Abstract
Human fecal matter contains a large number of viruses, and current bacterial indicators used for monitoring water quality do not correlate with the presence of pathogenic viruses. Adenoviruses and enteroviruses have often been used to identify fecal pollution in the environment; however, other viruses shed in fecal matter may more accurately detect fecal pollution. The purpose of this study was to develop a baseline understanding of the types of viruses found in raw sewage. PCR was used to detect adenoviruses, enteroviruses, hepatitis B viruses, herpesviruses, morbilliviruses, noroviruses, papillomaviruses, picobirnaviruses, reoviruses, and rotaviruses in raw sewage collected throughout the United States. Adenoviruses and picobirnaviruses were detected in 100% of raw sewage samples and 25% and 33% of final effluent samples, respectively. Enteroviruses and noroviruses were detected in 75% and 58% of raw sewage samples, respectively, and both viral groups were found in 8% of final effluent samples. This study showed that adenoviruses, enteroviruses, noroviruses, and picobirnaviruses are widespread in raw sewage. Since adenoviruses and picobirnaviruses were detected in 100% of raw sewage samples, they are potential markers of fecal contamination. Additionally, this research uncovered previously unknown sequence diversity in human picobirnaviruses. This baseline understanding of viruses in raw sewage will enable educated decisions to be made regarding the use of different viruses in water quality assessments.
Millions of viruses and bacteria are excreted in human fecal matter (5, 17, 82), and current methods of sewage treatment do not always effectively remove these organisms (74, 76-78). The majority of treated wastewater, as well as untreated sewage, drains into the marine environment (1) and has the potential to threaten environmental (e.g., nutrients and chemicals) (45) and public (e.g., pathogen exposure via swimming and seafood consumption) (1, 24, 28, 29, 33, 44, 57, 63) health. Currently, the U.S. Environmental Protection Agency (EPA) mandates the use of bacterial indicators such as fecal coliforms and enterococci to assess water quality (75). Although monitoring of these bacteria is simple and inexpensive, it has been shown that fecal-associated bacteria are not ideal indicators of fecal pollution.
Since fecal-associated bacteria are able to live in sediments in the absence of fecal pollution (18, 32, 55), their resuspension into the water column can result in false-positive results and mask correlations between their concentrations and the extent of recent fecal pollution. Another unfavorable characteristic of current bacterial indicators is their inability to predict or correlate with the presence of pathogenic viruses (25, 40, 41, 64, 80). Human-pathogenic viruses associated with feces are generally more robust than enteric bacteria and are not as easily eliminated by current methods of wastewater treatment (43, 80). For example, adenoviruses are more resilient to tertiary wastewater treatment and UV disinfection than are bacterial indicators of fecal pollution (74). Since bacterial indicators cannot accurately depict the risks to human health from fecal pollution, several studies have proposed the use of a viral indicator of wastewater contamination (35, 41, 61).
While it is impractical to monitor the presence of all viral pathogens related to wastewater pollution, the development of an accurate viral indicator of sewage contamination is needed for enhanced water quality monitoring. Enteric viruses (including viruses belonging to the families Adenoviridae, Caliciviridae, Picornaviridae, and Reoviridae) are transmitted via the fecal-oral route and are known to be abundant in raw sewage. These viruses have been used to identify fecal pollution in coastal environments throughout the world (27, 35, 39, 40, 48, 50, 56, 57, 63, 64, 67-69, 71, 80). To determine which viruses are effective indicators of fecal pollution, it is first necessary to establish a broad, baseline understanding of the many diverse groups of eukaryotic viruses in raw sewage. Several studies have identified adenoviruses, noroviruses, reoviruses, rotaviruses, and other enteroviruses (e.g., polioviruses, coxsackie viruses, and echoviruses) in raw sewage in Australia, Europe, and South Africa (30, 47, 58, 76-78). However, no broad baseline data on the presence of eukaryotic viruses in raw sewage in the United States currently exist.
This study determined the presence of 10 viral groups (adenoviruses, enteroviruses, hepatitis B viruses, herpesviruses, morbilliviruses, noroviruses, papillomaviruses, picobirnaviruses, reoviruses, and rotaviruses) in raw sewage samples collected throughout the United States. All viral groups that were detected in raw sewage were then examined further to determine if they were also present in final treated wastewater effluent. These 10 viral groups were chosen because of their potential to be transmitted via the fecal-oral route, suggesting that they might be found in raw sewage. Many of these viruses (excluding adenoviruses, enteroviruses, noroviruses, reoviruses, and rotaviruses) have not been studied in sewage despite their likely presence. Picobirnaviruses have been detected in individual fecal samples (12, 70, 79, 82); however, their presence has never been analyzed in collective waste, nor have they been proposed to be potential markers of fecal pollution. This study identified potential viral indicators of fecal pollution and will have important applications to water quality monitoring programs throughout the country.
MATERIALS AND METHODS
Concentration of viruses in wastewater samples.
Raw sewage and final effluent samples were collected in fall 2007 from 12 wastewater treatment facilities from the coastal United States. One raw sewage sample and one final effluent sample were collected from metropolitan areas in each of the following states: Alabama, California, Connecticut, Louisiana, Maine, Maryland, New Jersey, North Carolina, Oregon, and Washington. Two raw sewage and final effluent samples were collected from different wastewater treatment facilities in the state of Florida. All samples were vortexed prior to filtration to minimize the adhesion of viral particles to suspended solids. For each sample, 10 ml was filtered through a 0.45-μm polyether sulfone membrane filter cartridge (Millipore, Billerica, MA). The filtrate was concentrated to less than 200 μl using a combination of Centriplus YM-50 and Microcon Ultracel YM-30 centrifugal concentration devices (Millipore). Nucleic acid was extracted from the filtered, concentrated sample using the QIAamp MinElute virus spin kit (Qiagen, Valencia, CA). Immediately following nucleic acid extraction, cDNA was synthesized from the extracted RNA using a First-Strand Synthesis Superscript III reverse transcription kit (Invitrogen, Carlsbad, CA) with random hexamers.
PCR for eukaryotic viral groups.
PCR or reverse transcription-PCR for each of the targeted viral groups was executed using previously published primers and conditions (Table 1). All PCR mixtures had a total volume of 50 μl and contained 2 μl of target DNA, 1× REDTaq PCR buffer (10.0 mM Tris-HCl [pH 8.3], 50.0 mM KCl, 1.1 mM MgCl2, 0.01% gelatin; Sigma-Aldrich, St. Louis, MO), 0.25 mM each deoxynucleoside triphosphate, 1 μM of each primer, and 1 U REDTaq DNA polymerase (Sigma-Aldrich), unless otherwise noted.
TABLE 1.
Primer nucleotide sequences used to PCR amplify 10 viral groups
| Human viruses | Primer | Sequence (5′-3′)b | Sensitivity (no. of targets) | Amplicon size (bp) |
|---|---|---|---|---|
| Adenoviruses | AV-A1 | GCC GCA GTG GTC TTA CAT GCA CAT C | 100 | 300 |
| AV-A2 | CAG CAC GCC GCG GAT GTC AAA GT | |||
| AV-B1a | GCC ACC GAG ACG TAC TTC AGC CTG | 143 | ||
| AV-B2a | TTG TAC GAG TAC GCG GTA TCC TCG CGG TC | |||
| Enteroviruses | JP UP | TTA AAA CAG CCT GTG GGT TG | 100 | 600 |
| ENT DOWN | ACC GGA TGG CCA ATC | |||
| ENT UPa | CCT CCG CCC CTG AAT G | 154 | ||
| JP DOWNa | ATT GTC ACC ATA AGC GAC C | |||
| Hepatitis B viruses | HBS-1 | ATC AGG ATT CCT AGG ACC C | 10,000 | 310 |
| HBS-R1 | AGG ACA AAC GGG CAA CAA C | |||
| HBS-11a | GCG GGG TTT TTC TTG TTG AC | 241 | ||
| HBS-R11a | GAA CCA ACA AGA AGA TGA GGC | |||
| Herpesviruses | FP1 | GAY TTY GCI AGY YTI TAY CC | 10 | 800 |
| FP2 | TCC TGG ACA AGC AGC ARI YSG CIM TIA A | |||
| RP1 | GTC TTG CTC ACC AGI TCI ACI CCY TT | |||
| FP3a | TGT AAC TCG GTG TAY GGI TTY ACI GGI GT | 215-235 | ||
| RP2a | CAC AGA GTC CGT RTC ICC RTA IAT | |||
| Morbilliviruses | Up | ATG TTT ATG ATC ACA GCG GT | 100 | 429 |
| Down | ATT GGG TTG CAC CAC TTG TC | |||
| Noroviruses | P290 | GAT TAC TCC AAG TGG GAC TCC AC | 10,000 | 219 |
| P289 | TGA CAA TGT AAT CAT CAC CAT A | |||
| Papillomaviruses | FAP59 | TAA CWG TIG GIC AYC CWT ATT | 10,000 | 478 |
| FAP64 | CCW ATA TCW VHC ATI TCI CCA TC | |||
| Picobirnaviruses (genotype I) | PicoB25 | TGG TGT GGA TGT TTC | 1 | 200 |
| PicoB43 | ART GYT GGT CGA ACT T | |||
| Reoviruses | L1.rv5 | GCA TCC ATT GTA AAT GAC GAG TCT G | 100 | 416 |
| L1.rv6 | CTT GAG ATT AGC TCT AGC ATC TTC TG | |||
| L1.rv7a | GCT AGG CCG ATA TCG GGA ATG CAG | 344 | ||
| L1.rv8a | GTC TCA CTA TTC ACC TTA CCA GCA G | |||
| Rotaviruses (group A) | RV1 | GTC ACA TCA TAC AAT TCT AAT CTA AG | 1,000 | 1,059 |
| RV2 | CTT TAA AAG AGA GAA TTT CCG TCT G | |||
| RV3a | TGT ATG GTA TTG AAT ATA CCA C | 346 | ||
| RV4a | ACT GAT CCT GTT GGC CAW CC |
Internal primer set for a nested PCR.
H equals A/C/T, M equals A/C, R equals G/A, S equals G/C, V equals G/C/A, W equals A/T, and Y equals T/G. Note that inosine (I) has base-pairing bias for C, A, G, and then T.
Adenoviruses.
Nested PCR was used to amplify the hexon gene of 47 different adenovirus serotypes (2). Five microliters of the product from the first round of PCR was used as a template for the second reaction. Both rounds of PCR had an additional 0.4 mM MgCl2 in the reaction mixture. Both adenovirus PCR conditions were 4 min at 94°C, followed by 40 cycles of 92°C for 30 s, 60°C for 30 s, and 72°C for 1 min and a final incubation step at 72°C for 5 min.
Enteroviruses.
Nested PCR capable of detecting at least 25 different enteroviruses was used to amplify the 5′-untranslated region of the enterovirus genome (34). An additional 1.8 mM MgCl2 and an additional 1.4 mM MgCl2 were added to the first- and second-round PCR mixtures, respectively. The first-round PCR conditions were 40 cycles of 95°C for 30 s, 57.7°C for 30 s, and 72°C for 45 s, followed by 5 min at 72°C. One microliter of amplified PCR product was added to the second round of PCR, which was amplified by 40 cycles of 95°C for 30 s, 56.5°C for 30 s, and 72°C for 30 s, followed by 5 min at 72°C.
Hepatitis B virus.
Nested PCR was used to amplify the S gene of hepatitis B viruses (53). Both rounds of PCR had an additional 0.4 mM MgCl2 added to the mixture, and the PCR conditions were as follows: 5 min at 95°C, followed by 30 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 40 s. Two microliters of product from the first reaction was used as a template in the final PCR.
Herpesviruses.
Nested PCR was used to amplify the DNA polymerase genes of eight human herpesvirus strains (11). The second round of PCR used 5 μl of the first-round product. The conditions for both rounds of PCR were 2 min at 94°C, followed by 55 cycles of 94°C for 20 s, 46°C for 30 s, and 72°C for 30 s, followed by incubation at 72°C for 10 min.
Morbilliviruses.
The phosphoprotein gene of morbilliviruses was amplified by PCR (10). The reaction mixture contained an additional 0.4 mM MgCl2, and reaction conditions were 35 cycles of 94°C for 1.5 min, 25°C for 2 min, and 72°C for 2 min, followed by incubation at 72°C for 7 min.
Noroviruses.
The RNA polymerase genes of noroviruses were amplified by PCR (51). The reaction mixture contained an additional 0.4 mM MgCl2 and 100 μg/ml bovine serum albumin. The PCR conditions were as follows: 3 min at 94°C, followed by 40 cycles of 94°C for 30 s, 49°C for 80 s, and 72°C for 1 min, and a final extension step of 72°C for 10 min.
Papillomaviruses.
The L1 gene of papillomaviruses was amplified by PCR that was capable of detecting 87% of human papillomavirus strains (36). The reaction mixture contained an additional 1.4 mM MgCl2 and was incubated for 10 min at 94°C, followed by 45 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, followed by incubation at 72°C for 5 min.
Picobirnaviruses.
A portion of genomic segment 2 of genotype 1 picobirnaviruses was amplified by PCR (70). The picobirnavirus PCR mixture had an additional 0.9 mM MgCl2 and was incubated for 2 min at 94°C, followed by 40 cycles of 94°C for 1 min, 49°C for 2 min, and 72°C for 3 min and then incubation at 72°C for 7 min.
Reoviruses.
Nested PCR was used to amplify the L1 gene of reoviruses (54). The first PCR mixture contained an additional 1.4 mM MgCl2, and 1 μl of the first-round PCR product was added to the second PCR mixture, which contained an additional 0.9 mM MgCl2 and had a total volume of 25 μl. Both PCR mixtures were incubated for 1 min at 94°C, followed by 35 cycles of 94°C for 20 s, 50°C for 30 s, and 72°C for 30 s and then incubation at 72°C for 10 min.
Rotaviruses.
Nested PCR was used to amplify the VP7 gene of group A rotaviruses (38). The initial PCR mixture had an additional 0.4 mM MgCl2 and was incubated for 1 min at 94°C, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by incubation at 72°C for 3 min. The second reaction mixture, containing an additional 2.4 mM MgCl2 and 2 μg/ml bovine serum albumin, was incubated for 1 min at 94°C, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and then incubation at 72°C for 3 min.
Detection and sequencing of PCR positives.
All PCR products were visualized using agarose gel (2%) electrophoresis with ethidium bromide. The presence of inhibitors in each sample was examined by spiking samples with a positive control and analyzing 1:10 serial dilutions to determine if the sample inhibited amplification. To verify the identity of positive assays, PCR products were sequenced. Positive PCR products with one distinct band were purified using the UltraClean PCR Clean-Up kit (Mo Bio Laboratories, Inc., Carlsbad, CA). If more than one band was present, the UltraClean Gelspin kit (Mo Bio Laboratories, Inc.) was utilized to gel purify the positive PCR product of the correct size. All purified positive PCR products were sequenced with their respective forward primers. Sequences were trimmed using Sequencher (Gene Codes Corporation, Ann Arbor, MI), and the identities of the positive PCR products were confirmed by comparing the sequences against the GenBank nonredundant database using BLASTN (3, 4).
Wastewater viral concentration efficiency.
Known quantities of adenoviruses were added to a raw sewage sample and a final effluent sample to determine the efficiency of the methods employed to concentrate viruses. Twenty milliliters of raw sewage and final effluent was autoclaved for 15 min at 121°C to eliminate adenovirus particles while minimizing the alteration of particulates in the sample. Ten milliliters of these wastewater samples was spiked with adenovirus 20 at a final concentration of 8 × 105 viruses per milliliter. The concentration of adenovirus 20 (strain AV-931; ATCC VR-1097) particles suspended in cell culture supernatant was determined through SYBR gold staining and epifluorescent microscopy (66). No aggregation of adenovirus 20 particles was observed. The remaining (unspiked) 10 ml of autoclaved raw sewage and final effluent served as a negative control throughout the viral concentration, nucleic acid isolation, and viral detection processes. Viruses were concentrated from all four samples as described above. PCR for adenoviruses was performed on 1:10 serial dilutions of DNA extracted from spiked and control samples using nested PCR as described above.
Determination of assay sensitivity.
A standard curve of target DNA was created for each assay to allow determinations of assay sensitivity. Positive PCR products were purified using the UltraClean PCR Clean-Up kit (Mo Bio Laboratories, Inc.), and the amount of nucleic acid in the purified product was quantified using a NanoDrop ND-1000 apparatus (NanoDrop Technologies, Wilmington, DE). In the case of nested PCRs, the positive control from the first round of amplification was used to create the standard curve. The number of targets was back calculated using the measured concentration of DNA and amplicon size using the following equation, where Y is the concentration of DNA measured by the NanoDrop instrument and Z is the length of the amplicon:
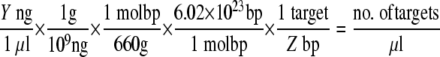 |
The purified PCR positive control was then serially diluted to 1 target/μl. The entire dilution series underwent the appropriate PCR or nested PCR for each viral group, with the exception that only 1 μl of target DNA was added to the reaction mixture. Sensitivity determination was performed at the same time as the sewage and effluent samples were assayed. PCR products were visualized on a 2% agarose gel stained with ethidium bromide, and the lowest concentration in which a band could be visualized defined the sensitivity of each assay.
Characterizing diversity of picobirnaviruses in raw sewage.
In order to gain a deeper understanding of the diversity of genotype I picobirnaviruses in raw sewage, positive PCR products from each state were cloned into pCR4-TOPO (Invitrogen), and transformants were screened for inserts by PCR with primers M13F and M13R. The UltraClean PCR Clean-Up kit (Mo Bio Laboratories, Inc.) was used to purify PCR products of the proper size, which were then sequenced with primer M13F. All sequences were trimmed using Sequencher (Gene Codes Corporation), and the identities of the positive PCR products were confirmed by comparing the sequences against the GenBank nonredundant database using BLASTN (3, 4).
All sequences with insignificant identities (E values of >0.001 and/or identities of less than 50 bp in length) to known picobirnaviruses were excluded from phylogenetic analyses. All of the remaining sequences were dereplicated to 99% sequence identity with gaps using the online program FastGroup II (81). The dereplication process and the representative sequences for each group were visually confirmed. FastGroup II was also used to calculate Chao1 (22, 23) and to perform rarefaction analyses (42, 46). The phylogenetic relationships of the picobirnaviruses detected in raw sewage to known picobirnaviruses were identified through aligning dereplicated raw sewage sequences with known sequences from GenBank using ClustalX v.1.8 (73). The resulting alignment was verified visually for accuracy and then imported into MEGA v. 4 for phylogenetic analyses (72).
The phylogenetic analysis of cloned picobirnavirus PCR products from raw sewage was executed in a manner similar to that used for previous picobirnavirus studies (12, 70). Prior to creating a phylogenetic tree, the average pairwise Jukes-Cantor distance was calculated to determine the appropriateness of creating a neighbor-joining tree. Since the average pairwise Jukes-Cantor distance was less than 1.0 (0.356), a neighbor-joining phylogenetic tree was calculated using a Jukes-Cantor model, and bootstrap analyses were performed with 2,000 replicates. Branches with insignificant bootstrap values (≤50%) were condensed to create the final phylogenetic tree.
Nucleotide sequence accession numbers.
The nucleotide sequences of 207 genotype I picobirnavirus amplicons from raw sewage have been deposited in the GenBank database under accession numbers EU938707 to EU938913.
RESULTS
Wastewater viral concentration efficiency.
The efficiencies of the centrifugal concentration devices used to concentrate raw sewage and final effluent were evaluated by spiking autoclaved wastewater samples with a final concentration of 8.48 × 105 adenovirus particles/ml. Spiked and unspiked samples were processed as described in Materials and Methods. PCR was performed on 1:10 serial dilutions of the extracted DNA and compared to an identical dilution series of DNA extracted directly from adenovirus particles. Using DNA extracted directly from adenovirus particles, the detection limit of the assay was determined to be 100 viruses. For both the spiked raw sewage and spiked final effluent samples, adenoviruses could be detected in a 10−3 dilution of extracted DNA, which was equivalent to ∼100 viruses. Negative (unspiked) controls were processed in parallel to the spiked samples, and the lack of PCR product in these controls ensured the elimination of preexisting adenovirus particles. Although this experiment was only semiquantitative, it demonstrated that the methods used here to concentrate wastewater samples were successful in concentrating viruses without major losses.
Viruses identified in wastewater samples.
The sensitivities of the assays used to identify the viruses under investigation ranged from 1 target to 100,000 targets and were identical to those reported previously (Table 1). The presence of 10 types of viruses was analyzed in raw sewage and final effluent samples from 12 wastewater treatment facilities located in 11 different states (Table 2). Adenoviruses and picobirnaviruses were found in 100% of the raw sewage samples. Enteroviruses were detected in 9 of 12 (75%) raw sewage samples, and noroviruses were detected in 7 of 12 (58%) raw sewage samples. Hepatitis B viruses, herpesviruses, morbilliviruses, papillomaviruses, reoviruses, and rotaviruses were not detected in any of the raw sewage samples; however, it is possible that they were present in concentrations below the detection limit of their assays (Table 1). Since adenoviruses, enteroviruses, noroviruses, and picobirnaviruses were detected in raw sewage, their presence was determined in final effluent samples collected from each wastewater treatment plant at the same time as raw sewage collection. Adenoviruses and picobirnaviruses were identified in 25% and 33% of the final effluent samples, respectively, while enteroviruses and noroviruses were identified in only one final effluent sample each.
TABLE 2.
Viruses detected in raw sewage and final effluent samples collected from throughout the coastal United States
| Virus | % Positive (no. of positive samples/total no. of samples)a
|
|
|---|---|---|
| Raw sewage | Final effluent | |
| Adenoviruses | 100 (12/12) | 25 (3/12) |
| Enteroviruses | 75 (9/12) | 8.3 (1/12) |
| Hepatitis B viruses | 0 | ND |
| Herpesviruses | 0 | ND |
| Morbilliviruses | 0 | ND |
| Noroviruses | 58.3 (7/12) | 8.3 (1/12) |
| Papillomaviruses | 0 | ND |
| Picobirnaviruses (genotype 1) | 100 (12/12) | 33 (4/12) |
| Reoviruses | 0 | ND |
| Rotaviruses (group A) | 0 | ND |
Positive results for targeted viruses were those with a PCR product of the expected size and BLASTN hit in GenBank with an E value of ≤0.001. ND, not determined.
Diversity of picobirnaviruses in raw sewage.
Since picobirnaviruses were found in all of the raw sewage samples, and limited knowledge exists regarding this virus, the diversity of picobirnaviruses in raw sewage was analyzed. A total of 288 (∼22 products per location) cloned picobirnavirus PCR products (200 bp) of the RNA-dependent RNA polymerase gene were sequenced by Agencourt (Beverly, MA), and 207 (72%) of these sequences had significant identities (E value of ≤0.001 and identities over 50 bp) to known picobirnaviruses in the GenBank database. The significant BLASTN sequence identities to known human and porcine picobirnaviruses ranged from 83% to 100% over regions ranging from 56 bp to 194 bp.
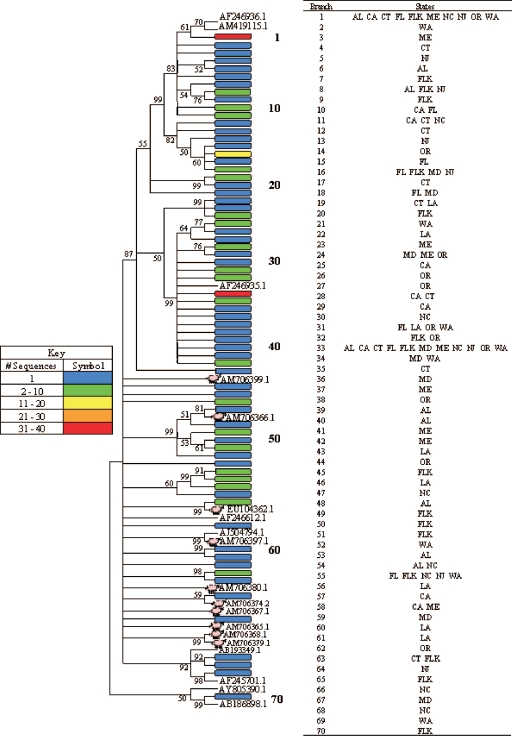
Prior to alignment, a total of 70 groups were created from the 207 raw sewage picobirnavirus sequences using FastGroup II (81), which was set to group sequences with 99% identity with gaps. While the majority of the groups were composed of one or two sequences, seven groups contained three or more sequences. The dereplicated sequences along with the top BLASTN hits from GenBank were aligned with ClustalX v.1.8 (73), and phylogenetic analyses were carried out with MEGA v. 4 (72). Since the average pairwise Jukes-Cantor distance was 0.356, a neighbor-joining tree was created using the Jukes-Cantor model with a bootstrap replication of 2,000. The final phylogenetic tree with insignificant (bootstrap value of <50) branches condensed is displayed in Fig. 1.
FIG. 1.
Condensed neighbor-joining (Jukes-Cantor model) phylogenetic tree of a ∼200-bp segment of the picobirnavirus RNA-dependent RNA polymerase gene from 207 U.S. raw sewage sequences. Each branch represents a sequence or a group of sequences (99% identical with gaps) depending upon the block color. Every colored block is numbered from top to bottom, and the number corresponds to the adjacent table, which explains the locations represented by each block (Alabama [AL], California [CA], Connecticut [CT], Florida [FL], Florida Keys [FL K], Louisiana [LA], Maine [ME], Maryland [MD], North Carolina [NC], New Jersey [NJ], Oregon [OR], and Washington [WA]). Reference sequences of human and porcine picobirnaviruses from previously published studies are identified by their GenBank accession numbers, and porcine picobirnaviruses are indicated with a pig (8, 9, 13, 20, 59, 70, 79; J. Buesa, R. Montava, C. Tellez, and J. M. Ribes, unpublished data).
Some of the picobirnavirus sequences from raw sewage had the greatest identities to porcine picobirnaviruses in the GenBank database, and this grouping is illustrated in the phylogenetic tree. The raw sewage picobirnavirus sequences do not have any discernible geographic distribution. The blocks on the phylogenetic tree representing more than one sequence show that identical (or >99% identical) sequences were recovered from multiple states. Furthermore, several unique sequences were identified from each state. For example, 8 of 10 sequences from North Carolina were unique (<99% identical).
In addition to dereplicating sequences, FastGroup II (81) was also used to calculate the Chao1 value, a richness estimator (22, 23), and to plot a rarefaction curve, which displays the number of unique sequences versus the number of clones sequenced (42, 46). Chao1 predicted a minimum of 200 unique picobirnaviruses in the raw sewage, but only 70 were sampled in this study; thus, more clones need to be sequenced to adequately describe the diversity of picobirnaviruses. Since the rarefaction curve is not yet approaching an asymptote (data not shown), more clones need to be sequenced in order to ensure a complete analysis of picobirnavirus diversity in raw sewage throughout the United States.
DISCUSSION
Viruses detected in wastewater.
This study used PCR to determine the presence of 10 types of viruses in raw sewage and final effluent collected from 12 wastewater treatment facilities in the United States. Four different types of viruses (adenoviruses, enteroviruses, noroviruses, and picobirnaviruses) were detected in at least 50% of the raw sewage samples. Interestingly, all of the viruses identified in wastewater were nonenveloped viruses, and these viruses tend to be more stable in the environment (31). Rotaviruses and reoviruses were not detected in any of the U.S. raw sewage samples even though their presence in raw sewage has been documented in other countries (30), and they have been used in prior fecal pollution studies (6, 7, 16, 19, 52, 62, 67). It is plausible that these viruses are not abundant in the United States or were simply not abundant on the day of sampling.
Adenoviruses and picobirnaviruses are potentially useful viral indicators of fecal pollution since they were found in 100% of raw sewage samples collected throughout the United States. Since these viruses were also detected in 25% and 33% of final effluent samples, respectively, future studies will need to determine how the presence of these viruses correlates with important human pathogens. Enteroviruses were detected in 75% of the raw sewage samples, and noroviruses were detected in 58% of the raw sewage samples; thus, the use of these viruses as markers of fecal pollution could potentially underestimate the extent of fecal contamination. The results of this study support prior findings regarding the prevalence of adenoviruses and enteroviruses in raw sewage and final effluent (15, 26, 37, 47, 58, 74, 77, 78). Furthermore, the use of adenoviruses and enteroviruses to indicate fecal contamination has been demonstrated by numerous studies in a variety of environments (reviewed in references 35 and 49) and experimentally tested in coastal ecosystems (35, 39, 40, 56, 64, 69, 80). This study is the first to demonstrate the widespread prevalence of picobirnaviruses in raw sewage and to suggest that these viruses can potentially be used as indicators of sewage contamination.
Since many viral types under investigation were not detected in this study, it is important to acknowledge the limitations of this research. It is expected that the viral load in the human population differs for each virus and that the abundance of these viruses fluctuates daily and seasonally in raw sewage. The samples analyzed in this study represent only a single time point at each treatment facility; therefore, it is possible that the types of viruses found could vary if samples were collected at different times of the year. In addition, the presence of viruses in the final effluent will also vary in response to fluctuations in treatment efficiency. Furthermore, the results of this study may have been biased by the differential recovery efficiency of the concentration and nucleic acid isolation methods for different viral groups. For example, viruses adhering to particles may have been lost in the filtration step. It is also possible that inefficient reverse transcription or the degradation of RNA could have skewed the results. Further studies need to be completed to verify the absence of undetected or underdetected viruses.
Similarly, it is possible that undetected viruses were present at concentrations below the assay detection limits on the day of sampling. To detect smaller concentrations of these viruses, whole-genome amplification using GenomiPhi V2 (GE Healthcare, Piscataway, NJ) was performed according to the manufacturer's instructions prior to viral PCR for each raw sewage sample and each final effluent sample as a means of increasing the concentration of target nucleic acid. Whole-genome amplification increases the total amount of DNA up to 600-fold (reviewed in reference 14), thus increasing the sensitivity of each assay by more than 2 orders of magnitude. After executing whole-genome amplification, papillomaviruses were identified in 2 of 12 raw sewage samples, and enteroviruses were detected in an additional 2 raw sewage samples, raising the total rate of enterovirus detection to 11 of 12 samples. This suggests that papillomaviruses and enteroviruses were present in those samples but at a lower abundance.
Diversity of picobirnaviruses in raw sewage.
Picobirnaviruses are currently an unclassified group of viruses even though they have recently been proposed to belong to the family Picobirnaviridae (9). As their name suggests, these viruses have a bisegmented double-stranded RNA genome (21, 70). Picobirnavirus particles are fairly small (35 nm), nonenveloped, and spherical. They have been found in the feces of a wide range of mammals including humans (9, 12, 60, 70, 79). They have not been successfully cultured in the laboratory, and their pathogenesis is unknown (21). These viruses have been implicated as possible enteric pathogens and have occasionally been associated with gastroenteritis (12, 21). This is the first study to examine the presence and diversity of picobirnaviruses in raw sewage collected throughout the United States.
Several important conclusions can be drawn from the phylogenetic analysis of the picobirnavirus sequences from raw sewage. First, it appears that a complete understanding of the sequence diversity of picobirnaviruses remains unknown given the large number of single sequences. This is further supported by the Chao1 and rarefaction analyses. The high level of diversity of genotype I human picobirnaviruses, based on the RNA-dependent RNA polymerase gene, has been previously reported (8, 12). Similarly, extensive diversity has been observed in genotype I porcine picobirnaviruses, and their existence as quasispecies has been postulated (9). To fully understand the sequence diversity of genotype I human picobirnaviruses in raw sewage, it would be necessary to sequence more clones until an asymptote is reached in the rarefaction curve.
While the majority of picobirnavirus sequences from raw sewage grouped with known human sequences, some raw sewage sequences grouped most closely with porcine picobirnaviruses. Since the presence of porcine picobirnaviruses in human raw sewage is doubtful and because strong sequence identity between human and porcine picobirnaviruses was previously observed (9), the close grouping of a few raw sewage picobirnavirus sequences to known porcine picobirnavirus sequences is likely the result of the conserved nature of the RNA-dependent RNA polymerase gene.
Potential of adenoviruses, enteroviruses, noroviruses, and picobirnaviruses as indicators of water quality.
To protect public health, it is necessary to identify a practical method for assessing fecal pollution in recreational waters. This is often accomplished through the detection of indicators, which are used to approximate the presence of pathogens. The current bacterial indicators of fecal contamination are not good indicators of wastewater pollution or human health risk in the marine environment (18, 32, 40, 41, 55, 80). The results of this study suggest that adenoviruses, enteroviruses, noroviruses, and picobirnaviruses have potential as viral indicators of fecal pollution because they were detected in the majority of raw sewage samples collected throughout the United States. Since they were detected in 100% of raw sewage samples, adenoviruses and picobirnaviruses are the most promising viral indicators of fecal pollution. Numerous previous studies (34, 35) have used adenoviruses to monitor water quality; however, this is the first study to suggest picobirnaviruses as possible indicator viruses. To assess the potential of picobirnaviruses as indicators to monitor water quality, future studies will need to determine how the presence, abundance, and stability of these viruses correlate with pathogens of concern throughout the wastewater treatment process and in contaminated coastal environments. It will also be necessary to determine if these viruses occur naturally in the environment in the absence of fecal pollution. Our preliminary data indicated that picobirnaviruses were not naturally present at detectable levels in seawater (data not shown); however, further research is needed.
In conclusion, adenoviruses, enteroviruses, noroviruses, and picobirnaviruses were found in the majority of raw sewage samples collected from 12 wastewater treatment facilities throughout the coastal United States. However, adenoviruses and picobirnaviruses were the only viruses detected in 100% of the raw sewage samples. Adenoviruses and picobirnaviruses were also detected in 25% and 33% of final effluent samples, respectively. While adenoviruses, enteroviruses, and noroviruses are known human pathogens and were proposed as markers of fecal pollution in previous studies, the results of this research demonstrate the potential use of picobirnaviruses as an indicator of fecal pollution. Further research will be needed to determine if these candidate viruses have the necessary characteristics of a microbial water quality indicator.
Acknowledgments
This work was supported by the U.S. Environmental Protection Agency (grant no. X7-96465507-0).
We give special thanks to the personnel from the wastewater treatment facilities that supplied raw sewage and final effluent samples. We thank Mike Gray (USGS, St. Petersburg, FL) for his assistance in the phylogenetic analysis of picobirnaviruses and John Paul (University of South Florida, St. Petersburg, FL) for his insight, positive controls for norovirus and rotavirus assays, and reviewing the manuscript.
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Alexander, L. M., A. Heaven, A. Tennant, and R. Morris. 1992. Symptomatology of children in contact with sea-water contaminated with sewage. J. Epidemiol. Commun. Health 46:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy-persons and patients with diarrhea by 2-step polymerase chain-reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apajalahti, J. H. A., A. Kettunen, P. H. Nurminen, H. Jatila, and W. E. Holben. 2003. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl. Environ. Microbiol. 69:5731-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aulicino, F. A., L. Mauro, M. Marranzano, M. Biondi, A. Ursino, and M. Carere. 2000. Microbiological quality of the Catania coastal sea water. Ann. Ig. 12:533-541. [PubMed] [Google Scholar]
- 7.Aulicino, F. A., P. Orsini, M. Carere, and A. Mastrantonio. 2001. Bacteriological and virological quality of seawater bathing areas along the Tyrrhenian coast. Int. J. Environ. Health Res. 11:5-11. [DOI] [PubMed] [Google Scholar]
- 8.Banyai, K., F. Jakab, G. Reuter, J. Bene, M. Uj, B. Melegh, and G. Szucs. 2003. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch. Virol. 148:2281-2291. [DOI] [PubMed] [Google Scholar]
- 9.Banyai, K., V. Martella, A. Bogdan, P. Forgach, F. Jakab, E. Meleg, H. Biro, B. Melegh, and G. Szucs. 2008. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J. Gen. Virol. 89:534-539. [DOI] [PubMed] [Google Scholar]
- 10.Barrett, T., I. K. G. Visser, L. Mamaev, L. Goatley, M. F. Vanbressem, and A. Osterhaus. 1993. Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology 193:1010-1012. [DOI] [PubMed] [Google Scholar]
- 11.Benson, K. A. S., C. A. Manire, R. Y. Ewing, J. T. Saliki, F. I. Townsend, B. Ehlers, and C. H. Romero. 2006. Identification of novel alpha- and gammaherpesviruses from cutaneous and mucosal lesions of dolphins and whales. J. Virol. Methods 136:261-266. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharya, R., G. C. Sahoo, M. K. Nayak, K. Rajendran, P. Dutta, U. Mitra, M. K. Bhattacharya, T. N. Naik, S. K. Bhattacharya, and T. Krishnan. 2007. Detection of genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect. Genet. Evol. 7:229-238. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya, R., G. C. Sahoo, M. K. Nayak, D. R. Saha, D. Sur, T. N. Naik, S. K. Bhattacharya, and T. Krishnan. 2006. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect. Genet. Evol. 6:453-458. [DOI] [PubMed] [Google Scholar]
- 14.Binga, E. K., R. S. Lasken, and J. D. Neufeld. 2008. Something from (almost) nothing: the impact of multiple displacement amplification on microbial ecology. Int. Soc. Microb. Ecol. J. 2:233-241. [DOI] [PubMed] [Google Scholar]
- 15.Bofill-Mas, S., N. Albinana-Gimenez, P. Clemente-Casares, A. Hundesa, J. Rodriguez-Manzano, A. Allard, M. Calvo, and R. Girones. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brassard, J., K. Seyer, A. Houde, C. Simard, and Y. L. Trottier. 2005. Concentration and detection of hepatitis A virus and rotavirus in spring water samples by reverse transcription-PCR. J. Virol. Methods 123:163-169. [DOI] [PubMed] [Google Scholar]
- 17.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brownell, M. J., V. J. Harwood, R. C. Kurz, S. M. McQuaig, J. Lukasik, and T. M. Scott. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747-3757. [DOI] [PubMed] [Google Scholar]
- 19.Caballero, S., F. X. Abad, F. Loisy, F. S. Le Guyader, J. Cohen, R. M. Pinto, and A. Bosch. 2004. Rotavirus virus-like particles as surrogates in environmental persistence and inactivation studies. Appl. Environ. Microbiol. 70:3904-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carruyo, G. M., G. Mateu, L. C. Martinez, F. H. Pujol, S. V. Nates, F. Liprandi, and J. E. Ludert. 2008. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J. Clin. Microbiol. 46:2402-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandra, R. 1997. Picobirnavirus, a novel group of undescribed viruses of mammals and birds: a minireview. Acta Virol. 41:59-62. [PubMed] [Google Scholar]
- 22.Chao, A. 1987. Estimating the population size for capture-recapture data and unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 23.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Statistics 11:783-791. [Google Scholar]
- 24.Cheung, W. H. S., K. C. K. Chang, R. P. S. Hung, and J. W. L. Kleevens. 1990. Health-effects of beach water-pollution in Hong-Kong. Epidemiol. Infect. 105:139-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi, S., and S. C. Jiang. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl. Environ. Microbiol. 71:7426-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crabtree, K. D., C. P. Gerba, J. B. Rose, and C. N. Haas. 1997. Waterborne adenovirus: a risk assessment. Water Sci. Technol. 35:1-6. [Google Scholar]
- 27.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 28.Donovan, E., K. Unice, J. D. Roberts, M. Harris, and B. Finley. 2008. Risk of gastrointestinal disease associated with exposure to pathogens in the water of the Lower Passaic River. Appl. Environ. Microbiol. 74:994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donovan, E. P., D. F. Staskal, K. M. Unice, J. D. Roberts, L. C. Haws, B. L. Finley, and M. A. Harris. 2008. Risk of gastrointestinal disease associated with exposure to pathogens in the sediments of the Lower Passaic River. Appl. Environ. Microbiol. 74:1004-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubois, E., F. LeGuyader, L. Haugarreau, H. Kopecka, M. Cormier, and M. Pommepuy. 1997. Molecular epidemiological survey of rotaviruses in sewage by reverse transcriptase seminested PCR and restriction fragment length polymorphism assay. Appl. Environ. Microbiol. 63:1794-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauquet, C. M., M. A. Mayo, M. A. Maniloff, J. Maniloff, J. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy, 8th ed. Elsevier Academic Press, Amsterdam, The Netherlands.
- 32.Ferguson, D. M., D. F. Moore, M. A. Getrich, and M. H. Zhowandai. 2005. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. J. Appl. Microbiol. 99:598-608. [DOI] [PubMed] [Google Scholar]
- 33.Fleisher, J. M., D. Kay, R. L. Salmon, F. Jones, M. D. Wyer, and A. F. Godfree. 1996. Marine waters contaminated with domestic sewage: nonenteric illnesses associated with bather exposure in the United Kingdom. Am. J. Public Health 86:1228-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fong, T. T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fong, T. T., and E. K. Lipp. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forslund, O., A. Antonsson, P. Nordin, B. Stenquist, and B. G. Hansson. 1999. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 80:2437-2443. [DOI] [PubMed] [Google Scholar]
- 37.Gerba, C. P., D. M. Gramos, and N. Nwachuku. 2002. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 68:5167-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilgen, M., D. Germann, J. Luthy, and P. Hubner. 1997. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 39.Griffin, D. W., K. A. Donaldson, J. H. Paul, and J. B. Rose. 2003. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 16:129-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. Bioscience 51:817-825. [Google Scholar]
- 42.Heck, K. L., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 43.Hejkal, T. W., F. M. Wellings, A. L. Lewis, and P. A. Larock. 1981. Distribution of viruses associated with particles in waste-water. Appl. Environ. Microbiol. 41:628-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henrickson, S. E., T. Wong, P. Allen, T. Ford, and P. R. Epstein. 2001. Marine swimming-related illness: implications for monitoring and environmental policy. Environ. Health Perspect. 109:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howarth, R. W. 1993. Managing wastewater in coastal urban areas: the role of nutrients in coastal waters. National Academy Press, Washington, DC.
- 46.Hurlbert, S. H. 1997. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 47.Irving, L. G., and F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang, S. C. 2006. Human adenoviruses in water: occurrence and health implications: a critical review. Environ. Sci. Technol. 40:7132-7140. [DOI] [PubMed] [Google Scholar]
- 50.Jiang, S. C., W. Chu, and J. W. He. 2007. Seasonal detection of human viruses and coliphage in Newport Bay, California. Appl. Environ. Microbiol. 73:6468-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83:145-154. [DOI] [PubMed] [Google Scholar]
- 52.Kittigul, L., S. Ekchaloemkiet, F. Utrarachkij, K. Siripanichgon, D. Sujirarat, S. Pungchitton, and A. Boonthum. 2005. An efficient virus concentration method and RT-nested PCR for detection of rotaviruses in environmental water samples. J. Virol. Methods 124:117-122. [DOI] [PubMed] [Google Scholar]
- 53.Koike, K., M. Kobayashi, M. Gondo, I. Hayashi, T. Osuga, and S. Takada. 1998. Hepatitis B virus DNA is frequently found in liver biopsy samples from hepatitis C virus-infected chronic hepatitis patients. J. Med. Virol. 54:249-255. [PubMed] [Google Scholar]
- 54.Leary, T. P., J. C. Erker, M. L. Chalmers, J. D. Wetzel, S. M. Desai, I. K. Mushahwar, and T. S. Dermody. 2002. Detection of reovirus by reverse transcription-polymerase chain reaction using primers corresponding to conserved regions of the viral L1 genome segment. J. Virol. Methods 104:161-165. [DOI] [PubMed] [Google Scholar]
- 55.Lee, C. M., T. Y. Lin, C. C. Lin, G. A. Kohbodi, A. Bhattl, R. Lee, and J. A. Jay. 2006. Persistence of fecal indicator bacteria in Santa Monica Bay beach sediments. Water Res. 40:2593-2602. [DOI] [PubMed] [Google Scholar]
- 56.Lipp, E. K., J. C. Futch, and D. W. Griffin. 2007. Analysis of multiple enteric viral targets as sewage markers in coral reefs. Mar. Pollut. Bull. 54:1897-1902. [DOI] [PubMed] [Google Scholar]
- 57.Lipp, E. K., S. A. Farrah, and J. B. Rose. 2001. Assessment and impact of microbial fecal pollution and human enteric pathogens in a coastal community. Mar. Pollut. Bull. 42:286-293. [DOI] [PubMed] [Google Scholar]
- 58.Lodder, W. J., and A. M. D. Husman. 2005. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 71:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez, L. C., M. O. Giordano, M. B. Isa, L. F. Alvarado, J. V. Pavan, D. Rinaldi, and S. V. Nates. 2003. Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology 46:207-213. [DOI] [PubMed] [Google Scholar]
- 60.Masachessi, G., L. C. Martinez, M. O. Giordano, P. A. Barril, B. M. Isa, L. Ferreyra, D. Villareal, M. Carello, C. Asis, and S. V. Nates. 2007. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch. Virol. 152:989-998. [DOI] [PubMed] [Google Scholar]
- 61.Metcalf, T. G., J. L. Melnick, and M. K. Estes. 1995. Environmental virology—from detection of virus in sewage and water by isolation to identification by molecular-biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 62.Muscillo, M., G. La Rosa, C. Marianelli, S. Zaniratti, M. R. Capobianchi, L. Cantiani, and A. Carducci. 2001. A new RT-PCR method for the identification of reoviruses in seawater samples. Water Res. 35:548-556. [DOI] [PubMed] [Google Scholar]
- 63.Nenonen, N. P., C. Hannoun, P. Horal, B. Hernroth, and T. Bergstrom. 2008. Tracing of norovirus outbreak strains in mussels collected near sewage effluents. Appl. Environ. Microbiol. 74:2544-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 65.Reference deleted.
- 66.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 67.Pianetti, A., W. Baffone, B. Citterio, A. Casaroli, F. Bruscolini, and L. Salvaggio. 2000. Presence of enteroviruses and reoviruses in the waters of the Italian coast of the Adriatic Sea. Epidemiol. Infect. 125:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rigotto, C., T. C. M. Sincero, C. M. O. Simoes, and C. R. M. Barardi. 2005. Detection of adenoviruses in shellfish by means of conventional-PCR, nested-PCR, and integrated cell culture PCR (ICC/PCR). Water Res. 39:297-304. [DOI] [PubMed] [Google Scholar]
- 69.Rose, M. A., A. K. Dhar, H. A. Brooks, F. Zecchini, and R. M. Gersberg. 2006. Quantitation of hepatitis A virus and enterovirus levels in the lagoon canals and Lido beach of Venice, Italy, using real-time RT-PCR. Water Res. 40:2387-2396. [DOI] [PubMed] [Google Scholar]
- 70.Rosen, B. I., Z. Y. Fang, R. I. Glass, and S. S. Monroe. 2000. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 277:316-329. [DOI] [PubMed] [Google Scholar]
- 71.Schets, F. M., J. H. van Wijnen, J. F. Schijven, H. Schoon, and A. M. de Roda Husman. 2008. Monitoring of waterborne pathogens in surface waters in Amsterdam, The Netherlands, and the potential health risk associated with exposure to Cryptosporidium and Giardia in these waters. Appl. Environ. Microbiol. 74:2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 73.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thompson, S. S., J. L. Jackson, M. Suva-Castillo, W. A. Yanko, Z. El Jack, J. Kuo, C. L. Chen, F. P. Williams, and D. P. Schnurr. 2003. Detection of infectious human adenoviruses in tertiary-treated and ultraviolet-disinfected wastewater. Water Environ. Res. 75:163-170. [DOI] [PubMed] [Google Scholar]
- 75.U.S. Environmental Protection Agency. 2006. Implementing the BEACH Act of 2000. Report to Congress. EPA-823-R-06-001. U.S. Environmental Protection Agency, Washington, DC.
- 76.van den Berg, H., W. Lodder, W. van der Poel, H. Vennema, and A. M. D. Husman. 2005. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol. 156:532-540. [DOI] [PubMed] [Google Scholar]
- 77.Van Heerden, J., M. M. Ehlers, W. B. Van Zyl, and W. O. K. Grabow. 2003. Incidence of adenoviruses in raw and treated water. Water Res. 37:3704-3708. [DOI] [PubMed] [Google Scholar]
- 78.Vantarakis, A., and M. Papapetropoulou. 1999. Detection of enteroviruses, adenoviruses and hepatitis A viruses in raw sewage and treated effluents by nested-PCR. Water Air Soil Pollut. 114:85-93. [Google Scholar]
- 79.Wakuda, M., Y. Pongsuwanna, and K. Taniguchi. 2005. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J. Virol. Methods 126:165-169. [DOI] [PubMed] [Google Scholar]
- 80.Wetz, J. J., E. K. Lipp, D. W. Griffin, J. Lukasik, D. Wait, M. D. Sobsey, T. M. Scott, and J. B. Rose. 2004. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. Mar. Pollut. Bull. 48:698-704. [DOI] [PubMed] [Google Scholar]
- 81.Yu, Y., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a Web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. L. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. J. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]