Abstract
Salmonella outbreaks from contaminated water and nonanimal foods (e.g., produce) are increasingly reported. To address the environment as a potential source of pathogenic Salmonella, we investigated levels of salmonellae and the geographic and temporal variation of Salmonella serotypes from surface waters in a region of Georgia (United States) with a history of high salmonellosis case rates. Monthly water samples were collected from six stations in the Little River (Upper Suwannee Basin) for 12 months (April 2005 to April 2006). Salmonellae were enumerated using a three-step most-probable-number (MPN) assay. Salmonellae were detected in 57 of the 72 water samples collected (79.2%). Monthly Salmonella densities ranged from an MPN of 2.5 liter−1 in April 2005 to 36.3 liter−1 in August 2005; concentrations were significantly higher in the summer months compared to other seasons (P < 0.05). Concentrations were not significantly different between stations. Levels of salmonellae were correlated with average daily watershed rainfall for the 1 and 2 days preceding each sample collection (r = 0.77 and 0.68, respectively; P < 0.005). Additionally, water temperature was also positively associated with total Salmonella levels (r = 0.44; P < 0.05). In total, 13 S. enterica serotypes were identified among 197 Salmonella isolates. Eighty (40.6%) were identified as S. enterica subsp. arizonae. Muenchen and Rubislaw were the most frequently identified serotypes of the remaining 117 isolates (28 and 26 isolates, respectively). Serotype diversity peaked in the summer, with 9 serotypes observed in August compared to only one serotype (S. enterica subsp. arizonae) observed in April (2005 and 2006) (P < 0.05). Furthermore, all samples collected in August (6/6) contained multiple serotypes (two to five per sample). The results of this study suggest that Salmonella abundance and diversity in the environment vary temporally and are strongly influenced by seasonal precipitation and water temperature.
The Salmonella genus consists of two separate species, Salmonella bongori and Salmonella enterica, and encompasses over 2,500 known serotypes, all of which are considered potential human pathogens (5); however, the majority of human and warm-blooded animal Salmonella infections (∼99%) are caused by relatively few serotypes of Salmonella enterica subsp. enterica (13). Additionally, most Salmonella serotypes must be introduced into human populations from exposure to contaminated foods, soil, water, or animal contact (6, 22, 29, 39, 52, 62).
Although salmonellosis traditionally has been thought of as an animal-origin food-borne disease, many cases and outbreaks have not been linked to any verifiable food sources (1, 19, 25, 33). In addition, recent outbreaks resulting from water and produce contaminated with Salmonella confirm environmental sources of Salmonella contribute to human illness (8, 40, 60) and are consequently now being increasingly investigated as a potentially significant reservoir of Salmonella transmission (52).
The ubiquitous nature of Salmonella and its widespread occurrence in both freshwaters and marine waters suggest that transmission in the aquatic environment from water consumption, contact, or the consumption of food treated with or harvested in contaminated water is highly probable (10, 34, 37, 52). The presence of clinically relevant Salmonella serotypes in natural waters has been well documented (12, 38, 56), and isolation patterns of serotypes in water follow patterns of infection among humans and local fauna, suggesting a local terrestrial origin (2, 7, 43). Sources may vary between wildlife (16, 28, 50), domesticated animals (30, 49), and humans (31), depending on the local land use and waste management facilities. Moreover, Salmonella has been demonstrated to remain viable for longer than many other enteric bacteria in freshwaters (17, 63), suggesting that the aquatic environment may represent a relatively stable environment for these bacteria, thus increasing the probability of environmental exposure.
Cases of salmonellosis follow typical seasonal patterns, with the highest rates noted in summer months (42). Weather or climate variability may be a factor in this seasonality. In particular, precipitation or storm events may mobilize pathogens from agricultural and urban environments as well as on-site sewage disposal systems and subsequently transport them into the aquatic environment (36, 38, 44). Thus, seasonal changes in weather (i.e., temperature and precipitation) along with seasonal changes in carriage rates of Salmonella in local human and fauna populations may influence waterborne Salmonella densities and ultimately exposure to Salmonella from the aquatic environment. Understanding the environmental parameters that influence Salmonella loading into aquatic ecosystems is important in predicting and preventing waterborne transmission of this pathogen. Here, we investigated temporal trends and drivers in Salmonella concentrations and serotype distribution and diversity in the coastal plain of southern Georgia, United States, a region that consistently leads the state in salmonellosis case rates (>50/100,000 in 2007) (26, 27).
MATERIALS AND METHODS
Sampling area.
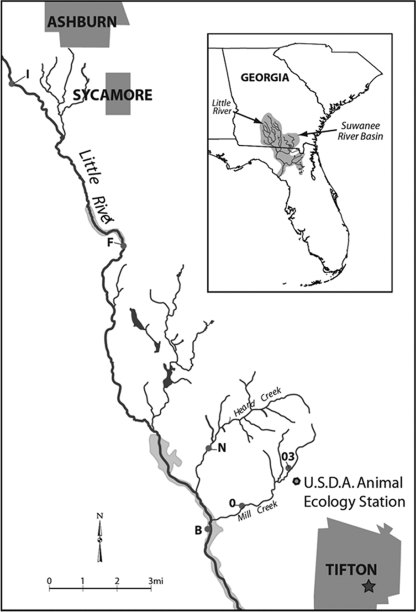
Water samples were collected within the 334-km2 Little River watershed spanning Tift, Turner, and Worth counties located within the Suwannee River Basin of the coastal plain of south Georgia (Fig. 1). The topography is generally flat with meandering streams (11). This watershed receives approximately 120 cm of rainfall each year (55). Precipitation is generally highest in the summer months, but stream flow is at its lowest during this time of year due to high levels of evapotranspiration from croplands (9). Soils in the region range from loamy sands to sandy loams (55). Surface water and shallow groundwater (an unconfined surficial aquifer that is present throughout the southeastern coastal plain) were described by Bosch (9) as being “substantially interconnected,” especially during and after periods of precipitation. The surficial aquifer ranges in thickness from 2 to 10 m in the Little River watershed and is underlain by the relatively impermeable Hawthorne Formation, which prevents the downward movement of surficial groundwater (3, 4). Surface water in this watershed is used for irrigation and recreation; shallow groundwater is used for domestic water supply and irrigation (55). Recreational activities in the Little River include fishing, swimming, and boating. The area is 44% forested, 25% cultivated, 15% pasture, 13% wetlands, and 3% urban (55). The watershed contains intensive row-crop corn, soybean, peanut, and cotton farms (20), as well as approximately 7,700 head of cattle, 500 swine, and two poultry houses that produce approximately 440,000 broiler chickens per year (55).
FIG. 1.
Map of sampling stations in the Little River watershed.
Samples were collected from six stations along the upper reaches of the Little River and its tributaries (Fig. 1). Sites I and F, the most upstream stations, were located on the Little River in Turner County near the city of Ashburn, which has a population of approximately 9,000, with 4,000 housing units, 1,500 septic tanks (38% of all households), and one wastewater treatment plant (57). Sites I and F had drainage areas of 50 km2 and 114.9 km2, respectively. Sites N, B, 03, and O were located in Tift County near Tifton, which has a population of approximately 38,000, with 15,000 housing units, 5,000 septic tanks (33% of all households), and one wastewater treatment plant (57). Site B, the most downstream station, was located on the Little River and had a drainage area of 334.3 km2. Site N was located on Heard Creek and has a drainage area of 15.7 km2. Sites O and 03 were located on Mill Creek. Site O had a drainage area of 15.9 km2. Site 03 was proximal to a University of Georgia research farm containing dairy cattle and crops that are fertilized with liquid dairy cattle manure from a pivot irrigation system. Pivot irrigation systems are common throughout the watershed.
Sample collection.
Water was collected monthly between April 2005 and April 2006 as grab samples (2 liters) in sterile polypropylene bottles. Samples were placed on ice immediately and kept at 5°C until microbiological analyses were completed. Water was analyzed in situ for conductivity (mS cm−1), temperature (°C), pH, dissolved oxygen (DO; mg liter−1), turbidity (nephelometric turbidity units), oxidation reduction potential (mV), fluorescence, and chlorophyll a (μg liter−1) with a YSI 6600 multiparameter sonde (Yellow Springs, OH) by the USDA Southeast Watershed Research Laboratory (Tifton, GA). The YSI probe was placed into the deepest section of the stream to record physicochemical parameters at the time of sample collection.
Microbiological analyses. (i) Fecal indicator bacteria.
Within 24 h of collection, samples were screened for Escherichia coli following U.S. EPA method 1603 (59). Ten milliliters and 1 ml were passed, in duplicate, through 0.45-μm-pore-size mixed cellulose ester membranes and placed on modified mTEC agar plates. Plates were initially incubated at 35 ± 0.5°C for 2 ± 0.5 h and then transferred to a water bath at 44.5 ± 0.2°C for 22 ± 2 h. E. coli cells were enumerated by counting the red and magenta colonies. Samples were also screened for enterococci by membrane filtration using mE agar (Becton Dickson) with indoxyl β-d-glucoside (Sigma-Aldrich, St. Louis, MO) following U.S. EPA method 1600 (58). Plates were incubated at 41 ± 0.5°C for 24 ± 2 h. Enterococci were enumerated by counting all colonies with a blue halo. All bacterial fecal indicators were reported as CFU 100 ml−1. The limit of detection for both indicators was 5 CFU 100 ml−1.
(ii) Salmonellae.
Within 24 h of sample collection, Salmonella spp. were enumerated using a three-step most-probable-number (MPN) assay involving pre-enrichment, enrichment, and selection. Five replicates of three 10-fold dilutions (1 to 100 ml) were pre-enriched with a 1% buffered peptone water solution (pH 7.2 ± 0.2) and incubated at 37°C for 18 to 24 h. One-hundred-microliter aliquots of the overnight enrichments were then added to individual 10-ml aliquots of Rappaport-Vassiliadis broth (Becton Dickinson, Franklin Lakes, NJ) and incubated at 43°C for 24 ± 2 h for selective enrichment of Salmonella. Ten microliters of the Rappaport-Vassiliadis broth enrichment was then streaked onto xylose-lysine-deoxycholate (Becton Dickinson) agar for isolation at 35°C for 24 ± 2 h. Colonies that appeared black were presumptively identified as Salmonella and further streaked to obtain a pure isolate. Positive (Salmonella enterica serotype Berta) and negative (no inoculate) controls were used to ensure the quality of culture results. The limit of detection for Salmonella was an MPN of 1.8 liter−1.
Suspected Salmonella isolates were identified to the genus level using Enterotube II (Becton Dickson) assay. Colonies that were positively identified as Salmonella were grown overnight in 10 ml of tryptic soy broth and then amended with glycerol (LabChem, Pittsburgh, PA) (20% final concentration) and dispensed into 1-ml aliquots before freezing at −80°C. Replicate cultures from each isolate were shipped to the Salmonella Reference Center (SRC; University of Pennsylvania New Bolton Center) for serotyping.
Rainfall data.
Daily precipitation data for each site were obtained from the USDA Southeast Watershed Research Laboratory precipitation archives (http://www.tifton.uga.edu/sewrl/archived_data.htm). Each station, except site 03, was equipped with a rainfall gauge that collected daily precipitation readings for the duration of the study. Site 03 precipitation data were estimated from site O data (USDA Southeast Watershed Research Laboratory, personal communication). The daily precipitation total for each of the 7 days prior to sample collection, the 1-, 2-, 3-, and 4-week totals, and the 4-week daily average prior to sample collection for each site were calculated. Precipitation data were also pooled to estimate the mean for the watershed.
Statistical analyses.
Data were analyzed using SAS release 8.2 (Cary, NC). For all bacterial counts, a value of zero was used for any sample with concentrations below the limit of detection. All microbial counts were log transformed to obtain a normal distribution and for statistical analyses; geometric means were calculated for presentation of data in the text and figures. Pearson linear correlation coefficients were determined to describe the relationships between the transformed Salmonella and fecal indicator bacterial concentrations and between the microbial densities and environmental parameters. Differences in mean microbial densities and environmental parameter means were determined using repeated-measures analysis of variance followed by Tukey's honestly significant differences test. We evaluated differences in the frequencies of recovered serotypes among sampling sites and seasons with Friedman's χ2 test followed by Tukey's honestly significant differences test. Binary logistic regressions were run to determine the relationship between environmental parameters and presence of Salmonella spp. and specific serotypes. For all measures of association, P values of ≤0.05 were considered significant. Only those environmental parameters that showed significant relationships with microbial levels were reported.
RESULTS
Spatial variability.
Twelve samples were collected from each site between April 2005 and April 2006 (72 samples total). E. coli cells were detected in all samples, but concentrations varied significantly by site (Fig. 2) (P < 0.05), with the highest mean E. coli concentrations observed at site 03 (geometric mean, 389 CFU 100 ml−1) and the lowest concentrations observed at site B (geometric mean, 45 CFU 100 ml−1). Similarly, enterococci were detected in all samples, while concentrations varied significantly by site (Fig. 2) (P < 0.05), with the highest levels at site 03 (geometric mean, 1,265 CFU 100 ml−1) and the lowest concentration at site B (geometric mean, 92 CFU 100 ml−1).
FIG. 2.
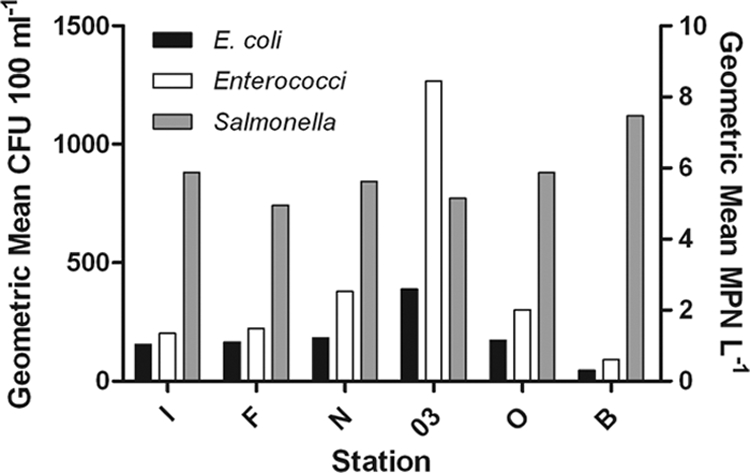
Geometric mean concentrations of fecal indicator bacteria and salmonellae by station. E. coli cells and enterococci are reported as CFU 100 ml−1, and salmonellae are reported as MPN liter−1.
Salmonellae were detected in 79.2% of the samples (57/72), and detection ranged from 75% positive (9/12) at stations I, F, and 03 to 83.3% (10/12) positive at stations N, O, and B. Although not significantly different, the highest geometric mean density was observed at site B (MPN of 7.46 1 liter−1) and the lowest was observed at site F (MPN of 4.96 1 liter−1) (Fig. 2).
Conductivity was the only water quality parameter that varied significantly by site and was significantly higher at site 03 than at all other stations (P < 0.05).
Temporal variability. (i) Environmental parameters.
Estimates of daily rainfall (cm) for each of the 3 days immediately preceding the sampling events were significantly correlated with microbial levels. Other precipitation measures (e.g., any rainfall measurements or totals for ≥4 days prior to the sampling) showed no significant relationships in this study; therefore, the discussion of rainfall effects is limited to data from ≤3 days preceding each sampling event. Total precipitation for the 1, 2, and 3 days preceding the sample collection differed significantly between sampling dates (P < 0.05) (Table 1). The total precipitation for the 1 and 2 days preceding sample collection in August (0.82 cm for both time points) was significantly higher than those of all other months (P < 0.05) (Table 1). The total precipitation for the 3 days preceding sample collection was significantly higher in both August (0.83 cm) and April 2006 (0.38 cm) than those of all other months (Table 1).
TABLE 1.
Mean cumulative precipitation for the 1, 2, and 3 days preceding sample collection and mean water temperature at the time of sample collection
| Sampling moa | Precipitation (cm) for day preceding collectionb
|
Water temp (°C)b | ||
|---|---|---|---|---|
| −1 | −2 | −3 | ||
| 2005 | ||||
| April | 0.00 B | 0.00 B | 0.05 B | 14.0 E |
| May | 0.00 B | 0.00 B | 0.00 C | 17.9 D |
| June | 0.10 B | 0.10 B | 0.26 BC | 22.2 B |
| July | 0.00 B | 0.00 B | 0.26 BC | 25.9 A |
| August | 0.58 A | 0.58 A | 0.83 A | 25.5 A |
| September | 0.00 B | 0.01 B | 0.01 C | 25.8 A |
| November | 0.00 B | 0.00 B | 0.07 C | NRc |
| December | 0.00 B | 0.00 B | 0.00 C | 8.6 G |
| 2006 | ||||
| January | 0.00 B | 0.00 B | 0.00 C | 10.4 F |
| February | 0.00 B | 0.00 B | 0.16 BC | 7.9 H |
| March | 0.00 B | 0.00 B | 0.02 C | 13.7 E |
| April | 0.00 B | 0.00 B | 0.38 AB | 20.3 C |
Samples were not collected in October 2005.
Values with the same letter in the same column were not significantly different.
NR, not recorded.
Mean water temperatures were significantly higher in July (25.9°C), August (25.5°C), and September (25.8°C) than all other months (P < 0.05) (Table 1). The lowest mean temperature was observed in February (7.9°C) and was significantly lower during this month than all other months (P < 0.05) (Table 1). Water temperature data were not recorded in November due to instrument failure. The other measured parameters (conductivity, pH, DO, turbidity, oxidation reduction potential, fluorescence, and chlorophyll a) did not vary significantly over the study period.
(ii) Fecal indicator bacteria.
E. coli concentrations varied significantly by month (P < 0.05). The highest geometric mean E. coli level was 558 CFU 100 ml−1 in June, while the lowest geometric mean level was 50 CFU 100 ml−1 in November. Geometric mean E. coli levels in June were significantly higher than levels in September and November (P < 0.05). Enterococcus levels also varied by month, with the highest geometric mean levels observed in July (712 CFU 100 ml−1), February (657 CFU 100 ml−1), and August (629 CFU 100 ml−1) and the lowest geometric mean levels observed in January (76 CFU 100 ml−1) (P < 0.05).
(iii) Salmonellae.
Salmonellae were detected in all study months; the percentage of samples positive ranged from a low of 33.3% (2/6) positive in April 2005 to 100% (6/6) positive in May, June, July, August, and December of 2005 and March and April of 2006 (Fig. 3). Salmonella densities varied significantly over the study period and were significantly higher in August than all other months except May, June, and March (P < 0.05) (Fig. 3). Over all samples collected, Salmonella densities ranged from nondetectable (MPN of <1.8 1 liter−1) (at least once at each station, excluding site B) to MPN of 68.6 1 liter−1 (site I, August 2005). For all stations, the highest Salmonella densities were observed in August, excluding site N where the highest densities were observed in March. Samples were not collected in October.
FIG. 3.
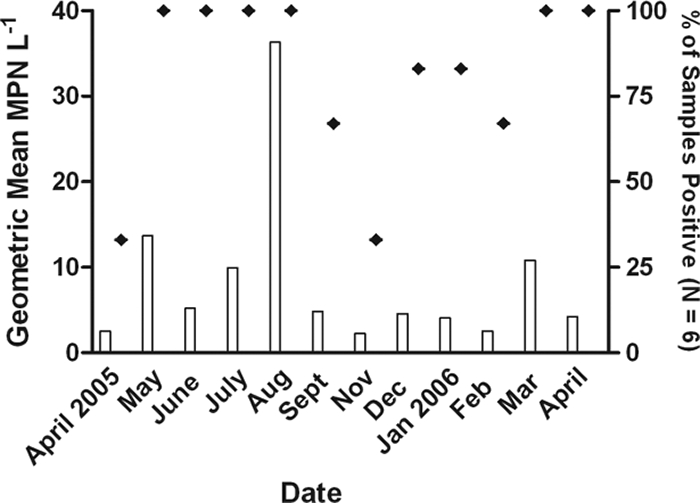
Frequency of detection (⧫) and geometric mean concentrations (bars) of salmonellae for all study sites by month (n = 6). Water samples were not collected in October 2005.
Mean watershed Salmonella densities (n = 12; with all stations averaged for each month) were significantly correlated with the total precipitation for the 1 day (r = 0.77, P = 0.0012) and 2 days (r = 0.68, P = 0.0014) preceding sample collection. When analyzed with all sample points individually (n = 72), Salmonella densities were also positively correlated with rainfall for the 1 day (r = 0.29, P = 0.0151), 2 days (r = 0.29, P = 0.0136), and 3 days (r = 0.17, P = 0.0279) preceding sample collection. Salmonella densities also were positively correlated with water temperature at the time of sample collection (r = 0.44, P < 0.05), which is consistent with its seasonal and monthly distribution.
Salmonella serotype distribution.
In total, 197 Salmonella enterica isolates, representing 13 different serotypes, were recovered from the Little River watershed during the 12-month sampling period (Table 2). More than one serotype was detected in 28 of the 72 water samples collected (38.9%), and more than 2 serotypes were detected in 18 of the samples (25%). In three samples (all collected in August), five serotypes were detected. Eighty of the 197 isolates (40.6%) were identified as Salmonella enterica subsp. arizonae by biochemical analysis. Of the remaining 117 isolates, 109 were identified as Salmonella enterica subsp. enterica and were further identified to serotype (Table 2); 8 isolates were either not typeable or were not typed (2 and 6 isolates, respectively). Muenchen (28 isolates, 14.2%), Rubislaw (26 isolates, 13.2%), and Mikawasima (12 isolates, 6.1%) were the three most frequently isolated serotypes from this watershed.
TABLE 2.
Frequency of isolation among Salmonella serotypes by month
| Serotype | No. (%) of isolates detected ina:
|
Friedman's χ2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2005
|
2006
|
Total | ||||||||||||
| April | May | June | July | August | September | November | December | January | February | March | April | |||
| S. enterica subsp. arizonae | 2 (2.5) | 17 (21.3) | 7 (8.8) | 1 (1.3) | 10 (11.3) | 5 (6.3) | 0 | 1 (1.3) | 10 (12.5) | 0 | 14 (17.5) | 13 (16.3) | 80 | P < 0.05 |
| Muenchen | 0 | 1 (3.6) | 3 (1.1) | 4 (1.3) | 5 (17.9) | 6 (21.4) | 1 (3.6) | 1 (3.6) | 1 (3.6) | 2 (7.1) | 4 (14.3) | 0 | 28 | P < 0.05 |
| Rubislaw | 0 | 3 (11.5) | 3 (11.5) | 9 (34.6) | 1 (3.8) | 3 (11.5) | 3 (11.5) | 2 (7.7) | 0 | 0 | 2 (7.7) | 0 | 26 | P < 0.05 |
| Mikawasima | 0 | 0 | 2 (16.7) | 0 | 5 (41.7) | 0 | 1 (8.3) | 4 (33.3) | 0 | 0 | 0 | 0 | 12 | NSb |
| Braenderup | 0 | 3 (25.0) | 0 | 3 (25.0) | 5 (41.7) | 0 | 0 | 0 | 1 (8.3) | 0 | 0 | 0 | 12 | NS |
| Saint Paul | 0 | 0 | 0 | 1 (11.1) | 4 (44.4) | 1 (11.1) | 0 | 3 (33.3) | 0 | 0 | 0 | 0 | 9 | NS |
| Bareilly | 0 | 2 (33.3) | 0 | 0 | 4 (66.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | NS |
| Liverpool | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 (100) | 0 | 0 | 6 | NS |
| I 4,[5]:b | 0 | 0 | 0 | 0 | 0 | 0 | 4 (100) | 0 | 0 | 0 | 0 | 0 | 4 | NS |
| Gaminara | 0 | 0 | 0 | 1 (33.3) | 2 (66.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | NS |
| Montevideo | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | NS |
| Anatum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 0 | 0 | 1 | NS |
| I 47:z4z23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (100) | 0 | 0 | 1 | NS |
| S. entericac | 0 | 0 | 0 | 0 | 2 (25.0) | 3 (38.0) | 1 (13.0) | 0 | 1 (13.0) | 0 | 1 (13.0) | 0 | 8 | NAd |
| Totale | 1 | 5 | 4 | 6 | 9 | 4 | 4 | 6 | 3 | 3 | 3 | 1 | ||
Samples were not collected in October 2005.
NS, not significantly different between months.
Not typeable (n = 2) or not typed (n = 6).
NA, not applicable.
Does not include those isolates that were untypeable or not typed.
A median of 32 isolates were recovered at each station over the study period and ranged between 27 isolates at station B and 43 isolates at station N. Among these, the number of serotypes found at each station did not differ significantly; as few as four serotypes were detected at station N and a maximum of 8 serotypes were detected at stations I and 03. The most frequently isolated serotypes were Muenchen at station I (15 of 43 isolates recovered) and Rublislaw at station B (9 of 27 isolates recovered); S. enterica subsp. arizonae was most commonly isolated serotype at all other stations.
Isolates recovered by month ranged from a low of 2 in April 2005 to a high of 39 in August. Likewise, most serotypes were detected between May and September and serotype diversity (total number of individual serotypes) was significantly higher in August (9 confirmed serotypes plus two isolates that were not typed) than all other months, as determined by Friedman's χ2 test (χ2 = 46.4, df = 11, P < 0.05) (Table 2). Additionally, in August at least two serotypes were detected in each sample and five serotypes were detected concurrently within three separate samples (stations I, F, and B). Serotype diversity was lowest in April (both 2005 and 2006), when only S. enterica subsp. arizonae was detected (Table 2). Among individual serotypes, the frequency of detection also varied monthly. This was particularly noted in serotypes Rubislaw and Muenchen, which were detected at their highest rates in July and September, respectively (Table 2). S. enterica subsp. arizonae isolation frequencies also differed significantly, with the highest isolation frequency in May (17 isolates) and the lowest in February and November (0 isolates) (P < 0.05) (Table 2). Overall, S. enterica subsp. arizonae showed the broadest temporal range in detection and could be found throughout the year (except for November and February) when few or no other serotypes were isolated.
Water quality parameters.
While salmonellae were detected 100% (10/10) of the time when E. coli thresholds were exceeded and 84% (32/38) of the time when enterococcus thresholds were exceeded (based on the U.S. EPA single-sample limit for infrequently used bathing waters [23]), the pathogen was also found at a high frequency below the threshold standards (76% [42/55] of samples were Salmonella positive at E. coli levels of less than 576 CFU 100 ml−1 and 71% [15/21] were positive at enterococcus levels below 151 CFU 100 ml−1). Furthermore, neither indicator provided significant predictive value by binary logistic regression for the presence of Salmonella (all serotypes) or the clinically relevant Salmonella serotypes (all Salmonella serotypes excluding S. enterica subsp. arizonae, I 4,[5]:b, Mikawasima, and 47:z4z23). E. coli varied inversely with oxidation reduction potential (r = −0.32, P < 0.05), while enterococci were positively correlated with conductivity (r = 0.54, P < 0.05). E. coli and enterococcus concentrations were not significantly correlated with each other or with any rainfall metric.
Salmonellae were not significantly correlated with other measured water quality parameters, except for DO, which was only significant at site O (r = −0.84, P < 0.05).
DISCUSSION
Although the majority of salmonellosis outbreaks have been linked to the consumption of contaminated foods (40), Salmonella is considered a leading cause of waterborne disease outbreaks (33) and is commonly isolated from environmental sources, including surface waters (18, 38, 47). Several large regional outbreaks of salmonellosis have been linked to contaminated water supplies (1, 25, 33) and exposure sources include recreational use, home water use, and consumption of produce irrigated with contaminated water (33, 45, 60).
The Little River watershed is located within Georgia Public Health District 8-1—a region that consistently reports salmonellosis case rates that exceed the mean rate for both the United States and the state of Georgia (14, 15). In 2007, this district had a case rate of 58.3 cases per 100,000, while the rate for the state of Georgia was 21.8 cases per 100,000 and that for the United States was 14.9 cases per 100,000 (14, 15, 26, 27). Salmonella exposure routes in this sparsely populated region are not well defined and frequently not epidemiologically linked to specific food items (Susan Lance, GA Division of Public Health, personal communication). Surface water in this study region is used for irrigation and recreation, and shallow groundwater is used for domestic water supplies and irrigation (55). Groundwater and surface water in this watershed have been described as being substantially interconnected due to the presence of loamy sand soils (9). Septic systems (up to 37% of all households in counties included in this study), pivot irrigation systems, as well as food animal agriculture and wildlife throughout the watershed may all be potential sources of Salmonella in the Little River area.
Within the study area, all stations were influenced by agriculture (25% cultivated land and 13% pastureland) and limited urban development (3% urban); however, station 03 was proximal to a research farm containing cattle and a pivot irrigation system. While significantly higher concentrations of both E. coli and enterococci were observed here, geometric mean Salmonella concentrations did not vary significantly between the six stations; however, the highest number of serotypes was recovered from this station (8, along with station I). This suggests that both close proximity to livestock and agricultural activities as well as non-point sources throughout a rural watershed contribute Salmonella loading to surface water.
The incidence of human salmonellosis peaks in summer months throughout the world (21, 24, 32, 42); although high case rates coincide with peak annual temperatures, it remains unclear which factors drive this seasonality regionally or how this pattern might relate to the presence of the pathogen itself in the environment. The occurrence and persistence of Salmonella in ambient waters may be governed, in part, by extrinsic parameters such as temperature and water chemistry (41, 51, 65) or by seasonal changes in loading from human (43) and animal hosts (53, 54), which may vary spatially and temporally with disease incidence. In the Georgia Coastal Plain, which includes the Little River watershed, rainfall annually peaks in the summer months. Over the study period, both rainfall and temperature were positively correlated with Salmonella levels in the watershed. This suggests that multiple environmental factors, likely including host shedding, enhanced environmental persistence of the pathogen at warm temperatures, and loading during storm events, facilitated the increased prevalence of Salmonella during summer months within this watershed.
In addition to seasonally high concentrations, samples collected in wet and warm summer months also resulted in the highest serotype diversity over the year, with multiple samples containing five different serotypes concurrently. Patterns in serotype prevalence may be related to variable persistence in the environment, with some better suited for survival during warmer months (23, 38, 46, 64). The increase in total serotypes detected may also be related to an overall increase in loading to the watershed during the months with the highest precipitation.
In total, 13 different Salmonella serotypes, including Salmonella enterica subsp. arizonae, were isolated from our study region. This is consistent with other studies of Salmonella spp. in surface waters which have typically isolated less than 20 of the greater than 2,500 known Salmonella serotypes (12, 38, 46, 61). This small range of the environmentally recovered serotypes may reflect the relatively small assortment of serotypes that commonly infect humans and animals as well as differential environmental persistence among serotypes at different temperature ranges. Of note, some of the more common human and animal clinical serotypes such as Enteritidis, Typhimurium, Heidelberg, and I 4,[5],12:i− were not recovered during our study period even though common hosts of these serotypes were present in the watershed (cattle, poultry, and swine) and could reflect lower survival rates of these serotypes in natural environments.
S. enterica subsp. arizonae was the most commonly recovered serotype in this study (40.6% of all isolates) and samples were found consistently throughout the year. Given that this region has a high density of wetlands, results suggest that local reptile populations may be a significant source of total Salmonella in this watershed (35).
Of the 12 remaining serotypes recovered, 9 were common clinical serotypes in Georgia (Anatum, Bareilly, Braenderup, Gaminara, Muenchen, Montevideo, Saint Paul, Rubislaw, and Liverpool) (15). In particular, Muenchen (28 isolates) and Rubislaw (26 isolates) were the most frequently recovered serotypes (following S. enterica subsp. arizonae) in this study and, respectively, represented the 6th and 15th most common serotypes among reported human cases in District 8-1 between 2000 and 2006. The relatively high prevalence of Muenchen in this watershed is of epidemiological significance given its increasing frequency in human clinical cases in Georgia over the past 10 years (34% increase from 1995 to 2004) (13). Furthermore, this serotype has been previously linked to non-animal product-associated outbreaks of salmonellosis (8, 48).
Conclusions.
Salmonella infections continue to cause significant morbidity worldwide, and outbreaks are increasingly associated with nonfood animal products. Defining the potential role of environmental reservoirs of Salmonella in the transmission pathways of human illness is critical to further reduce disease burden. Results from this study suggest that in mixed-land-use areas (such as the Coastal Plain) water could play an important role in the epidemiology of salmonellosis. Seasonal patterns in surface water loading and serotype distribution, influenced by elevated temperatures and rainfall patterns, coincide with increased summer case rates and support the notion of an environmental reservoir for Salmonella.
Acknowledgments
This work was funded by grant NA03OAR4310160 from the National Oceanic and Atmospheric Administration (NOAA) (with NSF, EPA, and EPRI) Joint Program on Climate Variability and Human Health.
This research was not subjected to NOAA review and therefore does not necessarily reflect the views of this agency.
We thank Richard Lowrance and Rex Blanchett for assistance with sample collection and Carrie Futch, Jeff Turner, and Ethell Vereen for laboratory and field assistance.
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Angulo, F., S. Tippen, D. Sharp, B. Payne, C. Collier, J. Hill, T. Barrett, R. Clark, E. Geldreich, H. Donnell, Jr., and D. Swerdlow. 1997. A community waterborne outbreak of salmonellosis and the effectiveness of a boil water order. Am. J. Public Health 87:580-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitidou, M., A. Papa, T. Constantinidis, V. Danielides, and V. Katsouyannopoulos. 1997. The occurrence of Listeria spp. and Salmonella spp. in surface waters. Microbiol. Res. 152:395-397. [DOI] [PubMed] [Google Scholar]
- 3.Asmussen, L. 1971. Hydrologic effects of quaternary sediments above the marine terraces in the Georgia coastal plain. Southeast. Geol. 12:189-201. [Google Scholar]
- 4.Asmussen, L. E., and A. W. Thomas. 1974. Computing phreatic groundwater storage. University of Georgia College of Agriculture Experiment Station Research Bulletin occasional paper 153. University of Georgia College of Agricultural and Environmental Sciences, Athens.
- 5.Baggesen, D. L., D. Sandvang, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barak, J. D., L. C. Whitehand, and A. O. Charkowski. 2002. Differences in attachment of Salmonella enterica serovars and Escherichia coli O157:H7 to alfalfa sprouts. Appl. Environ. Microbiol. 68:4758-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudart, J., K. Lemarchand, A. Brisabois, and P. Lebaron. 2000. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl. Environ. Microbiol. 66:1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boase, J., S. Lipsky, P. Simani, S. Smith, C. Skilton, and S. Greenman. 1999. Outbreak of Salmonella serotype Muenchen infections associated with unpasteurized orange juice—United States and Canada. MMWR Morb. Mortal. Wkly. Rep. 48:582-585. [PubMed] [Google Scholar]
- 9.Bosch, D., R. Lowrance, J. Sheridan, and R. Williams. 2003. Ground water storage effect on streamflow for a southeastern coastal plain watershed. Ground Water 41:903-912. [Google Scholar]
- 10.Brands, D. A., A. E. Inman, C. P. Gerba, C. J. Mare, S. J. Billington, L. A. Saif, J. F. Levine, and L. A. Joens. 2005. Prevalence of Salmonella spp. in oysters in the United States. Appl. Environ. Microbiol. 71:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cashion, J., V. Lakshmi, D. Bosch, and T. Jackson. 2005. Microwave remote sensing of soil moisture: evaluation of the TRMM microwave imager (TMI) satellite for the Little River Watershed, Tifton, Georgia. J. Hydrol. 307:242-253. [Google Scholar]
- 12.Catalao Dionisio, L., M. Joao, V. Soares Ferreiro, M. Leonor Fidalgo, M. García Rosado, and J. Borrego. 2000. Occurrence of Salmonella spp. in estuarine and coastal waters of Portugal. Antonie van Leeuwenhoek 78:99-106. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2005. Salmonella surveillance: annual summary, 2004. CDC, Atlanta, GA. http://www.cdc.gov/ncidod/dbmd/phlisdata/salmonella.htm#2004. Accessed 15 May 2006.
- 14.Centers for Disease Control and Prevention. 2007. Foodnet facts and figures—number of infections and incidence per 100,000 persons: all sites, 1996 to 2007. http://www.cdc.gov/FoodNet/factsandfigures/1.pdf. Accessed 10 July 2008.
- 15.Centers for Disease Control and Prevention. 2007. Foodnet facts and figures—number of infections and incidence per 100,000 persons: all sites, by site 2007. http://www.cdc.gov/FoodNet/factsandfigures/3.pdf. Accessed 10 July 2008.
- 16.Chambers, D. L., and A. C. Hulse. 2006. Salmonella serovars in the herpetofauna of Indiana County, Pennsylvania. Appl. Environ. Microbiol. 72:3771-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao, W., R. Ding, and R. Chen. 1987. Survival of pathogenic bacteria in environmental microcosms. Chin. J. Microb. Immunol. 20:339-348. [PubMed] [Google Scholar]
- 18.Cherry, W., B. Thomason, J. Gladden, N. Holsing, and A. Murlin. 1975. Detection of salmonellae in foodstuffs, feces, and water by immunofluorescence. Ann. N. Y. Acad. Sci. 254:350-368. [DOI] [PubMed] [Google Scholar]
- 19.Clark, R., E. Geldreich, K. Fox, E. Rice, C. Johnson, J. Goodrich, J. Barnick, and F. Abdesaken. 1996. Tracking a Salmonella serovar Typhimurium outbreak in Gideon, Missouri: role of contaminant propagation modelling. J. Water Supply Res. Technol. 45:171-183. [Google Scholar]
- 20.Crandall, C. 1996. Shallow ground-water quality in selected agricultural areas of south-central Georgia, 1994. Water-resources investigations report 96-4083. United States Geological Survey, Reston, VA.
- 21.D'Souza, R. M., N. G. Becker, G. Hall, and K. B. A. Moodie. 2004. Does ambient temperature affect foodborne disease? Epidemiology 15:86-92. [DOI] [PubMed] [Google Scholar]
- 22.Dutka, B., and J. Bell. 1973. Isolation of Salmonella from moderately polluted waters. J. Water Pollut. Control Fed. 45:316-324. [PubMed] [Google Scholar]
- 23.El-Gazzar, F., and E. Marth. 1992. Salmonellae, salmonellosis, and dairy foods: a review. J. Dairy Sci. 75:2327-2343. [DOI] [PubMed] [Google Scholar]
- 24.Fleury, M., D. F. Charron, J. D. Holt, O.B. Allen, and A. R. Maarouf. 2006. A time series analysis of the relationship of ambient temperature and common bacterial enteric infections in two Canadian provinces. Int. J. Biometeorol. 60:385-391. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Villanova Ruiz, B., A. Cueto Espinar, and M. Bolanos Carmona. 1987. A comparative study of strains of Salmonella isolated from irrigation waters, vegetables and human infections. Epidemiol. Infect. 98:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Georgia Department of Human Resources. 2008. Division of Public Health notifiable infectious diseases health statistics query. Salmonellosis. http://www.ph.dhr.state.ga.us:8090/ehi/owa/user_menu.main. Accessed 10 July 2008.
- 27.Georgia Department of Human Resources. 2008. Division of Public Health OASIS web query tool. Population. http://oasis.state.ga.us/.
- 28.Hart, R. P., S. D. Bradshaw, and J. B. Iveson. 1985. Salmonella infections in a marsupial, the quokka (Setonix brachyurus), in relation to seasonal changes in condition and environmental stress. Appl. Environ. Microbiol. 49:1276-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey, T. 2006. Public health aspects of Salmonella enterica in food production, p. 89-107. In D. Maskell and P. Mastreoni (ed.), Salmonella infections: clinical, immunological and molecular aspects. Cambridge University Press, Cambridge, United Kingdom.
- 30.Hutchison, M. L., L. D. Walters, S. M. Avery, F. Munro, and A. Moore. 2005. Analyses of livestock production, waste storage, and pathogen levels and prevalences in farm manures. Appl. Environ. Microbiol. 71:1231-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinde, H., M. Adelson, A. Ardans, E. Little, D. Willoughby, D. Berchtold, D. Read, R. Breitmeyer, D. Kerr, and R. Tarbell. 1997. Prevalence of Salmonella in municipal sewage treatment plant effluents in southern California. Avian Dis. 41:392-398. [PubMed] [Google Scholar]
- 32.Kovats, R. S., S. J. Edwards, D. Charron, J. Cowden, R. M. D'Souza, K. L. Ebi, C. Gauci, P. G. Smidt, S. Hajat, S. Hales, G. H. Pezzi, B. Kriz, K. Kutsar, P. McKeown, K. Mellou, B. Menne, S. O'Brien, W. van Pelt, and H. Schmidt. 2005. Climate variability and campylobacter infection: an international study. Int. J. Biometeorol. 49:207-214. [DOI] [PubMed] [Google Scholar]
- 33.Kramer, M., B. Herwaldt, G. Craun, R. Calderon, and D. Juranek. 1996. Surveillance for waterborne-disease outbreaks—United States, 1993-1994. MMWR Surveill. Summ. 12:1-33. [PubMed] [Google Scholar]
- 34.Lemarchand, K., and P. Lebaron. 2003. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: relationship with fecal indicators. FEMS Microbiol. Lett. 218:203-209. [DOI] [PubMed] [Google Scholar]
- 35.Libby, S. J., M. Lesnick, P. Hasegawa, M. Kurth, C. Belcher, J. Fierer, and D. G. Guiney. 2002. Characterization of the spv locus in Salmonella enterica serovar Arizona. Infect. Immun. 70:3290-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipp, E., N. Schmidt, M. Luther, and J. Rose. 2001. Determining the effects of El Niño-Southern Oscillation events on coastal water quality. Estuaries 24:491-497. [Google Scholar]
- 37.Martinez-Urtaza, J., E. Liebana, L. Garcia-Migura, P. Perez-Piñeiro, and M. Saco. 2004. Characterization of Salmonella enterica serovar Typhimurium from marine environments in coastal waters of Galicia (Spain). Appl. Environ. Microbiol. 70:4030-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Urtaza, J., M. Saco, J. de Novoa, P. Perez-Piñeiro, J. Peiteado, A. Lozano-Leon, and O. Garcia-Martin. 2004. Influence of environmental factors and human activity on the presence of Salmonella serovars in a marine environment. Appl. Environ. Microbiol. 70:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mermin, J., L. Hutwagner, D. Vugia, S. Shallow, P. Daily, J. Bender, J. Koehler, and R. Marcus. 2004. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin. Infect. Dis. 38:253-261. [DOI] [PubMed] [Google Scholar]
- 40.Mohle-Boetani, J., B. Werner, and M. Polumbo. 2002. Outbreak of Salmonella serotype Kottbus infections associated with eating alfalfa sprouts—Arizona, California, Colorado, and New Mexico, February-April 2001. MMWR Morb. Mortal. Wkly. Rep. 51:7-9. [PubMed] [Google Scholar]
- 41.Mouslim, C., F. Hibert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019-1027. [DOI] [PubMed] [Google Scholar]
- 42.Naumova, E. N., J. S. Jjagai, B. Matyas, A. DeMaria, I. B. MacNeill, and J. K. Griffiths. 2006. Seasonality in six enterically transmitted diseases and ambient temperature. Epidemiol. Infect. 135:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen, S., R. Bishop, F. Brenner, T. Roels, N. Bean, R. Tauxe, and L. Slutsker. 2001. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987-1997. J. Infect. Dis. 183:753-761. [DOI] [PubMed] [Google Scholar]
- 44.O'Shea, M., and R. Field. 1992. An evaluation of bacterial standards and disinfection practices used for the assessment and treatment of stormwater. Adv. Appl. Microbiol. 37:21-40. [DOI] [PubMed] [Google Scholar]
- 45.Placha, I., J. Venglovsky, N. Sasakova, and I. Svoboda. 2001. The effect of summer and winter seasons on the survival of Salmonella Typhimurium and indicator microorganisms during the storage of solid fraction of pig slurry. J. Appl. Microbiol. 91:1036-1043. [DOI] [PubMed] [Google Scholar]
- 46.Polo, F., M. Figueras, I. Inza, J. Sala, J. Fleisher, and J. Guarro. 1999. Prevalence of Salmonella serotypes in environmental waters and their relationships with indicator organisms. Antonie van Leeuwenhoek 75:285-292. [DOI] [PubMed] [Google Scholar]
- 47.Polo, F., M. Figueras, I. Inza, J. Sala, J. Fleisher, and J. Guarro. 1998. Relationship between presence of Salmonella and indicators of faecal pollution in aquatic habitats. FEMS Microbiol. Lett. 160:253-256. [DOI] [PubMed] [Google Scholar]
- 48.Proctor, M. E., M. Hamacher, M. L. Tortorello, J. R. Archer, and J. P. Davis. 2001. Multistate outbreak of Salmonella serovar Muenchen infections associated with alfalfa sprouts grown from seeds pretreated with calcium hypochlorite. J. Clin. Microbiol. 39:3461-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray, K., L. Warnick, R. Mitchell, J. Kaneene, P. Ruegg, S. Wells, C. Fossler, L. Halbert, and K. May. 2006. Antimicrobial susceptibility of Salmonella from organic and conventional dairy farms. J. Dairy Sci. 89:2038-2050. [DOI] [PubMed] [Google Scholar]
- 50.Refsum, T., K. Handeland, D. L. Baggesen, G. Holstad, and G. Kapperud. 2002. Salmonellae in avian wildlife in Norway from 1969 to 2000. Appl. Environ. Microbiol. 68:5595-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schutze, G., J. Sikes, R. Stefanova, and M. Cave. 1999. The home environment and salmonellosis in children. Pediatrics 103:1-5. [DOI] [PubMed] [Google Scholar]
- 53.Smith, W. A., J. A. Mazet, and D. C. Hirsh. 2002. Salmonella in California wildlife species: prevalence in rehabilitation centers and characterization of isolates. J. Zoo Wildl. Med. 33:228-235. [DOI] [PubMed] [Google Scholar]
- 54.Srikantiah, P., J. C. Lay, S. Hand, J. A. Crump, J. Campbell, M. S. Van Duyne, R. Bishop, R. Middendor, M. Currier, P. S. Mead, and K. Mølbak. 2004. Salmonella enterica serotype Javiana infections associated with amphibian contact, Mississippi, 2001. Epidemiol. Infect. 132:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suttles, J., G. Vellidis, D. Bosch, R. Lowrance, J. Sheridan, and E. Usery. 2003. Watershed-scale simulation of sediment and nutrient loads in Georgia coastal plain streams using the annualized AGNPS model. Trans. ASAE 46:1325-1335. [Google Scholar]
- 56.Thomason, B. M., J. W. Biddle, and W. B. Cherry. 1975. Detection of salmonellae in the environment. Appl. Environ. Microbiol. 30:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.U.S. Census Bureau. 1990. Sewage disposal for housing units. <http://factfinder.census.gov/home/saff/main.html?_lang=en>. Accessed 15 May 2006.
- 58.U.S. Environmental Protection Agency. 2002. Method 1600: enterococci in water by membrane filtration using membrane Enterococcus indoxyl-β-d-glucoside agar (mEI). EPA-821-R-02-022. U.S. Environmental Protection Agency, Washington, DC.
- 59.U.S. Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA-821-R-02-023. U.S. Environmental Protection Agency, Washington, DC.
- 60.Van Houten, R., D. Farberman, J. Norton, J. Ellison, J. Kiehlbach, T. Morris, and P. Smith. 1998. Plesiomonas shigelloides and Salmonella serotype Hartford infections associated with a contaiminated water supply—Livingston County, New York. MMWR Morb. Mortal. Wkly. Rep. 47:394-396. [PubMed] [Google Scholar]
- 61.Wilson, I., and J. Moore. 1996. Presence of Salmonella spp. and Campylobacter spp. in shellfish. Epidemiol. Infect. 116:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright, R. 1989. The survival patterns of selected faecal bacteria in tropical fresh waters. Epidemiol. Infect. 103:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yam, W., C. Chan, S. Ho Bella, T. Tam, C. Kueh, and T. Lee. 2000. Abundance of clinical enteric bacterial pathogens in coastal waters and shellfish. Water Res. 34:51-56. [Google Scholar]
- 65.Zhuang, R.-Y., L. R. Beuchat, and F. J. Angulo. 1995. Fate of Salmonella montevideo on and in raw tomatoes as affected by temperature and treatment with chlorine. Appl. Environ. Microbiol. 61:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]