presence/absence data) of the eukaryotic microbial communities based on morphological (only protozoa) and molecular (all microbial eukaryotes) data. Cluster analysis was used only on the 30 DGGE community fingerprints. As a preliminary DCA using detrending by segments had revealed a strong turnover in species composition between the samples in both data sets (length of gradient of >6 standard deviations) (29), we used this unimodal indirect ordination method for our analyses. After replicate samples were pooled and negative and outlier samples and taxa that occurred only once were omitted, the final DCA data set (morphological data) was reduced to 66 taxa and 70 samples. Farm C (the only farm with enough positive samples for all three sampling phases) was analyzed separately to assess whether the species composition of the protozoan communities changed during the course of the sampling period (46 samples and 59 taxa). For DCA of the molecular data, omitting negative samples left 23 samples and 47 bands to be analyzed.
RESULTS
Bacterial status of the farms.
All farms were Campylobacter positive; C. jejuni was isolated from cecal droppings in farm B (broiler houses X1 and X2) while C. coli was isolated from farm A (X1 and X2) and farm C (X1). Salmonella-positive birds were found only in farm A (X1) (Table 1).
Occurrence of protozoa in commercial broiler houses.
Samples from farm A were more often negative for protists than samples of farm C, both in the microscopic (only protozoa; 48 versus 81%, respectively) and the DGGE analyses (all protists; 32 versus 86%, respectively). Water samples tested more often positive than dry samples, especially using microscopy (Table 2).
Morphology-based diversity assessment of enrichment cultures.
In total, 91 protozoan taxa could be distinguished on the basis of morphology, 60 of which could be identified to the genus or species level (see Table S1 in the supplemental material). Thirty-one forms could not be identified and were assigned to one of the main protozoan morphogroups (ciliate, flagellate, or amoeba). An overview of all taxa, together with their frequencies of occurrence in the data set and their habitat preferences, is given in Table S1 in the supplemental material. Almost half of all organisms were amoeboid (Amoebozoa and amoeboid with unknown affinity), about one-quarter were ciliates (Chromalveolata), and the remainder were flagellates (Excavata, Opisthokonta, Rhizaria [Cercozoa]) (Fig. 2A). The order Euamoebida was represented by the highest number of taxa (18 taxa), followed by the orders Colpodida (4), Schizopyrenida (4), Hymenostomatida (3), and Kinetoplastida (3). The highest number of species was observed in the genera Vannella and Colpoda (4 different species), Hartmannella (3), Chilodonella, Cyclidium, and Vahlkampfia (2). The most frequently encountered taxa were the kinetoplastids Pleuromonas jaculans and Bodo saltans (36 and 30 occurrences, respectively), flagellate 4 in the group of flagellates with unknown affinity (24 occurrences) (see Table S1 in the supplemental material), and the amoeba Vannella sp./Platyamoeba sp. (20 occurrences). Twenty-three taxa were widely distributed and occurred in all three farms; no taxon, however, was common to all nine microhabitats sampled. Twenty-six of all taxa were observed only once. Farm C had the highest overall diversity, with 76 taxa being observed during the whole study period, almost double the number of taxa found at farms A and B (45 and 38, respectively). Ciliates especially were more diverse and common at farm C (Fig. 3A). In all three farms, amoebae were the most diverse group (49 to 56%), followed by flagellates in farms A and B (31 to 39%, respectively) and ciliates (29%) in farm C. While water samples usually contained representatives of the three main morphogroups, ciliates were largely absent from the dry samples from farms A and B.
Differences in average microhabitat and total habitat diversity between the broiler houses within a farm (two-way ANOVA) (see Table S3 in the supplemental material) and also between farms were not significant (RM ANOVA) (see Table S4 in the supplemental material). Both parameters, however, were consistently significantly higher in the water than in the dry samples (RM ANOVA and two-way ANOVA) (see Tables S3 and S4 in the supplemental material). Despite the fact that total habitat diversity seemed to display temporal trends (gradual increase with time in both habitats in farm A and, to a lesser degree, in farm B, and decrease in the dry samples in farm C) (Fig. 3A), these were not significant. Only in farm C was a significant decrease in average microhabitat diversity in the dry samples observed between sampling phases 1 to 3 (paired t tests and RM ANOVA) (see Table S4 in the supplemental material).
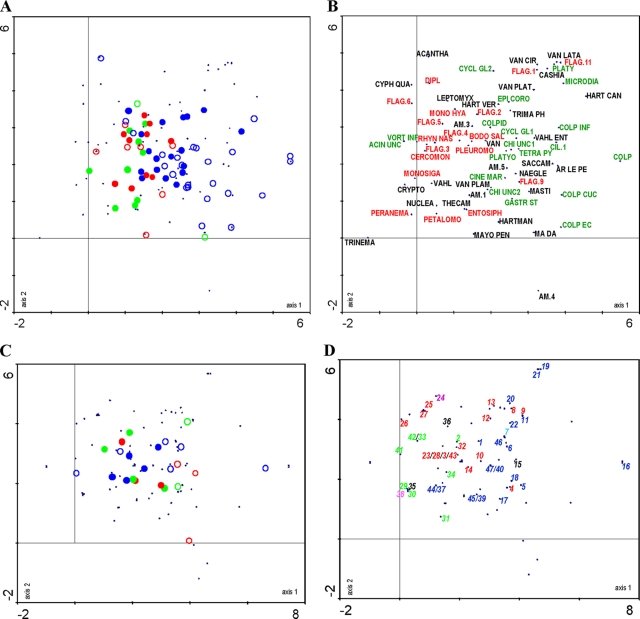
Variation in species composition was investigated using DCA. The first and second axes (Fig. 4A) (70 samples) together explained about 12% of the total variation in the species data. Overall, variation between broiler houses, habitat, and sampling phases within farms was less pronounced in farms A and B than in farm C. Along the first axis, the samples of farms A and B (Fig. 4A, left side) are more or less separated from the samples of farm C (right side). This separation is less pronounced in the water samples, suggesting more similarity in species composition in this habitat type among the three farms. In contrast, variation in species composition in the dry samples is much more pronounced. This is mainly due to the samples of farm C, as the dry samples of farms A and B were often protozoa negative. The positioning of the farm C samples appears to be largely due to the presence of various ciliates in these samples (Fig. 4B). In all three farms, flagellates appear to be largely confined to the water samples, while amoebae do not seem to show any preference for a particular habitat (Fig. 3A and 4A and B). Variation along the second axis could not be interpreted; there was no correlation with temperature (data not shown). This was confirmed by a separate DCA analysis of 46 samples of farm C (see Fig. S1 in the supplemental material). This ordination, which explains 14% of the variation in the data along two axes, mainly confirmed the results of the analysis of all three farms and showed that although slight shifts in species composition did occur during the sampling cycle, these were subordinate to habitat-related differences.
FIG. 4.
DCA ordination plots (axes 1 and 2) summarizing differences and changes in eukaryotic microbial community composition at the three farms, based on morphology (samples in panel A and morphotaxa in panel B) and DGGE (samples in panel C and phylotypes in panel D). In plots A and C, water samples and dry samples are indicated by closed and open circles, respectively, and the sources of the samples are indicated by green, red, and blue for farms A, B, and C, respectively. For taxon abbreviations (plot B) and phylotype numbers (plot D), see Table S1 and Table S2, respectively, in the supplemental material. Ciliates, flagellates, and amoebae are represented in green, red, and black, respectively. In panel D, fungi are indicated in blue, diatoms are in purple, plantae are in turquoise, and Apicomplexa are in pink.
Molecular community profiling using DGGE.
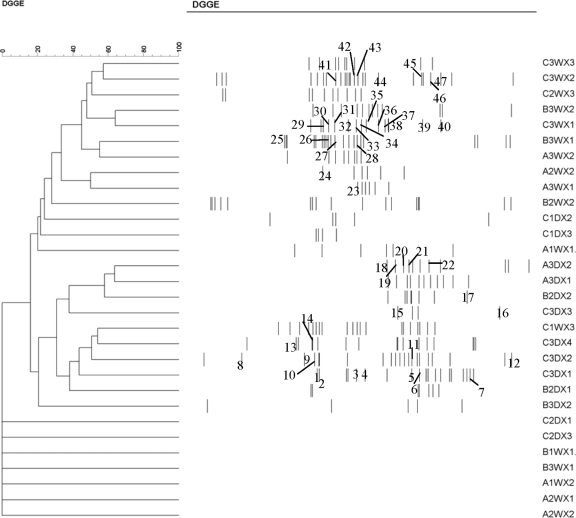
DGGE analyses of 30 (pooled) samples yielded a total of 78 different bands (phylotypes) (Fig. 5); seven samples yielded no bands. Forty-seven bands were excised and sequenced (Fig. 5; see also Table S2 in the supplemental material). Most bands showed high similarity (>97%) to known sequences; almost half of these had a 100% match with an EMBL sequence (see Table S2 in the supplemental material). In total, 41 unique sequences were recovered, belonging to the following groups (see Table S2 in the supplemental material): Amoebozoa (2 sequences), Excavata (2), Chromalveolata (16), fungi (15), plantae (1), and Rhizaria (5). Twenty-two sequences appeared to be affiliated to free-living protozoa (6 ciliates, 12 flagellates, and 4 amoebae), with the majority belonging to the Chromalveolata and Rhizaria (Fig. 2B). Different bands isolated from similar positions in the gradient usually yielded the same sequence. Some slightly different sequences (derived from different positions in the DGGE gels) yielded the same affiliation (e.g., Spumella-like flagellate) (see Table S2 in the supplemental material); these may represent variation at the specific or intraspecific level. The most common phylotypes had sequence affinities with Cyclidium glaucoma, Candida fennica, Chrysophyta sp., Chilodonella uncinata, Spumella-like flagellate, Heteromita globosa, Paraconiothyrium sporulosum, and Candida membranifaciens (Fig. 5; see also Table S2 in the supplemental material). As in the morphology-based analyses, the dry samples of farms A and B contained few or no eukaryotes, in contrast to farm C, where more phylotypes were present (Fig. 3B). Temporal trends in diversity followed those of the morphology-based assessment, but due to the low number of samples, no statistical analyses could be made.
FIG. 5.
Dendrogram generated by an UPGMA cluster analysis comparison of DGGE patterns from direct amplification of samples collected from three farms. In the sample code, A, B, and C refer to the farms, and the numbers 1, 2, and 3 immediately following indicate the sampling phases. W and D refer to water or dry habitat, respectively, while X1, X2, X3, and X4 represent the different broiler houses at one farm. Similarity is expressed as a percentage value of the Dice correlation coefficient. The numbers refer to the excised bands (see Table S2 in the supplemental material).
UPGMA clustering revealed two distinct clusters, one comprising most of the water samples and another containing most of the dry samples. Most of the bands present in water samples showed up in the upper part of the DGGE gels while bands present in dry samples were located mainly in the lower part of the gels. The latter bands correspond mainly to fungal sequences that have a higher GC content (Fig. 5; see Table S2 in the supplemental material). Samples did not cluster according to farm, broiler house, or sampling phase. The first and second axes in a DCA analysis (23 samples) explained about 17% of the total variation in the DGGE data. Similar to results in the morphology-based DCA analysis and confirming the cluster analysis, variation along the first DCA axis was related mainly to habitat type (water versus dry), with water samples being more similar than dry samples (Fig. 4C). Differentiation between farms, however, was less pronounced. The dry samples were characterized mainly by fungi (Fig. 4D), which were not included in the morphology-based analyses. Both flagellates and ciliates appeared to be more typical for the water samples. The number of amoebic sequences was too low to interpret.
DISCUSSION
The involvement of protozoa in the survival and spread of pathogenic bacteria is increasingly recognized (27, 28, 34) and may be especially important for microaerobic prokaryotes like Campylobacter (6). Using a combination of microscopy of enrichment cultures and molecular fingerprinting (DGGE) of environmental samples, we investigated the occurrence and diversity of protozoa during a rearing cycle in commercial broiler houses. Despite the use of stringent disinfection measures between rearing cycles, more than half of all samples tested positive for the presence of protozoa by both methods. In total, 91 different protozoan morphotaxa and more than 22 unique protozoan phylotypes were distinguished that belong to all main morphogroups (amoebae, ciliates, and flagellates) and eukaryotic supergroups (sensu Adl et al.) (1) (see Tables S1 and S2 in the supplemental material). The most frequently encountered genera in both the enrichment cultures (e.g., Bodo, Pleuromonas, Vannella, and Colpoda) and the DGGE analyses (Cyclidium, Chilodonella, Heteromita, and Spumella-like flagellate) are typical bacterivorous components of (organically enriched) aquatic benthic environments and/or soils worldwide (see Table S1 in the supplemental material) (11, 36). This observation and the fact that the protozoan communities differed between farms (compare farms A and B to farm C) suggest that poultry houses do not harbor specialized protozoan faunas. Instead, the communities appear to be selected from local natural environments and may also be strongly determined by farm-specific factors (e.g., the ciliate dominance in farm C) (see below).
That a generally lower number of protozoa was detected using the molecular method than with the microscopic analyses may be due not only to the lower number of samples analyzed using DGGE but also to the use of a general eukaryotic primer set for DGGE. This led to the amplification of many nonprotozoan sequences, especially fungi, which may interfere with the amplification of (rarer) protozoan DNA. Snelling et al. (42), in a molecular study of the eukaryotic microbial diversity in the drinking water supply of broiler farms, also observed mainly fungal sequences. Both methods, however, also displayed other marked differences with respect to the nature of the observed biodiversity. The most striking differences are the almost complete absence of amoebozoan sequences in the DGGE analyses and the absence of heterotrophic flagellates related to Spumella and Heteromita from the microscopic analyses. In addition, the diversity of some groups, notably the ciliates, was significantly underestimated using DGGE. In effect, only a limited number of species (Chilodonella uncinata and Cyclidium glaucoma) and genera (Colpoda sp., Platyophrya sp., Naegleria sp., and Acanthamoeba sp.) was identified with both methods. These discrepancies can at least partly be attributed to well-known biases of both methods: culturing selection and lack of taxonomic resolution and/or expertise in microscopic analyses (38), on the one hand, and DNA extraction, primer and/or PCR bias, and incompleteness of the existing sequence databases (18S ribosomal DNA), on the other (21, 38, 49). The rarity and/or the cryptic nature of a significant part of the protozoan diversity, however, may also play an important role. Many protozoa can exist in a cryptic and/or resting state and become active only when environmental conditions become favorable (e.g., in relation to food availability) (see reference 12). It has recently been shown that certain protozoan cysts, like those of Acanthamoeba, are very resistant to routinely used DNA extraction methods (22) and may therefore be underestimated or even missed in routine molecular surveys. The use of enrichment cultures can activate this cryptic and/or resting part of protozoan diversity (12) and may therefore, in combination with the analysis of the original samples, prove to be essential for obtaining a more complete representation of the true diversity of protozoan communities. Molecular techniques, on the other hand, may be more suitable for detecting fine-scale genetic variability in the region amplified by the primer sets (e.g., in the Spumella-like flagellate group). This other form of (semi)cryptic diversity (13) is virtually impossible to detect during routine microscopic analyses. Our results illustrate that an integrated approach combining molecular techniques and microscopic observations may therefore yield a more realistic picture of microbial diversity (see references 2 and 38).
Variation in diversity and composition of the protozoan communities was mainly related to habitat type (water versus dry). Samples from the water supplies in all farms were more often protozoa positive and had a significantly higher diversity (both average microhabitat and total habitat diversity) than samples from dry microhabitats (damp surfaces or litter, e.g.). In addition, community composition was different between the water and dry samples, with flagellates and ciliates being characteristic of water and dry samples, respectively, and with amoebae appearing more or less indifferent to habitat type. In contrast, the DGGE data indicate that only fungi appear to show a clear preference for habitat (dry samples), with ciliates and flagellates being detected in both water and dry samples. These differences, however, may at least partly be related to the above-described methodological particularities. Overall, the water samples were more similar to one another with respect to species composition, both within and between farms, in contrast to the dry samples, which had a more variable species composition, especially when different farms were compared. Temporal trends in diversity and species composition were subordinate to variation related to habitat type. While no significant differences in diversity were found among the three farms, the ordination analyses show that the species composition at farm C was clearly different from those at the other farms, especially with respect to the samples from the dry habitats, which were dominated by highly variable ciliate assemblages. This difference may be related to the specific type of litter used in farms A and B versus that used in farm C, namely, commercial packed wood shavings and natural straw, respectively. Protozoa, including many ciliates, are common, often as cysts, in natural litter layers (11) and could therefore have been introduced this way into the broiler houses.
To date, very little information is available on the occurrence and diversity of protozoa in commercial poultry houses or in farms in general. The only dedicated study to date (42) applied molecular tools (temperature gradient gel electrophoresis and cloning) on mainly water samples from farms and, for the reasons outlined above, may have missed a substantial part of the protozoan (and especially amoebal) biodiversity present.
There have been more extensive investigations of the presence of free-living protozoa, with a focus on pathogens and associations between protozoa and pathogenic bacteria, in water supplies, especially in anthropogenic environments (10, 46) and recently also in natural environments (10, 34). Our study indicates not only that protozoan communities in commercial broiler houses are highly diverse but also that they include many potentially pathogenic forms, such as members of the genera Acanthamoeba, Hartmannella, Vahlkampfia, and Naegleria (see Tables S1 and S2 in the supplemental material), which can cause infections such as keratitis (50) and also potentially life-threatening infections (25, 45) in humans. In addition, our study revealed the presence of members of the Tetrahymena pyriformis complex and the genus Acanthamoeba, which are known to act as hosts for C. jejuni (5, 30, 41). Likewise, Naegleria and Hartmannella species have also been shown to harbor pathogens like Salmonella and L. pneumophila (24, 51). As all farms tested positive for Campylobacter and farm A also had a Salmonella-positive status, it is not unlikely that these protozoa may act as vehicles that allow the bacteria to persist in the farm environment.
On the basis of our results, some preliminary recommendations for the improvement of biosecurity measures in commercial poultry houses can be formulated. First, it is clear that despite the use of stringent and highly toxic cleansing and disinfection programs in all three farms (Table 1), highly diverse free-living protozoan communities could soon be detected, with many forms being present even before the new flocks were introduced into the broiler houses. At least for farm C, this appears to be related to the use of untreated straw as litter. Given the fact that many protozoa from natural environments may harbor pathogenic microbes (34), the use of natural straw should be avoided. Second, water samples appeared to harbor a consistently more diverse protozoan community than dry samples. Biosecurity measures should therefore be targeted primarily at treating the water supply, which should also be an easier target for treatment.
In conclusion, our study, which to our knowledge constitutes the first comprehensive multimethod inventory of protozoan communities in commercial poultry houses, underscores the need for further studies into the dynamics and, hence, potential contamination pathways of these microorganisms in livestock environments. More specifically, further research is needed to determine to what degree protozoan communities are newly recruited from the surrounding natural environments (e.g., soil, local water supplies, and litter) or whether they can become endemic in the longer term (across multiple rearing cycles). In addition, the in situ occurrence of pathogenic bacteria and, more specifically, Campylobacter inside protozoan cells and cysts needs to be assessed.
Supplementary Material
Acknowledgments
This work was supported by a Ph.D. grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen, Brussels, Belgium) to J.B. and by the Research Fund of Ghent University (BOF, Ghent, Belgium).
We thank Andy Vierstraete for his help with the sequencing and Pieter Vanormelingen for advice on the statistical analyses. Many thanks are also due to the farmers for their valuable cooperation during this study.
Footnotes
Published ahead of print on 5 January 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, Ø. Moestrup, S. E. Mozley-Standrigde, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera, A., F. Gómez, E. Lospitao, and R. Amils. 2006. A molecular approach to the characterization of the eukaryotic communities of an extreme acidic environment: methods for DNA extraction and denaturing gradient gel electrophoresis analysis. Syst. Appl. Microbiol. 29:593-605. [DOI] [PubMed] [Google Scholar]
- 3.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axelsson-Olsson, D., J. Waldenström, T. Broman, B. Olsen, and M. Holmberg. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson-Olsson, D., P. Ellström, J. Waldström, P. D. Haemig, L. Brudin, and B. Olsen. 2007. Acanthamoeba-Campylobacter coculture as a novel method for enrichment of Campylobacter species. Appl. Environ. Microbiol. 73:6864-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, D. A. 1993. Enrichment, isolation, and culture of free-living heterotrophic flagellates, p. 77-89. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 8.Cirillo, J. D., S. Falkow, and L. S. Tompkins. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo, J. D., S. Falkow, L. S. Tompkins, and L. E. Bermudez. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Declerck, P., J. Behets, V. van Hoef, and F. Ollevier. 2007. Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 41:3159-3167. [DOI] [PubMed] [Google Scholar]
- 11.Fenchel, T. 1987. Ecology of protozoa. The biology of free-living phagotrophic protists. Science Tech Publishers, Inc., Berlin, Germany.
- 12.Fenchel, T., G. F. Esteban, and B. J. Finlay. 1997. Local versus global diversity of microorganisms: cryptic diversity of ciliated protozoa. Oikos 80:220-225. [Google Scholar]
- 13.Finlay, B. J. 1998. The global diversity of protozoa and other small species. Int. J. Parasitol. 28:29-48. [DOI] [PubMed] [Google Scholar]
- 14.Foissner, W., H. Blatterer, H. Berger, and F. Kohmann. 1991. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band I: Cyrtophorida, Oligotrichida, Hypotrichia, Colpodea. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, Heft 1/91 Bartels und Wernitz Druck, München, Germany.
- 15.Foissner, W. 1991. Basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa. Eur. J. Protistol. 27:313-330. [DOI] [PubMed] [Google Scholar]
- 16.Foissner, W., H. Berger, and F. Kohmann. 1992. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band II: Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft. Bartels und Wernitz Druck, München, Germany.
- 17.Foissner, W., H. Berger, and F. Kohmann. 1994. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band III: Hymenostomata, Protomatida, Nassulida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft. Bartels und Wernitz Druck, München, Germany.
- 18.Foissner, W., H. Berger, H. Blatterer, and F. Kohmann. 1995. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft. Bartels und Wernitz Druck, München, Germany.
- 19.Foissner, W., and H. Berger. 1996. A user-friendly guide to the ciliates (Protozoa, Ciliophora) commonly used by hydrobiologists as bioindicators in rivers, lakes, and waste water, with notes on their ecology. Freshw. Biol. 35:375-482. [Google Scholar]
- 20.Foissner, W., and F. Wenzel. 2004. Life and legacy of an outstanding ciliate taxonomist, Alfred Kahl (1877-1946), including a facsimile of his forgotten monograph from 1943. Acta Protozool. 43(Suppl.):3-69. [Google Scholar]
- 21.Gast, R. J., M. R. Dennett, and D. A. Caron. 2004. Characterization of protistan assemblages in the Ross Sea, Antarctica, by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2028-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldschmidt, P., S. Degorge, C. Saint-Jean, H. Year, F. Zekhnini, L. Batellier, L. Laroche, and C. Chaumeil. 2008. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br. J. Ophthalmol. 92:112-115. [DOI] [PubMed] [Google Scholar]
- 23.Hald, B., A. Wedderkopp, and M. Madsen. 2000. Thermophilic Campylobacter spp. in Danish broiler production: a cross-sectional survey and a retrospective analysis of risk factors for occurrence in broiler flocks. Avian Pathol. 29:123-131. [DOI] [PubMed] [Google Scholar]
- 24.Harb, O. S., L.-Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 25.Hausmann, K., N. Hülsmann, and R. Radek. 2003. Protistology. Schweizerbart'sche Verlagbuchhandlung, Stuttgart, Germany.
- 26.Humphrey, T., S. O'Brien, and M. Madsen. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 3:237-257. [DOI] [PubMed] [Google Scholar]
- 27.Huws, S. A., A. W. Smith, M. C. Enright, P. J. Wood, and M. R. W. Brown. 2006. Amoebae promote persistence of epidemic strains of MRSA. Environ. Microbiol. 8:1130-1133. [DOI] [PubMed] [Google Scholar]
- 28.Huws, S. A., R. J. Morley, M. V. Jones, M. R. W. Brown, and A. W. Smith. 2008. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiol. Lett. 282:258-265. [DOI] [PubMed] [Google Scholar]
- 29.Jongman, R. H. G., C. J. F. ter Braak, and O. F. R. Van Tongeren. 1995. Data analysis in community and landscape ecology. Cambridge University Press, Cambridge, United Kingdom.
- 30.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matz, C., and S. Kjelleberg. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302-307. [DOI] [PubMed] [Google Scholar]
- 32.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page, F. C. 1988. A new key to freshwater and soil Gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 34.Pagnier, I., D. Raoult, and B. La Scola. 2008. Isolation and identification of amoeba-resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ. Microbiol. 10:1135-1144. [DOI] [PubMed] [Google Scholar]
- 35.Patterson, D. J., and J. Larsen. 1991. The biology of free-living heterotrophic flagellates. Oxford University Press, New York, NY.
- 36.Patterson, D. J. 1998. Free-living freshwater protozoa: a colour guide. Manson Publishing Ltd, London, United Kingdom.
- 37.Rasschaert, G., K. Houf, J. Van Hende, and L. De Zutter. 2007. Investigation of the concurrent colonization with Campylobacter and Salmonella in poultry flocks and assessment of the sampling site for status determination at slaughter. Vet. Microbiol. 123:104-109. [DOI] [PubMed] [Google Scholar]
- 38.Savin, M. C., J. L. Martin, M. LeGresley, M. Giewat, and J. Rooney-Varga. 2004. Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microb. Ecol. 48:51-65. [DOI] [PubMed] [Google Scholar]
- 39.Sherr, E. B., and B. F. Sherr. 1993. Preservation and storage of samples for enumeration of heterotrophic protists, p. 207-212. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, FL.
- 40.Smirnov, A., and S. Brown. 2004. Guide to the methods of study and identification of soil gymnamoebae. Protistology 3:148-190. [Google Scholar]
- 41.Snelling, W. J., J. P. McKenna, D. M. Lecky, and J. S. G. Dooley. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 71:5560-5571. (Author's correction, 71:7631.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snelling, W. J., J. P. McKenna, C. J. Hack, J. E. Moore, and J. S. G. Dooley. 2006. An examination of the diversity of a novel Campylobacter reservoir. Arch. Microbiol. 186:31-40. [DOI] [PubMed] [Google Scholar]
- 43.Snelling, W. J., J. E. Moore, J. P. McKenna, D. M. Lecky, and J. S. G. Dooley. 2006. Bacterial-protozoa interactions; an update on the role these phenomena play towards human illness. Microbes Infect. 8:578-587. [DOI] [PubMed] [Google Scholar]
- 44.ter Braak, C. J. F., and P. Smilauer. 1998. Canoco reference manual and user's guide to Canoco for Windows: software for canonical community ordination (version 4). Microcomputer Power, Ithaca, NY.
- 45.Tsvetkova, N., M. Schild, S. Panaiotov, R. Kurdova-Mintcheva, B. Gottstein, J. Walochnik, A. Horst, M. S. Lucas, and N. Müller. 2004. The identification of free-living environmental isolates of amoebae from Bulgaria. Parasitol. Res. 92:405-413. [DOI] [PubMed] [Google Scholar]
- 46.Vaerewijck, M. J. M., K. Sabbe, J. Baré, and K. Houf. 2008. Microscopic and molecular studies of the diversity of free-living protozoa in meat-cutting plants. Appl. Environ. Microbiol. 74:5741-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Giessen, A. W., J. J. H. C. Tilburg, W. S. Ritmeester, and J. Van Der Plas. 1998. Reduction of Campylobacter infections in broiler flocks by application of hygiene measures. Epidemiol. Infect. 121:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Hannen, E. J., M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 49.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Walochnik, J., A. Obwaller, and H. Aspöck. 2000. Correlations between morphological, molecular biological, physiological characteristics in clinical and non-clinical isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:4408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wildschutte, H., D. M. Wolfe, A. Tamewitz, and J. G. Lawrence. 2004. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc. Natl. Acad. Sci. USA 29:10644-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwart, G., R. Huismans, M. P. van Agterveld, Y. Van de Peer, P. De Rijk, H. Eenhoorn, G. Muyzer, E. J. van Hannen, H. J. Gons, and H. J. Laanbroek. 1998. Divergent members of the bacterial division Verrucomicrobiales in a temperate freshwater lake. FEMS Microbiol. Ecol. 25:159-169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.