Abstract
Mycobacterium abscessus is a rapidly growing mycobacterial species that can be involved in pulmonary and disseminated infections in immunosuppressed or young cystic fibrosis patients. It is an emerging pathogen and has attracted recent attention due to the numerous cases of infection; furthermore, genomic tools have been developed for this species. Nevertheless, the study of this species has until now been limited to spontaneous variants. We report here a comparison of three different mutagenesis systems—the ts-sacB, the phage, and the recombineering systems—and show that there are important differences in their efficiency for the construction of allelic-exchange mutants. We show, using the mmpL4b gene of the glycopeptidolipid pathway as a target, that allelic-exchange mutants can be constructed with a reasonable efficiency (∼7%) using the recombineering system. These observations will facilitate genetic and cellular microbiology experiments involving the construction and use of well-defined mutants to study the virulence determinant of this emerging pathogen.
The mycobacterial genus contains plethora of species that are pathogenic for either humans or animals. The most well-known are undoubtedly Mycobacterium leprae, M. tuberculosis, and M. ulcerans, the etiologic agents of human leprosy, tuberculosis, and Buruli ulcer, respectively (47-49). M. avium subsp. paratuberculosis, responsible for Johnes disease in ruminants, is also a serious health concern since it is suspected to be a threat to human via infected milk (9, 10). M. abscessus is an emerging pathogen involved in pulmonary and disseminated infection in young cystic fibrosis patients (26, 36). M. abscessus can cause nosocomial infections of skin and soft tissues in immunosuppressed patients (28, 35). It is also able to cross the blood-brain barrier and to cause meningoencephalitis (42). M. abscessus is phylogenetically related to M. chelonae and, indeed, these species have long been grouped together under the designation of the “M. abscessus-chelonae complex” (6). M. abscessus is a rapid grower that forms colonies in 5 days. Like other mycobacterial species, M. abscessus is equipped with a robust waxy cell wall that, as in other species, probably contributes to virulence (12). The emerging and growing interest in M. abscessus has led to its genome being sequenced (accession no. NC010397) (F. Ripoll et al., unpublished data) and to the development of DNA microarrays (Jean-Yves Coppée, unpublished data).
The availability of genomic resources and animal models (32) makes M. abscessus a very attractive system. However, there is no defined mutagenesis system for this species and, to the best of our knowledge, no defined mutants have been constructed thus far. The consequence is that the study of this organism has been restricted to spontaneous variants. Utilization of spontaneous mutants has, nevertheless, allowed the characterization of morphotypically rough isolates that are hypervirulent both in vitro and in vivo (7, 8, 17). These rough isolates are low glycopeptidolipid producers. Glycopeptidolipid is an extractable lipid found at the surface of the bacilli (4, 11, 13). However, its role in the virulence process is currently unknown. The lack of a suitable genetic system is certainly responsible for the rarity of studies on this species (fewer than 500 references in Medline, whereas there are more than 32,000 for M. tuberculosis). Other mycobacterial species, especially M. tuberculosis, have been genetically intractable for many years (15, 18, 24). This has forced researchers to develop dedicated systems for the construction of allelic-exchange mutants. Three major systems have mainly been used thus far in M. tuberculosis and in other mycobacteria: (i) a thermosensitive counterselectable plasmid based on sucrose sensitivity (21-23), (ii) a thermosensitive mycobacteriophage (2) and, more recently, (iii) a mycobacterial recombinase-based system (43, 44). These three systems are effective in M. tuberculosis, M. smegmatis, and other refractory species, including M. avium subsp. avium, and allow straightforward construction of both marked and unmarked mutants.
The aim of the present study was to compare the three main mutagenesis systems available for mycobacteria and to determine which system is best adapted to M. abscessus. To this end, we used mmpL4b as a target gene and the three genetic tools described above. The mmpL4b gene is involved in glycopeptidolipid synthesis (29, 40) and is a good model target because its mutation results in a rough phenotype that can be visually distinguished. We show here that there are large differences in efficacy between the three systems and that the mycobacterial recombinase-based system is the most efficient. For an unknown reason, allelic exchange is much less frequent in M. abscessus than in other species, including M. tuberculosis; this complicates the construction of defined mutants. The availability of a suitable genetic system will undoubtedly facilitates the characterization of the virulence determinants in this emerging pathogen.
MATERIALS AND METHODS
Bacterial strains and media.
Strains used in the present study are listed in Table S1 in the supplemental material. Escherichia coli DH5α and HB101 strains were used for cloning and grown as previously described (34). M. smegmatis mc2155 (39) and M. abscessus CIP104536T (8) strains were grown in LB or 7H9 (Difco) medium. When required, antibiotics were added to the medium at the following final concentration: kanamycin, 25 μg/ml; zeocin, 25 μg/ml; gentamicin, 20 μg/ml for E. coli and M. smegmatis mc2155; and 100 μg/ml of each for M. abscessus CIP104536T.
Construction of the allelic-exchange substrate of mmpL4b gene.
The plasmids and primers used in the present study are listed in Table 1 and Table S2 in the supplemental material, respectively. The marked mutation construct of mmpL4b gene (MAB_4115c) was generated by PCR, using genomic DNA of M. abscessus CIP104536T as a template. Primer pair mmpL4B1_F and mmpL4B1_Rv, containing EcoRI and SalI sites, respectively, were used to amplify a fragment (fragment 1) encoding N-terminal amino acids 1 to 3, along with a 908-bp sequence upstream of the mmpL4b gene. Similarly, the primers mmpL4B2_F and mmpL4B2_Rv, containing the SalI and ApaI sites, respectively, were used to amplify a fragment (fragment 2) encoding amino acids 985 to 987, along with a 950-bp region downstream of the mmpL4b gene. Fragment 1 and fragment 2 were digested by either EcoRI/SalI or SalI/ApaI (Biolabs) and cloned in a EcoRI-ApaI-digested pBluescript II SK(+) (Stratagene) to generate pBSK-ΔmmpL4b. The kanamycin cassette was extracted from the pUC4K vector (Pharmacia biotech) by SalI digestion, purified, and cloned into the SalI of the pBSK-ΔmmpL4b to generate the pBSK-ΔmmpL4b::Km. This allelic-exchange substrate was used to perform mutagenesis in M. abscessus and M. smegmatis. It should be noted that the identity at the DNA level between the mmpL4a genes of M. abscessus and M. smegmatis is 76% (fragment 1), whereas it is 78% for fragment 2 (31).
TABLE 1.
Plasmids used in this study
| Plasmid | Featuresa | Source or reference |
|---|---|---|
| pBSK | Cloning vector; Ampr | Stratagene |
| pBSK-ΔmmpL4b | pBSK containing upstream and downstream regions of the mmpL4b gene | This study |
| pBSK-ΔmmpL4b::Km | pBSK containing the allelic-exchange substrate | This study |
| pPR27 | Mycobacterial replicating shuttle vector, containing the sacB gene and the Gmr cassette | 21 |
| pPR27-ΔmmpL4b::Km | pPR27 containing the allelic-exchange substrate | This study |
| pJSC347 | Vector containing PacI site and λ-cos | 33 |
| pJSC347-ΔmmpL4b::Km | pJSC347 containing the allelic-exchange substrate | This study |
| pJV53 | chec9 genes gp60_61 under the control of the acetamidase promoter | 44 |
| pJV53-zeo | pJV53, which the Kmr cassette replaced with a Zeor cassette | This study |
| pLYG204.Zeo | Plasmid encoding a Zeor cassette | 14 |
| pUC4K | Plasmid encoding a Kmr cassette | Pharmacia Biotech |
Ampr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Zeor, zeocin resistance.
Mutagenesis procedures. (i) ts-sacB system.
The ts-sacB system has been described previously (22, 23). The derived mycobacterial temperature-sensitive replicating plasmid pPR27-ΔmmpL4b::Km used in the present study was constructed as follows. The allelic-exchange substrate for the mmpL4b gene was obtained by PCR amplification using the mmpL4B1_F (XbaI) and mmpL4B2_Rv (XbaI) primer pair and pBSK-ΔmmpL4b::Km as a template. The purified PCR fragment was digested with XbaI and cloned into the unique XbaI site of dephosphorylated pPR27 vector to generate pPR27-ΔmmpL4b::Km. This construct was used to electroporate M. abscessus CIP104536T and M. smegmatis mc2155. Transformants were selected on LB agar plates containing kanamycin and gentamicin at 30°C, colony purified, and inoculated into LB containing kanamycin and gentamicin at 30°C for 5 days. Cells were then plated on LB agar plates containing kanamycin and 10% (wt/vol) sucrose, followed by incubation for 4 days at 39°C. Plasmid pPR27-ΔmmpL4b::Km was extracted from the transformants of M. abscessus by electroduction (3). The frequency of counterselection was calculated as described previously (22, 23).
(ii) Mycobacteriophage system.
The allelic-exchange substrate for the mmpL4b gene was extracted by XbaI restriction of pPR27-ΔmmpL4b::Km and cloned at the compatible SpeI and XbaI sites of the cosmid vector pJSC347 (33), generating pJSC347-ΔmmpL4b::Km. To generate each specialized transducing phage, the PacI-digested pJSC347-ΔmmpL4b::Km was cloned in PacI-digested phAE87 DNA and packaged in vitro by using a MaxPlax packaging extracts kit (Tebu-Bio) as previously reported (19). E. coli HB101 was transduced and plated on selective media containing kanamycin. Phagemid DNA was prepared from a pool of antibiotic-resistant transductants and electroporated into M. smegmatis mc2155 to produce recombinant phage particles. All transducing phages were plaque purified and tested for their temperature-sensitive phenotype. M. smegmatis mc2155 and M. abscessus CIP104536T were grown in LB medium (with 0.5% of Tween 80) to an optical density at 600 nm of 1. A 10-ml portion of the culture was centrifuged, resuspended in 10 ml of washing medium 7H9 broth (Difco) with 0.2% glycerol-1× albumin dextrose saline and incubated as standing culture for 24 h at 37°C to remove all traces of Tween 80 detergent. After incubation, bacterial cells were centrifuged and resuspended in 10 ml of 7H9-albumin dextrose saline and then mixed with the specialized transducing phage at a multiplicity of infection of 10. The cell-phage mix was incubated at a nonpermissive temperature for 30 min for M. smegmatis mc2155 and for 2 h for M. abscessus, centrifuged, resuspended in 1 ml of phosphate-buffered saline containing 0.1% Tween 80, and plated on selective medium containing kanamycin.
(iii) Recombineering system.
This system is based on the use of a plasmid allowing the expression of an inducible phage recombinase, which is more effective at promoting homologous recombination than the endogenous recombinase. The strain containing the plasmid is then electroporated with a linear DNA fragment containing the mutant allele and plated on selective medium.
The kanamycin cassette of pJV53 (44) was replaced with the zeocin cassette, which was PCR amplified using primer pair Zeo_F and Zeo_Rv and pLYG204.Zeo as a template (14). Plasmid pJV53 was SpeI and NheI digested, blunted, and ligated to the zeocin PCR product to generate pJV53-zeo. pJV53-zeo was electroporated into M. smegmatis mc2155 and M. abscessus CIP104536T. The allelic-exchange substrate of mmpL4b gene of M. abscessus was extracted by XbaI digestion of pPR27-ΔmmpL4b::Km and purified. The strains containing the pJV53-zeo construct were cultured overnight at 37°C in LB medium containing zeocin and 0.2% succinate. At an optical density at 600 nm of ∼0.5, acetamide (0.2%) was added, and electrocompetent cells were prepared after 3 h of induction as described previously (44). The competent cells were transformed with 100 ng of allelic-exchange substrate of mmpL4b gene and treated as previously described (44).
Southern blot analysis.
The genomic DNA of M. smegmatis and M. abscessus strains was extracted, digested with SalI (for M. abscessus) or PstI (for M. smegmatis), run on a 1% agarose gel, and electrotransferred. The nitrocellulose membrane was probed with a fragment corresponding to the upstream region of mmpL4b (908 bp). In the case of M. smegmatis, the genomic DNA was probed with a fragment corresponding to the downstream region of mmpL4b (1,093 bp).
Biochemical analysis.
Lipids were extracted from bacterial cell pellets with a mixture of chloroform and methanol as previously described (46). The extracts were dried under vacuum and partitioned between water and chloroform at 1:1 (vol/vol). The organic phases were extensively washed with distilled water and evaporated to dryness. The lipid extracts were dissolved in chloroform and analyzed by thin-layer chromatography (TLC) on silica gel Durasil 25-precoated plates (0.25-mm thickness; Macherey-Nagel). The GPLs were resolved in chloroform-methanol at 9:1 (vol/vol) and visualized by spraying the plates with iodine.
RESULTS
mmpL4b as a target gene: characterizing the system.
We chose the mmpL4b gene, which is not essential and belongs to the glycopeptidolipid pathway, as a target for several reasons. First, this pathway has been identified in silico in M. abscessus (31), which allows the prediction of the banding pattern in Southern blot hybridization and thus the precise identification of the molecular event responsible for the formation of the various colonies. Second, mutation in the glycopeptidolipid biosynthesis pathway leads to rough mutants that can be easily distinguished by visual inspection of colonies growing on petri dishes (5, 40). Third, the mmpL4b gene is the last gene of an operon, and the downstream gene is in the opposite orientation, which reduces the risk of a polar effect. Fourth, since glycopeptidolipid is suspected to be involved in virulence (7, 8, 17), any mutants constructed will be useful for subsequent studies.
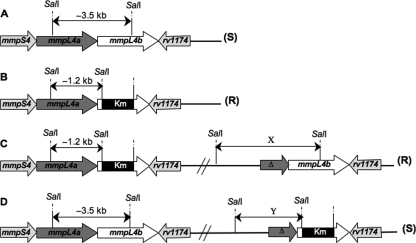
A construct containing a kanamycin cassette flanked by ∼1 kb of mmpL4a on the 5′ end and ∼1 kb on the 3′ end was made to promote homologous recombination at the mmpL4b locus. Knowledge on the M. abscessus GPL locus allows us to predict that the hybridization pattern after a SalI restriction using the upstream region of mmpL4b as a probe will give a band of ∼3.5 kb in the case of a wild-type gene (Fig. 1A) and a band of ∼1.2 kb in the case of an allelic-exchange event (Fig. 1B). In the case of homologous recombination with a single crossover, two bands would be obtained: (i) a mutant band (∼1. 2 kb) plus a variable band (depending on the system) if the crossover is located upstream from the kanamycin cassette (Fig. 1C) or (ii) a wild-type band (∼3. 5 kb) plus a variable band (depending on the system) if the crossover is located downstream from the kanamycin cassette (Fig. 1D). Furthermore, only the allelic exchange and single crossover upstream from the kanamycin cassette should lead to a rough colony morphology and low GPL production (5, 40). In contrast, both the wild type and the mutant resulting from a single crossover mutant downstream from the kanamycin cassette will display a smooth morphotype and produce wild-type amounts of GPL, since they both contain a wild-type copy of the mmpSL operon (Fig. 1).
FIG. 1.
Schematic representation of the wild-type mmpSL4 operon in M. abscessus and the various genetic events leading to its disruption. (A) Wild-type mmpSL4 operon; (B) allelic-exchange mutant; (C) single crossover event upstream from the kanamycin cassette; (D) single crossover event downstream from the kanamycin cassette. The predicted phenotype is indicated in parentheses. R, rough; S, smooth. X and Y indicate the variable bands of the single crossover upstream or downstream of the kanamycin cassette. The sizes of the X variable bands are 4, 4.5, and 4.1 kb for the ts-sacB, mycobacteriophage, and recombineering systems, respectively. The sizes of the Y variable bands are 1.7, 2.2, and 2.5 kb for the ts-sacB, mycobacteriophage, and recombineering systems, respectively. Δ, 5′-truncated mmpL4a gene.
This construct was inserted into the ts-sacB system and the mycobacteriophage system and used to transform a strain expressing the phage recombinase. M. smegmatis strain mc2155 strain was used as a positive control to construct allelic-exchange mutants with this heterologous construct in all experiments. M. smegmatis strain mc2155 is smooth and thus allows the easy identification of rough clones when the GPL locus is mutated (29, 40). The average transformation efficiency of M. smegmatis mc2155 was approximately 5 × 105 transformants/μg of DNA, using a replicative plasmid, and that of the M. abscessus strain was 103. The transformation efficiencies of other isolates were similar to that of the M. abscessus CIP104536T type strain (data not shown), and we therefore used this type strain for the experiments described below. We further characterized the frequency of a spontaneous kanamycin-resistant mutation in both M. smegmatis and M. abscessus. To this end, both species were transformed with a kanamycin-resistant replicative plasmid (pJV53) and plated onto selective medium at 37°C. More than 100 kanamycin-resistant colonies of both M. smegmatis and M. abscessus were analyzed by PCR for the presence of the plasmid. This analysis showed that 100% of the M. smegmatis clones contained the plasmid, whereas ca. 40% of the M. abscessus clones contained it. This means that, in the case of M. abscessus, ca. 60% of the kanamycin-resistant clones did not harbor the plasmid and correspond to spontaneous kanamycin-resistant mutants.
Comparative studies of allelic-exchange mutant construction. (i) ts-sacB system.
The ts-sacB system has been widely used and is based on a thermosensitive plasmid bearing the Bacillus subtilis sacB gene (23). In this plasmid (pPR27), the presence of the sacB gene allows counterselection of clones generated by single crossover events, and counterselection efficiency is very high at nonpermissive temperature in the presence of sucrose (21-25). This system has been used with success in many mycobacterial species, notably M. smegmatis, M. tuberculosis, M. avium subsp. avium, and M. marinum.
The pPR27-ΔmmpL4b::Km plasmid was introduced into both M. smegmatis and M. abscessus, and transformants were selected on kanamycin and gentamicin at permissive temperatures (23, 30). One isolate of each was used to inoculate liquid medium, grown at a permissive temperature, and then serially diluted and spread onto agar plates containing kanamycin and sucrose at a nonpermissive temperature (23).
Although the counterselection efficiency for the M. smegmatis strain was high and similar to what previously reported (23), there was no reduction in the colony count in the case of M. abscessus (Table 2). All of the M. smegmatis colonies were rough, whereas all of the M. abscessus colonies displayed the wild-type smooth phenotype. Five isolates of each M. smegmatis and M. abscessus were analyzed for their GPL content by TLC and iodine staining. All five of the M. smegmatis clones were almost devoid of GPL as expected, whereas all five of the M. abscessus clones contained as much GPL as did the wild-type strain (data not shown). Genomic DNA was prepared from these five rough M. smegmatis colonies and was analyzed by Southern blotting: all had undergone an allelic-exchange event (data not shown). In the case of M. abscessus, no rough colonies were observed, a finding consistent with the absence of counterselection by sucrose and temperature; these smooth M. abscessus colonies were not investigated by Southern blotting since they could not have been subject to an allelic-exchange event. Mutations in the pPR27-ΔmmpL4b::Km could explain this lack of counter selection. Therefore, the plasmid was extracted from M. abscessus (from an aliquot of a culture kept at a permissive temperature) and transferred into M. smegmatis; counterselection on plates containing kanamycin and sucrose at a nonpermissive temperature was very high (10−5), thus demonstrating that the plasmid was not bearing mutations in either ori-ts or the sacB gene. We tested whether the nonfunctionality was a particularity of the type strain by testing the ts-sacB system in several other M. abscessus clinical isolates; the system was similarly unsuccessful in all of the strains tested, suggesting that the system is not functional in this species.
TABLE 2.
Comparative counterselection efficiency of the ts-sacB system in M. smegmatis mc2155 and M. abscessus CIP104536T strainsa
| Transformant | CFU/ml in:
|
Frequency of counterselection (SD) | |
|---|---|---|---|
| LB (30°C) | LB + 10% sucrose (39°C) | ||
| M. smegmatis (pPR27-ΔmmpL4b) | 2 × 106 | 130 | 6 × 10−5 (9.8 × 10−6) |
| M. abscessus (pPR27-ΔmmpL4b) | 5 × 107 | 2 × 107 | 0.4 (0.06) |
Experiments were performed five times.
Since the temperature and the presence of sucrose have an additive counterselective effect (23) and no reduction was observed in M. abscessus, it is likely that neither ori-ts nor sacB is functional in this species. First, the nonpermissive temperature we used was 39°C rather than the classical 40°C, simply because M. abscessus does not grow above this temperature (the optimal temperature is 28°C). The very slow multiplication of the bacterium combined with a potential leakage of replication of the plasmid at 39°C might affect the efficiency of counterselection by temperature. Second, M. abscessus might lack a sucrose transport system. The absence of a sucrose import system would explain the lack of toxicity in the presence of sucrose, even if the sacB gene is expressed. Whatever the explanation, this set of experiments demonstrates that the ts-sacB system in its present form is not functional in M. abscessus.
(ii) Mycobacteriophage system.
This system has been developed in parallel with the ts-sacB system. It is based on a thermosensitive mycobacteriophage that is used to deliver a mutated allele (2). We tested this system because it does not rely on the counterselection by sucrose in the presence of the sacB gene and also because it is very effective in various phylogenetically diverse mycobacterial species. We produced recombinant mycobacteriophages carrying the mmpL4b mutated allele and used them to transduce both M. smegmatis and M. abscessus. Transductants were selected at nonpermissive temperature (38°C) on kanamycin plates: similar numbers of clones were obtained in both cases (Table 3). All of the kanamycin-resistant M. smegmatis transductants were rough. In contrast, most (94%) of the kanamycin-resistant M. abscessus transductants were smooth (Table 3). We further investigated only rough transductants, the only isolates that may correspond to an allelic-exchange event. Several independent transduction experiments were performed with M. abscessus to obtain a sufficient number of independent rough colonies. Twenty rough transductants of each species were analyzed by Southern blotting using the upstream region of mmpL4b as a probe. All 20 rough M. smegmatis clones (100%) had undergone an allelic-exchange event; all 20 rough M. abscessus clones (100%) had undergone homologous recombination with a single crossover upstream of the kanamycin cassette (data not shown). Although the single crossover suggests that the entire phagemid (∼45 kb) is integrated, it is possible that in some cases the initial single crossover is followed by recombination and deletion events that suppress most of the phagemid DNA. Five rough clones of both M. smegmatis and M. abscessus were analyzed by TLC for their GPL content. All contained much less GPL than the parental wild-type strain, as expected (data not shown). Therefore, although it is possible to construct mutants with the desired phenotype (lack of GPL), none of the M. abscessus clones were the result of a double crossover event. The difference with M. smegmatis for which all of the clones corresponded to the double crossover event was striking. M. abscessus was less susceptible than M. smegmatis to an infection with the phage used (phAE87). Indeed, ∼105 times more phages were needed with M. abscessus to produce the same number of plaques as in M. smegmatis; we thought that the lower susceptibility to infection could be due to the presence of Tween 80 that could be metabolized less efficiently by M. abscessus than by M. smegmatis. Thus, the higher residual amount of this detergent would inhibit the infection by phages. We then performed transduction experiments using bacterial cells grown in the absence of Tween 80, but the results were not different from the previous experiments conducted in the presence of detergent. Tween 80 is thus not responsible of the lack of functionality of this system in M. abscessus. We next hypothesized that it might be feasible to isolate mutant mycobacteriophages that could be more infectious for M. abscessus. Phages were recovered from an infected M. abscessus culture and used to reinfect M. abscessus, but progenies were not more infectious than the parental strain. It is possible that the phage receptor is less accessible in M. abscessus, explaining the difference in phage susceptibility between the two species. It is also possible that this difference in susceptibility explains in part the inefficacy of this mutagenesis system in M. abscessus.
TABLE 3.
Comparative efficiency of the mycobacteriophage system in M. smegmatis mc2155 and M. abscessus CIP104536T strainsa
| Strain | Recovered coloniesb
|
Rough colonies
|
||
|---|---|---|---|---|
| Mean no. | SD | Mean no. | SD | |
| M. smegmatis | 50 | 3.7 | 50 | 3.7 |
| M. abscessus | 50 | 6.1 | 3 | 0.5 |
Experiments were performed five times.
That is, the mean number of kanamycin-resistant clones per experiment.
(iii) Recombineering system.
This system was developed very recently. It exploits the conditional expression of phage recombinases to promote the very efficient integration of a construct via homologous recombination. The recombinant strain is transformed with a linear DNA fragment carrying the mutation; this has the advantage of reducing background due to single crossover events (44). This potential of this tool has been illustrated in M. tuberculosis and in M. smegmatis, although it has been less extensively assessed than the two other systems discussed here due to its recent development. Nevertheless, this recombineering system has been used in M. smegmatis to produce unmarked mutants, demonstrating its high efficiency (43).
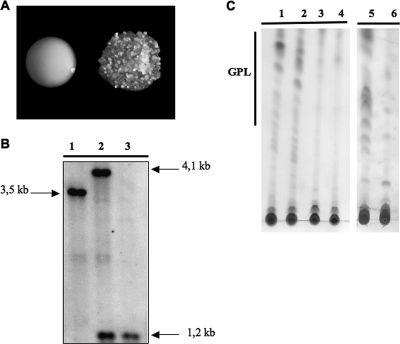
A zeocin-resistant derivative of the replicative plasmid pJV53 was constructed and used to transform both M. smegmatis and M. abscessus. Both recombinant strains were also transformed with a cassette containing the mmpL4b mutated allele and selected on kanamycin plates. In the case of M. smegmatis, all of the kanamycin-resistant transformants were rough; by contrast, only ∼10% of the M. abscessus kanamycin-resistant transformants were rough clones (Table 4 and Fig. 2A). The transformation experiment was repeated several times to obtain sufficient rough M. abscessus transformants for subsequent analysis. We further analyzed 20 rough clones by Southern blotting using the upstream region of mmpL4b as a probe. We further investigated only rough transformants, the only isolates that may correspond to an allelic-exchange event. The 20 rough M. smegmatis clones (100%) had all undergone an allelic-exchange event. Nineteen of the twenty rough M. abscessus transformants (95%) had undergone a homologous recombination event involving a single crossover upstream from the kanamycin cassette; one transformant (5%) had undergone an allelic-exchange event (Fig. 2B). Therefore, as only 1 in 20 rough kanamycin-resistant transformants had undergone an allelic-exchange event, this means that only ∼0.5% of the kanamycin resistant transformants were allelic-exchange mutants.
TABLE 4.
Comparative efficiency of recombineering system in M. smegmatis mc2155 and M. abscessus CIP104536T strainsa
| Strain | Recovered coloniesb
|
Rough colonies
|
Allelic exchangec | ||
|---|---|---|---|---|---|
| Mean no. | SD | Mean no. | SD | ||
| M. smegmatis | 43 | 1.5 | 43 | 1.5 | 43 |
| M. abscessus (cohesive ends) | 22 | 0.6 | 2 | 0.2 | 1 |
| M. abscessus (incompatible ends) | 22 | 2.1 | 3 | 0.3 | 8 |
Experiments were performed five times.
That is, the mean number of kanamycin-resistant clones per experiment.
That is, the mean number of allelic-exchange events with respect of the kanamycin-resistant clones.
FIG. 2.
(A) Phenotypic appearance of wild-type smooth and rough M. abscessus mutant strains. (B) Southern blot analysis of the wild-type strain (lane 1), the single crossover (upstream from the kanamycin cassette) (lane 2), and the allelic-exchange mutant (lane 3) in M. abscessus using the upstream part of mmpL4b as a probe. (C) TLC analysis of the crude lipid fractions of the wild-type strain, (lane 1), a smooth kanamycin transformant (lane 2), an allelic-exchange mutant (lane 3), and the single crossover (upstream from the kanamycin cassette, lane 4) mutants of M. abscessus; TLC analyses of M. smegmatis wild type (lane 5) and mmpL4b mutant (lane 6) are also shown. TLC was performed using chloroform-methanol (9:1 [vol:vol]) as the solvent system and developed with iodine (250 μg each deposit).
The observation of single crossover events in M. abscessus was surprising since the construct used was linear. To obtain this type of event, the linear construct would need to recircularize in the bacilli. The extremities of the construct were compatible, and this may have favored recircularization and thereby reduced the frequency of double crossover events. We therefore designed a new construct with incompatible ends and used it to transform M. smegmatis and M. abscessus. Again, 100% of the M. smegmatis kanamycin-resistant transformants were rough. In the case of M. abscessus, 15% of the transformants were rough with this new construct. We obtained 20 rough M. abscessus transformants (several independent transformations were required) and analyzed them by Southern blotting with the upstream region of mmpL4b as a probe. Eight (∼40%) corresponded to allelic exchange, and twelve (∼60%) corresponded to homologous recombination with a single crossover upstream from the kanamycin cassette (Table 4). Therefore, ca. 7% of the kanamycin-resistant transformants (including both rough and smooth) were allelic-exchange mutants; this is substantially better than the 0.5% for the system with the compatible ends. Nevertheless, the system is still much less efficient than in M. smegmatis, in which the corresponding value is 100%. We tried to increase the efficiency of the system further by treating the recombinogenic substrate with an alkaline solution, since this has been shown in other model systems to increase the frequency of allelic-exchange events (16). However, in our model system, alkaline treatment did not increase the frequency of allelic-exchange events, and the ratio between rough and smooth transformants was unaffected. Nevertheless, the recombineering system was the only one of the three systems tested that, in our hands, could be successfully used to construct allelic-exchange mutants in M. abscessus.
Several rough and smooth M. abscessus transformants were analyzed for their GPL content by TLC (Fig. 2C). The GPL content of the smooth transformants was indistinguishable from that in the parental wild-type strain; the rough mutants (both those corresponding to allelic exchange or to a single crossover upstream from the kanamycin cassette) contained much less GPL than the wild-type parental strain, a finding consistent with the mmpL4b mutation.
DISCUSSION
Allelic exchange is a powerful method for generating targeted mutants and thereby studying the contribution of a particular gene or amino acid to a particular phenotype or function (30). Work on mycobacteria has long lagged behind work on other genera (18), but effective experimental tools have now been developed for several species, including M. tuberculosis and M. smegmatis (41). M. abscessus is an emerging pathogen that has received very little attention until recently. We report here for the first time the construction of a defined mutant in this species. We also show that the ts-sacB system cannot be used in M. abscessus, for an unknown reason. The best of the three systems we tested was the recombineering system (Table 5). The use of mmpL4b as a target gene greatly facilitated the identification of mutants and quantification of the various genetic events, since the mutation gives rise to rough colonies, a visually observable phenotype (29, 40). All kanamycin-resistant M. smegmatis clones were allelic-exchange mutants, but even in the best conditions only 7% of the kanamycin-resistant M. abscessus clones were the result of that type of event. It is worth noting that 100% of the M. smegmatis clones corresponded to allelic exchange, whatever the system used, despite the fact that a heterologous construct was used. There were indeed only 78 and 76% identities at the DNA level on the right and left arms of the construct, respectively. This illustrates the power of these three genetic tools in this species. We found that if the recombinogenic substrate contains cohesive ends, it may circularize in vivo and thereby generate single crossover events in M. abscessus (Table 4). We did not analyze the smooth transformants or transductants of M. abscessus since they could not be the result of allelic exchange. It is likely that these clones correspond to a mixture of spontaneous kanamycin-resistant clones and mutants originating from illegitimate recombination and homologous recombination with a single crossover downstream from the kanamycin cassette.
TABLE 5.
Summary of the efficiencies of the ts-sacB, mycobacteriophage, and recombineering genetic systems examined in this studya
| Strain | Efficiency (mean %)
|
|||||
|---|---|---|---|---|---|---|
| ts-sacB
|
Mycobacteriophage
|
Recombineering
|
||||
| Rough clones | Allelic-exchange mutants | Rough clones | Allelic-exchange mutants | Rough clones | Allelic-exchange mutants | |
| M. smegmatis | 100 | 100 | 100 | 100 | 100 | 100 |
| M. abscessus | 0 | 0 | ∼6 | 0 | ∼15 | ∼7 |
The mean percentages of rough clones and allelic-exchange mutants expressed of all kanamycin-resistant clones are given.
The reason for the lower efficiency of the recombineering system in M. abscessus than in M. smegmatis and M. tuberculosis is unclear. The transformation of M. abscessus is ∼2 orders of magnitude less efficient than the transformation of M. smegmatis mc2155 or M. tuberculosis, but this cannot completely explain the low efficiency of the recombineering system in this species. The phage recombinases (genes gp60_61) that are overexpressed in the recipient strain are from a mycobacteriophage (Che9c) that was originally selected because it infects M. smegmatis (44). It is not known whether this phage efficiently infects M. abscessus. Possibly, these recombinogenic enzymes may be much more active in M. smegmatis than in M. abscessus, such that the frequency of double crossover events in M. abscessus is low. It may therefore be useful to isolate phages that infect M. abscessus efficiently (20), since they could be used to adapt the system to this species by overexpressing appropriate recombinases. Another possibility for improving this system would be to construct an M. abscessus ligD mutant strain: this gene promotes DNA circularization by ligation in vivo (27) and may be responsible for the high background due to single crossover events. In addition, the high frequency of spontaneous kanamycin-resistant clones obviously decreased the potency of selection. In the future, this recombineering system could be adapted to M. massiliense, another emerging opportunistic pathogen that is closely related to M. abscessus and M. chelonae (1, 37, 38, 45).
Although further improvement and adaptation of this system for M. abscessus is feasible and may be desirable, we show here that it is already possible to construct defined mutants in this species. The present study thus constitutes the first step toward the development of new therapeutic and preventive strategies against this emerging pathogen.
Supplementary Material
Acknowledgments
We thank T. Weisbrod and W. Jacobs for providing us with the phage-based system (phAE87) and for their expert advice. We thank J. van Kessel and G. Hatfull for providing the recombineering system and for valuable advice. We thank L. Kremer for kindly providing pJSC347. We thank Jean-Louis Gaillard for stimulating discussion. We thank B. Heyme for preparing and providing M. abscessus clinical isolates. We thank D. Euphrasie for lipid preparation and TLC analysis. We thank R. Manganelli for discussing the cohesive-end hypothesis. We thank C. Jeanneau and M. Bertili for bacterial medium preparation.
H.M. is funded by a doctoral grant from Vaincre la Mucoviscidose. We gratefully acknowledge Vaincre la Mucoviscidose and INSERM for funding this project.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adekambi, T., M. Reynaud-Gaubert, G. Greub, M. J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulard, A., C. Jourdan, A. Mercenier, and C. Locht. 1992. Rapid mycobacterial plasmid analysis by electroduction between Mycobacterium spp. and Escherichia coli. Nucleic Acids Res. 20:4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billman-Jacobe, H. 2004. Glycopeptidolipid synthesis in mycobacteria. Curr. Sci. 86:11-114. [Google Scholar]
- 5.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 6.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd, T. F., and C. R. Lyons. 1999. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67:4700-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catherinot, E., J. Clarissou, G. Etienne, F. Ripoll, J. F. Emile, M. Daffe, C. Perronne, C. Soudais, J. L. Gaillard, and M. Rottman. 2007. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect. Immun. 75:1055-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerf, O., M. Griffiths, and F. Aziza. 2007. Assessment of the prevalence of Mycobacterium avium subsp. paratuberculosis in commercially pasteurized milk. Foodborne Pathog. Dis. 4:433-447. [DOI] [PubMed] [Google Scholar]
- 10.Chacon, O., L. E. Bermudez, and R. G. Barletta. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329-363. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell Mol. Life Sci. 58:2018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 13.Deshayes, C., D. Kocincova, G. Etienne, and J. M. Reyrat. 2008. Glycopeptidolipids: a complex pathway for small pleiotropic molecules. ASM Press, Washington, DC.
- 14.Gao, L. Y., R. Groger, J. S. Cox, S. M. Beverley, E. H. Lawson, and E. J. Brown. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glickman, M. S., and W. R. Jacobs, Jr. 2001. Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104:477-485. [DOI] [PubMed] [Google Scholar]
- 16.Hinds, J., E. Mahenthiralingam, K. E. Kempsell, K. Duncan, R. W. Stokes, T. Parish, and N. G. Stoker. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145(Pt. 3):519-527. [DOI] [PubMed] [Google Scholar]
- 17.Howard, S. T., E. Rhoades, J. Recht, X. Pang, A. Alsup, R. Kolter, C. R. Lyons, and T. F. Byrd. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152:1581-1590. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, W. R., Jr. 2000. Mycobacterium tuberculosis: a once genetically intractable organism. ASM Press, Washington, DC.
- 19.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 20.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 21.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J. Bacteriol. 178:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 24.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1998. Genetic advances for studying Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 28:413-420. [DOI] [PubMed] [Google Scholar]
- 25.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Positive selection of allelic exchange mutants in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 144:161-166. [DOI] [PubMed] [Google Scholar]
- 26.Pierre-Audigier, C., A. Ferroni, I. Sermet-Gaudelus, M. Le Bourgeois, C. Offredo, H. Vu-Thien, B. Fauroux, P. Mariani, A. Munck, E. Bingen, D. Guillemot, G. Quesne, V. Vincent, P. Berche, and J. L. Gaillard. 2005. Age-related prevalence and distribution of nontuberculous mycobacterial species among patients with cystic fibrosis. J. Clin. Microbiol. 43:3467-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitcher, R. S., L. M. Tonkin, J. M. Daley, P. L. Palmbos, A. J. Green, T. L. Velting, A. Brzostek, M. Korycka-Machala, S. Cresawn, J. Dziadek, G. F. Hatfull, T. E. Wilson, and A. J. Doherty. 2006. Mycobacteriophage exploit NHEJ to facilitate genome circularization. Mol. Cell 23:743-748. [DOI] [PubMed] [Google Scholar]
- 28.Prinz, B. M., S. Michaelis, N. Kettelhack, B. Mueller, G. Burg, and W. Kempf. 2004. Subcutaneous infection with Mycobacterium abscessus in a renal transplant recipient. Dermatology 208:259-261. [DOI] [PubMed] [Google Scholar]
- 29.Recht, J., A. Martinez, S. Torello, and R. Kolter. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182:4348-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyrat, J. M., V. Pelicic, B. Gicquel, and R. Rappuoli. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 66:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripoll, F., C. Deshayes, S. Pasek, F. Laval, J. L. Beretti, F. Biet, J. L. Risler, M. Daffe, G. Etienne, J. L. Gaillard, and J. M. Reyrat. 2007. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics 8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rottman, M., E. Catherinot, P. Hochedez, J. F. Emile, J. L. Casanova, J. L. Gaillard, and C. Soudais. 2007. Importance of T cells, gamma interferon, and tumor necrosis factor in immune control of the rapid grower Mycobacterium abscessus in C57BL/6 mice. Infect. Immun. 75:5898-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambandamurthy, V. K., X. Wang, B. Chen, R. G. Russell, S. Derrick, F. M. Collins, S. L. Morris, and W. R. Jacobs, Jr. 2002. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat. Med. 8:1171-1174. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 35.Sanguinetti, M., F. Ardito, E. Fiscarelli, M. La Sorda, P. D'Argenio, G. Ricciotti, and G. Fadda. 2001. Fatal pulmonary infection due to multidrug-resistant Mycobacterium abscessus in a patient with cystic fibrosis. J. Clin. Microbiol. 39:816-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sermet-Gaudelus, I., M. Le Bourgeois, C. Pierre-Audigier, C. Offredo, D. Guillemot, S. Halley, C. Akoua-Koffi, V. Vincent, V. Sivadon-Tardy, A. Ferroni, P. Berche, P. Scheinmann, G. Lenoir, and J. L. Gaillard. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg. Infect. Dis. 9:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin, D. M., C. S. Yang, J. M. Yuk, J. Y. Lee, K. H. Kim, S. J. Shin, K. Takahara, S. J. Lee, and E. K. Jo. 2008. Mycobacterium abscessus activates the macrophage innate immune response via a physical and functional interaction between TLR2 and dectin-1. Cell. Microbiol. 10:1608-1621. [DOI] [PubMed] [Google Scholar]
- 38.Simmon, K. E., J. I. Pounder, J. N. Greene, F. Walsh, C. M. Anderson, S. Cohen, and C. A. Petti. 2007. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J. Clin. Microbiol. 45:1978-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 40.Sonden, B., D. Kocincova, C. Deshayes, D. Euphrasie, L. Rhayat, F. Laval, C. Frehel, M. Daffe, G. Etienne, and J. M. Reyrat. 2005. Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface. Mol. Microbiol. 58:426-440. [DOI] [PubMed] [Google Scholar]
- 41.Stoker, N. G., P. Sander, and J. M. Reyrat. 2005. Gene replacement systems. ASM Press, Washington, DC.
- 42.Talati, N. J., N. Rouphael, K. Kuppalli, and C. Franco-Paredes. 2008. Spectrum of CNS disease caused by rapidly growing mycobacteria. Lancet Infect. Dis. 8:390-398. [DOI] [PubMed] [Google Scholar]
- 43.van Kessel, J. C., and G. F. Hatfull. 2008. Mycobacterial recombineering. Methods Mol. Biol. 435:203-215. [DOI] [PubMed] [Google Scholar]
- 44.van Kessel, J. C., and G. F. Hatfull. 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4:147-152. [DOI] [PubMed] [Google Scholar]
- 45.Viana-Niero, C., K. V. Lima, M. L. Lopes, M. C. Rabello, L. R. Marsola, V. C. Brilhante, A. M. Durham, and S. C. Leao. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J. Clin. Microbiol. 46:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villeneuve, C., G. Etienne, V. Abadie, H. Montrozier, C. Bordier, F. Laval, M. Daffe, I. Maridonneau-Parini, and C. Astarie-Dequeker. 2003. Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. J. Biol. Chem. 278:51291-51300. [DOI] [PubMed] [Google Scholar]
- 47.World Health Organization. 2007. Buruli ulcer disease. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs199/en/.
- 48.World Health Organization. 2005. Leprosy. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs101/en/.
- 49.World Health Organization. 2007. Tuberculosis. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs104/en/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.