Abstract
CsrA is a global posttranscriptional regulator of numerous physiological processes, such as glycogenesis and glycolysis. Here, we show that the csrA gene of Escherichia coli is essential for growth on LB and on synthetic medium containing glycolytic carbon sources. However, csrA is not necessary for growth on synthetic medium containing pyruvate, showing that the Krebs cycle is functional in the csrA::cat deletion mutant. Deletion of the glgCAP operon in the csrA::cat mutant restored the ability to grow on LB and on synthetic medium containing glycolytic carbon sources, showing that growth inhibition is due to an excess of glycogen synthesis.
The Escherichia coli csrA (carbon storage regulator A) gene encodes a global regulator controlling a wide variety of biological processes, such as glycogen synthesis, glycolysis, and motility as well as cyclic-di-GMP synthesis and biofilm formation (2, 7, 14, 19, 21, 22, 26, 28). CsrA regulates gene expression both positively and negatively at the posttranscriptional level by modulating translation. It binds as a homodimer to a specific sequence (consensus sequence, ACANGGANGA) on both its mRNA targets and its small regulator RNAs (1, 2, 7, 18, 23, 26). Binding of CsrA to the leader sequence of the mRNA target blocks ribosome binding on the Shine-Dalgarno sequence (2, 7, 26) and most likely leads to its rapid degradation (18). As a positive regulator, CsrA enhances translation efficiency by stabilizing its mRNA targets (28).
Homologues of csrA have been identified, and their role in several bacterial species has been studied (for a review, see reference 15). These csrA homologues share regulatory networks with those identified in E. coli, such as motility (17) and biofilm formation (8). These homologues also appear to regulate other processes, such as virulence in Salmonella enterica, Legionella pneumophila, Helicobacter pylori, Proteus mirabilis, and Yersinia pseudotuberculosis (3, 9, 13, 17, 24); quorum sensing in Erwinia carotovora and Vibrio cholerae (5, 16); and oxidative stress resistance in Campylobacter jejuni (8).
In E. coli, the csrA gene was identified by transposon mutagenesis in a screen for mutants with altered glycogen accumulation. The kanamycin transposon insertion site is located at codon 51 in the csrA coding sequence (which encodes a protein of 61 amino acids). In addition to an increased glycogen accumulation, the csrA::kan mutant is affected for motility, biofilm formation, and glycolysis (2, 19, 21, 22, 28). In association with an rpoS defective mutation, the csrA::kan mutation led to an extended lag phase upon growth on acetate (27). Suppressor mutations restoring growth of the double mutant on acetate were mapped in the glg locus (27). This indicates that the growth defect presented by the double csrA::kan rpoS mutant was associated with a high level of glycogen accumulation (27). Acetate sensitivity appears to require both the csrA::kan and the rpoS mutations since single mutants were not sensitive to acetate and did not evolve suppressor mutations (27).
The E. coli csrA gene is essential under certain growth conditions.
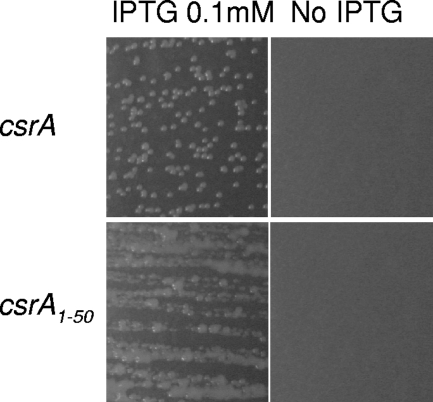
In an attempt to construct a csrA deletion mutant of E. coli, large series of homologous recombination experiments were carried out with the DJ624 strain (MG1655 lacX74 malP::lacIq) (12) to replace the csrA open reading frame (ORF) with a chloramphenicol resistance cassette (6). Despite many attempts under various conditions, no deletion mutants were obtained, suggesting that csrA is essential for growth. To test this hypothesis, various rescue plasmids were constructed. The csrA gene was cloned in the pBAD33 (11) and pKK223-3 (4) expression vectors. The expression of the rescue copy of csrA was induced in LB medium by the addition of arabinose (0.4%) and isopropyl β-d-1-thiogalactopyranoside (IPTG) (1 mM). These constructs were checked by sequencing, and their ability to complement a csrA::kan mutation was tested. Using these plasmids as csrA rescue copy, attempts to delete csrA in DJ624 were not successful, indicating that high expression levels of CsrA might be deleterious under these conditions. To bypass this problem, the csrA gene was cloned in the low-copy-number pWSK129 vector under the control of the lac promoter (25). DJ624 was transformed by this construct, and csrA expression was induced by the addition of 0.1 mM IPTG in the culture medium. Under these conditions, csrA deletion mutants were obtained on LB plates containing chloramphenicol (20 μg/ml) and kanamycin (50 μg/ml) at 37°C (ΔcsrA::cat). The replacement of the csrA ORF with the chloramphenicol resistance cassette was checked by PCR using primers flanking the csrA ORF. The ΔcsrA::cat mutant strain containing the pWSK-csrA plasmid was able to grow in the presence of 0.1 mM IPTG. No growth was observed on LB plates without IPTG, showing that the csrA gene is essential for growth in LB medium (Fig. 1, upper panels).
FIG. 1.
The csrA gene is essential in E. coli. The ΔcsrA::cat mutant containing either the pWSK-csrA (upper panels) or the pKK-csrA1-50 (lower panels) plasmids were grown on LB plates containing the appropriate antibiotics with 0.1 mM IPTG (left panels) and without IPTG (right panels).
The CsrA1-50 peptide is partially active.
The kan transposon is inserted at codon 51 in the csrA gene (21). Since this insertion mutant is viable on LB medium, it leaves the possibility that the 50 amino-terminal amino acids might retain some partial activity as proposed by Romeo and coworkers (21). To test this hypothesis, the first 150 5′ nucleotides of the csrA coding sequence, followed by a stop codon, were cloned in pKK223-3. The csrA deletion mutant containing the pWSK-csrA plasmid was transformed with the pKK-csrA1-50 plasmid. To displace the pWSK-csrA plasmid, we used the incompatibility properties of pWSK129 and pMLO59 (both plasmids have a pSC101 replication origin but different antibiotic resistance genes; pWSK129 confers resistance to kanamycin and pMLO59 to spectinomycin). The ΔcsrA::cat pWSK-csrA pKK-csrA1-50 strain was transformed by the pMLO59 vector, and the resulting transformants were plated on LB plates containing spectinomycin (100 μg/ml), chloramphenicol (20 μg/ml), and 0.1 mM IPTG to induce the expression of the csrA1-50 polypeptide. Transformants were checked for kanamycin sensitivity (loss of the pWSK-csrA plasmid). The ΔcsrA::cat mutant containing the pKK-csrA1-50 plasmid was able to form colonies on LB plates containing 0.1 mM IPTG but not on plates without IPTG (Fig. 1, lower panels), showing that the CsrA1-50 peptide is functional. On IPTG plates, the colonies had an abnormal phenotype (forming heterogeneous and “sticky” colonies). The molecular mechanisms underlying this phenotype are unknown. This phenotype does not appear to be dependent on the CsrA1-50 amount, since it has also been observed at higher IPTG concentrations (data not shown). As the carboxy-terminal part of CsrA (amino acids 51 to 54) is involved in RNA binding (20), the truncated form of CsrA might have a reduced affinity for RNA (another RNA binding site has been mapped in the amino-terminal region of CsrA). This might provide an explanation for the phenotype of the csrA::kan mutant.
An excess of glycogen accumulation impairs the growth of the csrA::cat mutant.
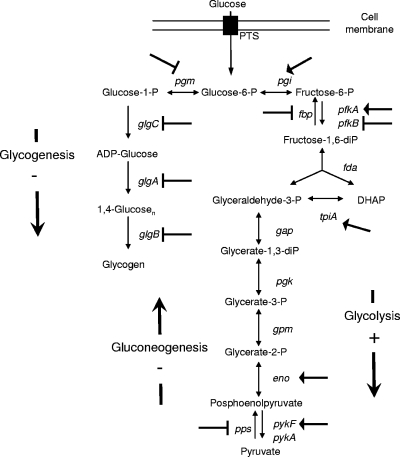
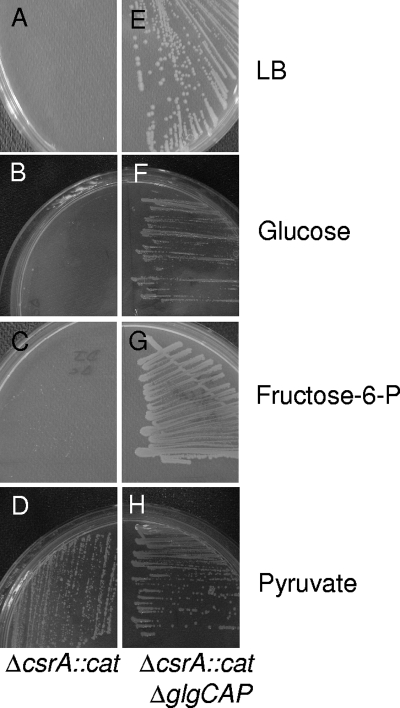
CsrA controls the intracellular carbon flux by positively regulating the glycolysis pathway and negatively regulating the gluconeogenesis pathway and glycogen synthesis (Fig. 2) (22). In the absence of CsrA, glycogen synthesis is strongly favored. Indeed, the csrA::kan insertion mutation resulted in 20-fold-higher levels of glycogen (21). The csrA::cat mutant might thus be unable to direct the carbon flux to the Krebs cycle, because of a reduced glycolysis and an enhanced glycogenesis. To test this hypothesis, the ΔcsrA::cat mutant was plated on Ceria synthetic medium (10) containing amino acids and glycolytic carbon sources. The ΔcsrA::cat mutant was unable to grow on glucose and on fructose-6-phosphate as sole carbon sources (Fig. 3B and C). Interestingly, it was able to grow on pyruvate, which is the carbon source at the junction between glycolysis and the Krebs cycle (Fig. 3D). On fructose-6-phosphate plates, very small colonies appeared after 16 h at 37°C (too small to be seen in Fig. 3C). These data indicate that the early steps of glycolysis are not fully functional in the ΔcsrA::cat mutant. With glucose and fructose-6-phosphate as sole carbon sources, the carbon flux might be essentially directed to glycogen synthesis, thereby impairing glycolysis and the Krebs cycle. We tested whether the deletion of the glgCAP operon, which is involved in the first two steps of glycogen synthesis, would restore the ability to grow on glycolytic carbon sources. The glgCAP operon was deleted in the DJ624 strain by using the method described in reference 6. The csrA::cat deletion was then transduced by P1 in the ΔglgCAP strain containing the pWSK-csrA plasmid in the presence of 0.1 mM IPTG. Transductants were obtained and checked by PCR with primers flanking the csrA ORF. Interestingly, the double ΔglgCAPΔcsrA::cat pWSK-csrA mutant was able to form colonies on LB plates without IPTG (Fig. 3E) as well as on glucose and fructose-6-phosphate plates (Fig. 3F and G), unlike the ΔcsrA::cat pWSK-csrA mutant (Fig. 3A, B, and C). Both strains were able to grow in the presence of pyruvate as the sole carbon source (Fig. 3D and H). Our data show that in the csrA::cat mutant, glycogen synthesis is favored and that glycogen accumulation impairs viability. In the absence of the glycogen synthesis pathway, the carbon flux is most likely redirected to glycolysis and/or pentose phosphate pathways, thereby enabling the Krebs cycle to be functional.
FIG. 2.
CsrA-mediated regulation of the central carbon flux. CsrA positively regulates glycolysis and negatively regulates gluconeogenesis and glycogen synthesis. Negative and positive regulations of target genes are indicated by blocked arrows ( ) and arrows, respectively. Adapted from reference 22.
) and arrows, respectively. Adapted from reference 22.
FIG. 3.
Excessive glycogen accumulation in the ΔcsrA::cat mutant impairs viability on glycolytic carbon sources. The ΔcsrA::cat (left panels) and the ΔcsrA::cat ΔglgCAP (right panels) mutants containing the pWSK-csrA plasmid were streaked without IPTG on LB medium (A and E) or on minimal medium containing amino acids, with either glucose (B and F), fructose-6-phosphate (C and G), or pyruvate (D and H) as the sole carbon source.
Conclusion.
We have shown that the csrA gene is essential in E. coli under growth conditions that lead to an excess of glycogen accumulation. Deletion of the glycogen synthesis pathway restored the growth of the ΔcsrA::cat mutant, most likely by forcing the carbon flux to enter glycolysis and the Krebs cycle.
Acknowledgments
We thank all the members of the laboratory for discussions and suggestions and Nadim Majdalani for critical reading of the manuscript. We also thank Tony Romeo for providing us with the csrA::kan mutant.
This work was supported by Fonds de la Recherche Scientifique Médicale (3.4510.02), the Fonds Brachet, and the Fondation Van Buuren. J.T. is supported by the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture.
Footnotes
Published ahead of print on 19 December 2008.
REFERENCES
- 1.Babitzke, P., and T. Romeo. 2007. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10156-163. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. S., I. Morozov, K. Suzuki, T. Romeo, and P. Babitzke. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol. Microbiol. 441599-1610. [DOI] [PubMed] [Google Scholar]
- 3.Barnard, F. M., M. F. Loughlin, H. P. Fainberg, M. P. Messenger, D. W. Ussery, P. Williams, and P. J. Jenks. 2004. Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 5115-32. [DOI] [PubMed] [Google Scholar]
- 4.Brosius, J., and A. Holy. 1984. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl. Acad. Sci. USA 816929-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui, Y., A. Chatterjee, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J. Bacteriol. 1775108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubey, A. K., C. S. Baker, K. Suzuki, A. D. Jones, P. Pandit, T. Romeo, and P. Babitzke. 2003. CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J. Bacteriol. 1854450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields, J. A., and S. A. Thompson. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 1903411-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsbach-Birk, V., T. McNealy, C. Shi, D. Lynch, and R. Marre. 2004. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int. J. Med. Microbiol. 29415-25. [DOI] [PubMed] [Google Scholar]
- 10.Glansdorff, N. 1965. Topography of cotransducible arginine mutations in Escherichia coli K-12. Genetics 51167-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen, A. M., H. Lehnherr, X. Wang, V. Mobley, and D. J. Jin. 2003. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 481621-1631. [DOI] [PubMed] [Google Scholar]
- 13.Heroven, A. K., K. Bohme, M. Rohde, and P. Dersch. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 681179-1195. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapouge, K., M. Schubert, F. H. Allain, and D. Haas. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67241-253. [DOI] [PubMed] [Google Scholar]
- 16.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 581186-1202. [DOI] [PubMed] [Google Scholar]
- 17.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2003. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J. Med. Microbiol. 5219-28. [DOI] [PubMed] [Google Scholar]
- 18.Liu, M. Y., and T. Romeo. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 1794639-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, M. Y., H. Yang, and T. Romeo. 1995. The product of the pleiotropic Escherichia coli gene csrA modulates glycogen biosynthesis via effects on mRNA stability. J. Bacteriol. 1772663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercante, J., K. Suzuki, X. Cheng, P. Babitzke, and T. Romeo. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. J. Biol. Chem. 28131832-31842. [DOI] [PubMed] [Google Scholar]
- 21.Romeo, T., M. Gong, M. Y. Liu, and A. M. Brun-Zinkernagel. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 1754744-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabnis, N. A., H. Yang, and T. Romeo. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J. Biol. Chem. 27029096-29104. [DOI] [PubMed] [Google Scholar]
- 23.Schubert, M., K. Lapouge, O. Duss, F. C. Oberstrass, I. Jelesarov, D. Haas, and F. H. Allain. 2007. Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat. Struct. Mol. Biol. 14807-813. [DOI] [PubMed] [Google Scholar]
- 24.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 1857257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 26.Wang, X., A. K. Dubey, K. Suzuki, C. S. Baker, P. Babitzke, and T. Romeo. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 561648-1663. [DOI] [PubMed] [Google Scholar]
- 27.Wei, B., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 1821632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40245-256. [DOI] [PubMed] [Google Scholar]