Abstract
The Brachyspira hyodysenteriae B204 genome sequence revealed three VSH-1 tail genes, hvp31, hvp60, and hvp37, in a 3.6-kb cluster. The location and transcription direction of these genes relative to those of the previously described VSH-1 16.3-kb gene operon indicate that the gene transfer agent VSH-1 has a noncontiguous, divided genome.
Gene transfer agents (GTAs) have been described for diverse bacterial species (2, 5, 13, 19, 22). These GTAs resemble small tailed bacteriophages in ultrastructure and package short, random fragments of their bacterial host genomes. Gene transfer ability and “avirulence,” i.e., an inability of GTA particles to lyse bacteria or form plaques, are GTA traits (1, 11, 14, 20, 21, 24).
VSH-1 is a GTA produced by the anaerobic, pathogenic spirochete Brachyspira hyodysenteriae strain B204 (11). VSH-1 particles contain 7.5-kb fragments of B. hyodysenteriae DNA and transfer genes, including antibiotic resistance genes, between strains of the spirochete (11, 15, 20, 21). VSH-1-like elements are widespread in Brachyspira species (3, 4, 22, 26).
Previously, head (capsid) and tail proteins of VSH-1 particles were sequenced, and the sequences were used to establish the VSH-1 genome (15). VSH-1 structural and escape (lysis) proteins are encoded by contiguous clusters of VSH-1 head, tail, and lysis genes (Fig. 1; Table 1) occupying 16.3 kb of the B204 chromosome (15). In those studies, the gene encoding Hvp60, a tail-associated protein with the sequence N′-M_K_MPYHFLRNKIYKLPPAPYINE was not found within the 16.3-kb VSH-1 genome cluster (15). Two additional tail-associated proteins, Hvp31 and Hvp37, with sequence ambiguities at several amino acid positions, also could not be located (unpublished observations).
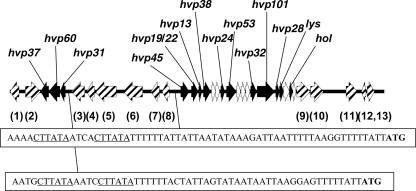
FIG. 1.
VSH-1 gene map based on this and previous studies. VSH-1 genes (black arrows), B. hyodysenteriae genes (striped arrows), and ORFs greater than 240 bp with no GenBank homologs (white arrows) are oriented according to their direction of transcription. Descriptions of identified or putative proteins encoded by the genes are included in Table 1. At the bottom of the figure, sequences immediately upstream of the VSH-1 16.3-kb and 3.6-kb gene clusters are given. (For convenience, the reverse complement sequence upstream of the 3.6-kb gene cluster is provided.) ATG represents start codons of the respective genes encoding Hvp31 and Hvp45 proteins. CTTATA hexanucleotide repeat sequences representing potential transcription control regions are underlined.
TABLE 1.
Genes and proteins of VSH-1 and B. hyodysenteriae B204 identified in this and previous studiesa
| Gene(s) | ORF in B204 draft genome | Protein identity | Identification basis | Closest protein match (GenBank accession no.) and E value |
|---|---|---|---|---|
| VSH-1 | ||||
| hvp45 to hvp24 | 02481 to 02474 | Head proteins | Protein sequencing | None |
| hvp53 to hvp28 | 02472 to 02467 | Tail proteins | Protein sequencing | None |
| lys/hol | 02465/02463 | Endolysin/holin lysis proteins | Enzyme activity/predicted properties | Salmonella phage ε15 endolysin (AAO06088); E = 5 × 10−29 |
| hvp31 to hvp37 | 02494 to 02497 | Tail proteins | Protein sequencing | None |
| B. hyodysenteriae B204b | ||||
| 1 | 02501 | AdSS | Conserved domains | Thermosinus carboxydivorans adenylosuccinate synthase (ZP_01666816.1); E = 10−146 |
| 2 | 02498 | Unknown | Paramecium TPR protein (XP_001427675); E = 7 × 10−50 | |
| 3 | 02493 | Mcp | Conserved domains | B. hyodysenteriae methyl-accepting chemosensory protein (AAAX81982); E = 10−92 |
| 4 | 02491 | Adh | Conserved domains | Caldicellulosiruptor saccharolyticus Fe-alcohol dehydrogenase (YP_001179521); E = 10−116 |
| 5 | 02490 | Unknown | Tetrahymena TPR protein (XP_001026739); E = 10−22 | |
| 6 | 02487 | Trep | Treponema denticola unknown function protein (NP_972591); E = 8 × 10−34 | |
| 7 | 02485 | AA transport | Conserved domains | Fusobacterium nucleatum Ser/Thr transporter (NP_604045); E = 10−105 |
| 8 | 02484 | Fe-S oxidoreductase | Conserved domains | Clostridium kluyveri Fe-S oxidoreductase (YP_001393555); E = 10−39 |
| 9 | 02461 | Mcp | Conserved domains | B. hyodysenteriae methyl-accepting chemosensory protein (AAP58978); E = 4 × 10−94 |
| 10 | 02460 | Mcp | Conserved domains | B. hyodysenteriae methyl-accepting chemosensory protein (AAP58978); E = 3 × 10−112 |
| 11 | 01437 | NhaC | Conserved domains | Clostridium leptum Na+/H+ antiporter (ZP_02079635); E = 8 × 10−101 |
| 12 | 01435c | Unknown | Conserved domains | Ruminococcus gnavus unknown function protein (ZP_02040322); E = 3 × 10−58 |
| 13 | 01433c | Adh | Conserved domains | B. hyodysenteriae WA-1 alcohol dehydrogenase (ABS12704); E = 0.0 |
Described previously by Matson et al. (15). ORF number designations in column 2 refer to positions in the B. hyodysenteriae B204 draft genome. Closest protein matches in column 5 are all hypothetical proteins.
B. hyodysenteriae B204 genes (bacterial gene homologs) are labeled, for convenience, according to their positions in Fig. 1.
ORFs 01433 and 01435 have adjacent gene homologs on the Ruminococcus gnavus genome map.
A draft sequence of the B. hyodysenteriae B204 genome was recently generated in our laboratory by pyrosequencing (Roche FLX). (The B. hyodysenteriae B204 draft genome is currently undergoing closure and annotation. During completion of this effort, web access to a pseudogenome assembly of the DNA contigs will be provided upon request. The VSH-1 genes described in this paper are contained within contig00046 in the draft assembly.) One open reading frame (ORF) was predicted to encode a 62.5-kDa protein with an N-terminal sequence matching the Hvp60 sequence. This ORF, designated hvp60, was flanked by two ORFs, designated hvp31 and hvp37 (Fig. 1). The predicted sequences of proteins encoded by these ORFs, N′-MAFDFKLIVRDKYSEEIFDI… and N′-MNNNLNKNISDTNKELQNIE…, were discernible in the ambiguous sequences previously obtained for Hvp31 and Hvp37, respectively (unpublished observations). Predicted tail proteins Hvp31, Hvp60, and Hvp37, encoded by a 3.6-kb gene cluster, share no significant amino acid identity with any of the proteins in the GenBank database.
The VSH-1 3.6-kb and 16.3-kb gene clusters are oriented in opposite transcriptional directions (Fig. 1). Six genes (gene numbers 3 to 8 [Fig. 1]) encoding putative proteins with various predicted functions (Table 1) comprise a 16.7-kb region that separates the 3.6-kb and 16.3-kb clusters. None of the six hypothetical proteins resemble other GTA or phage-related proteins, and several are likely to have bacterial physiological functions.
The sequence ACTTATA was previously identified as a heptanucleotide repeat upstream of hvp45, the first gene in the VSH-1 16.3-kb cluster, and was proposed as a possible binding site for proteins involved in transcription regulation of VSH-1 expression (16). The repeat is more likely to be a hexanucleotide sequence, in that a matching six-nucleotide repeat sequence (… CTTATAAATCCTTATATTTTTTA… [repeat sequence is underlined]) was also detected upstream of the VSH-1 3.6-kb cluster (Fig. 1). A string of six T's follows the second CTTATA repeat upstream of both VSH-1 gene clusters. Direct-repeat recognition sequences have been identified for both bacterial and bacteriophage promoters (7, 8, 17, 23). In the draft B204 genome, similar “recognition” sequences are present upstream of two other single ORFs at least 100 kb from the VSH-1 genes. The identities of hypothetical proteins encoded by those genes are uncertain. Their possible involvement in VSH-1 production is currently being investigated.
Quantitative real-time PCR was used to measure transcription of VSH-1 genes hvp38 and hvp60 and putative bacterial gene numbers 4 and 7 (Fig. 1) during the induction of VSH-1 by the antimicrobial carbadox. Quantitative real-time PCR conditions and specific primers for hvp38 have been described (20). Primers for hvp60 mRNA were 5′-ACAAATAACAATGTCATTCAGCG and 5′-TATCCGTCAAAATCTACTCCCC, and the quencher-reporter (Q-R) probe was 5′-CGAGGTAGAAGAAAGTTTATCAGATACTTGGTGC. Primers for gene number 4 mRNA were 5′-GGATCATGTAATGGAAGGTGCTGC and 5′-TTCCTTTACCAGTACCGCCGAA, and the Q-R probe was 5′-AGGAGGAGGAAGCAGTATGGACTCTT. The primers for gene number 7 were 5′-CTGCTGCAACTTTGCCTGTAGCTT and 5′-ATAGTGCCAATAGCAGGAAGCTGG, and the Q-R probe was 5′-GGGAGCAACTATACACTTATGCGGATCAGT.
Transcription of both VSH-1 genes hvp38 and hvp60 increased by 200- to 300-fold within 5.5 h after B. hyodysenteriae cultures were treated with 0.5 μg carbadox/ml. In contrast, transcription of bacterial gene number 4 decreased 10- to 20-fold to undetectable levels in treated versus control cultures, and transcription of bacterial gene number 7 was undetectable in both treated and control cultures. These results and the finding of common upstream sequences with identical hexanucleotide repeat sequences suggest that the VSH-1 16.3-kb and 3.6-kb gene clusters, although noncontiguous, are regulated by a common mechanism. Transcription of the intervening bacterial-type genes is not detectably induced and, thus, those genes are not essential for VSH-1 production.
VSH-1 particles contain 7.5-kb fragments of DNA (11). The newly discovered 3.6-kb cluster of VSH-1 tail genes together with the previously identified 16.3-kb VSH-1 gene cluster spans 36.6 kb of DNA in the B. hyodysenteriae B204 genome. It is highly unlikely, therefore, that a single VSH-1 particle carries sufficient DNA (i.e., five 7.5-kb fragments) to encode proteins involved in structural and escape-from-host functions. This genome arrangement strongly suggests that it is impossible for VSH-1 to have an independent, self-propagating lifestyle and explains the avirulence of VSH-1 particles.
The discovery of noncontiguous genes, i.e., a “divided” genome for VSH-1, has several implications for GTAs and prophage-like gene clusters in Brachyspira and other bacterial species. First, the contiguity or dispersion patterns of VSH-1 genes in other Brachyspira genomes could provide insight into the origin, evolution, and spread of VSH-1-like elements among Brachyspira species. Second, gene proximity is not essential for VSH-1 production. A contiguous genome, therefore, may not be a requirement for the production of GTAs by other bacteria. No recognizable lysis genes are associated with the gene cluster of RcGTA, GTA of Rhodobacter capsulatus, and these and other essential genes could be positioned at other undiscovered locations in the R. capsulatus genome (12). Additionally, certain Rickettsiales species contain multiple gene homologs of RcGTA dispersed within their genomes (13). Techniques for identifying GTAs (19) might be applied to detect functional GTAs in these bacteria. Finally, gene clusters for incomplete prophages have been found in bacterial chromosomes (6, 10, 18, 25). Although clearly defective as self-propagating prophages, perhaps some of these gene clusters can interact with distal genes to form effective GTAs. While there is no direct evidence to support this speculation, it is worth considering in view of the widespread distribution of GTA genes among alphaproteobacteria (13). Indeed, the possibility that some “defective” prophages serve as GTAs may provide an alternative explanation for the occasional failure (9) to demonstrate that mitomycin C-induced “bacteriophages” are capable of lytic (plaque-forming) growth.
Acknowledgments
We thank Tom Casey for providing an insightful, in-depth review of the manuscript.
Any mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 19 December 2008.
REFERENCES
- 1.Bertani, G. 1999. Transduction-like gene transfer in the methanogen Methanococcus voltae. J. Bacteriol. 1812992-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biers, E. J., K. Wang, C. Pennington, R. Belas, F. Chen, and M. A. Moran. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 742933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderaro, A., G. Dettori, L. Collini, P. Ragni, R. Grillo, P. Cattani, G. Fadda, and C. Chezzi. 1998. Bacteriophages induced from weakly β-haemolytic human intestinal spirochaetes by mitomycin C. J. Basic Microbiol. 38323-335. [PubMed] [Google Scholar]
- 4.Calderaro, A., G. Dettori, R. Grillo, P. Plaisant, G. Amalfitano, and C. Chezzi. 1998. Search for bacteriophages spontaneously occurring in cultures of haemolytic intestinal spirochaetes of human and animal origin. J. Basic Microbiol. 38313-322. [PubMed] [Google Scholar]
- 5.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49277-300. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S., and R. W. Hendrix. 2005. Bacteriophages and the bacterial genome, p. 39-52. In N. P. Higgins (ed.), The bacterial chromosome. ASM Press, Washington, DC.
- 7.Datta, A. B., S. Panjikar, M. S. Weiss, P. Chakrabarti, and P. Parrack. 2005. Structure of lambda CII: implications for recognition of direct-repeat DNA by an unusual tetrameric organization. Proc. Natl. Acad. Sci. USA 10211242-11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31117-131. [DOI] [PubMed] [Google Scholar]
- 9.Gemski, P., A. D. O'Brien, and J. A. Wohlhieter. 1978. Cellular release of heat-labile enterotoxin of Escherichia coli by bacteriophage induction. Infect. Immun. 19:1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 962192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey, S. B., T. B. Stanton, N. S. Jensen, and R. L. Zuerner. 1997. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J. Bacteriol. 179323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang, A. S., and J. T. Beatty. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 97859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang, A. S., and J. T. Beatty. 2007. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 1554-62. [DOI] [PubMed] [Google Scholar]
- 14.Marrs, B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 71971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matson, E. G., M. G. Thompson, S. B. Humphrey, R. L. Zuerner, and T. B. Stanton. 2005. Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J. Bacteriol. 1875885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson, E. G., R. L. Zuerner, and T. B. Stanton. 2007. Induction and transcription of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. Anaerobe 1389-97. [DOI] [PubMed] [Google Scholar]
- 17.Ni, X., and J. Westpheling. 1997. Direct repeat sequences in the Streptomyces chitinase-63 promoter direct both glucose repression and chitin induction. Proc. Natl. Acad. Sci. USA 9413116-13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 19.Stanton, T. B. 2007. Prophage-like gene transfer agents—novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 1343-49. [DOI] [PubMed] [Google Scholar]
- 20.Stanton, T. B., S. B. Humphrey, V. K. Sharma, and R. L. Zuerner. 2008. Collateral effects of antibiotics: carbadox and metronidazole induce VSH-1 and facilitate gene transfer among Brachyspira hyodysenteriae strains. Appl. Environ. Microbiol. 742950-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanton, T. B., E. G. Matson, and S. B. Humphrey. 2001. Brachyspira (Serpulina) hyodysenteriae gyrB mutants and interstrain transfer of coumermycin A1 resistance. Appl. Environ. Microbiol. 672037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanton, T. B., M. G. Thompson, S. B. Humphrey, and R. L. Zuerner. 2003. Detection of bacteriophage VSH-1 svp38 gene in Brachyspira spirochetes. FEMS Microbiol. Lett. 224225-229. [DOI] [PubMed] [Google Scholar]
- 23.Walker, S. A., and T. R. Klaenhammer. 1998. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage φ31. J. Bacteriol. 180921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wall, J. D., P. F. Weaver, and H. Gest. 1975. Genetic transfer of nitrogenase-hydrogenase activity in Rhodopseudomonas capsulata. Nature 258630-631. [DOI] [PubMed] [Google Scholar]
- 25.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 712775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuerner, R. L., T. B. Stanton, F. C. Minion, C. Li, N. W. Charon, D. J. Trott, and D. J. Hampson. 2004. Genetic variation in Brachyspira: chromosomal rearrangements and sequence drift distinguish B. pilosicoli from B. hyodysenteriae. Anaerobe 10229-237. [DOI] [PubMed] [Google Scholar]