Abstract
MfpAMt and QnrB4 are two newly characterized pentapeptide repeat proteins (PRPs) that interact with DNA gyrase. The mfpAMt gene is chromosome borne in Mycobacterium tuberculosis, while qnrB4 is plasmid borne in enterobacteria. We expressed and purified the two PRPs and compared their effects on DNA gyrase, taking into account host specificity, i.e., the effect of MfpAMt on M. tuberculosis gyrase and the effect of QnrB4 on Escherichia coli gyrase. Whereas QnrB4 inhibited E. coli gyrase activity only at concentrations higher than 30 μM, MfpAMt inhibited all catalytic reactions of the M. tuberculosis gyrase described for this enzyme (supercoiling, cleavage, relaxation, and decatenation) with a 50% inhibitory concentration of 2 μM. We showed that the D87 residue in GyrA has a major role in the MfpAMt-gyrase interaction, as D87H and D87G substitutions abolished MfpAMt inhibition of M. tuberculosis gyrase catalytic reactions, while A83S modification did not. Since MfpAMt and QnrB4 have been involved in resistance to fluoroquinolones, we measured the inhibition of the quinolone effect in the presence of each PRP. QnrB4 reversed quinolone inhibition of E. coli gyrase at 0.1 μM as described for other Qnr proteins, but MfpAMt did not modify M. tuberculosis gyrase inhibition by fluoroquinolones. Crossover experiments showed that MfpAMt also inhibited E. coli gyrase function, while QnrB4 did not reverse quinolone inhibition of M. tuberculosis gyrase. In conclusion, our in vitro experiments showed that MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase and that these effects are protein and species specific.
The pentapeptide repeat protein (PRP) family includes more than 500 proteins in the prokaryotic and eukaryotic kingdoms (45). PRPs are characterized by the repetition of the pentapeptide repeat motif [S,T,A,V][D,N][L,F][S,T,R][G] (6), which results in a right-handed β-helical structure (8, 17). The functions of the majority of the members of this large and heterogeneous family remain unknown, but three PRPs, McbG (from Escherichia coli), MfpAMt (from Mycobacterium tuberculosis), and Qnr (from Klebsiella pneumoniae and other enterobacteria) were reported to interact with DNA gyrase, at least with the E. coli enzyme (17, 33, 35, 44). McbG was shown to protect E. coli DNA gyrase from the toxic action of microcin B17 (33). Qnr and MfpAMt were involved in resistance to fluoroquinolones, which are synthetic antibacterial agents prescribed worldwide for the treatment of various infectious diseases, including tuberculosis (7).
DNA gyrase is an essential ATP-dependent enzyme that transiently cleaves a segment of double-stranded DNA, passes another piece of DNA through the break, and reseals it (12). DNA gyrase is unique in catalyzing the negative supercoiling of DNA in order to facilitate the progression of RNA polymerase. Most eubacteria, such as E. coli, have two type II DNA topoisomerases, i.e., DNA gyrase and topoisomerase IV, but a few, such as M. tuberculosis, harbor only DNA gyrase (11).
Quinolones target type II topoisomerases, and their activity is measured by the inhibition of supercoiling by gyrase or decatenation by topoisomerase IV and stabilization of complexes composed of topoisomerase covalently linked to cleaved DNA (16). The DNA gyrase active enzyme is a GyrA2GyrB2 heterotetramer. The quinolone-gyrase interaction site in gyrase is thought to be located at the so-called quinolone resistance-determining regions (QRDR) in the A subunit (amino acids 57 to 196 in GyrA) and the B subunit (amino acids 426 to 466 in GyrB), which contain the majority of mutations conferring quinolone resistance (19). The GyrB QRDR is thought to interact with the GyrA QRDR to form a drug-binding pocket (18). Resistance to quinolones is usually due to chromosomal mutations either in the structural genes encoding type II topoisomerases (QRDR) (19, 22) or in regulatory genes producing decreased cell wall permeability or enhancement of efflux pumps (36). The recent emergence of plasmid-borne resistance genes, such as qnr (9, 13, 31, 38, 46), aac(6′)-Ib-cr (32, 39) and qepA (34, 47), renewed interest in quinolone resistance, and especially interest in the new Qnr-based mechanism. Three qnr determinants have been identified so far: qnrA (variants A1 to A6), qnrB (variants B1 to B19), and qnrS (variants S1 and S2) (15, 21, 23, 27). Qnr confers a new mechanism of quinolone resistance by mediating DNA gyrase protection (42): in vitro, QnrA1 and QnrB1 protect E. coli DNA gyrase and topoisomerase IV from the inhibitory effect of fluoroquinolones in a concentration-dependent manner (23, 42-44). Although Qnr was shown to bind GyrA and GyrB and compete with DNA binding, the consequences of Qnr binding for enzyme performance are not yet clear.
mfpA, a chromosomal gene that encodes a 192-amino-acid PRP, is an intrinsic quinolone resistance determinant of Mycobacterium smegmatis (29). A similar gene, mfpAMt, was found in the M. tuberculosis genome, and MfpAMt shows 67% identity with MfpA. Recent crystallography analysis of MfpAMt showed that its atomic structure displays size, shape, and electrostatic similarity to B-form DNA, and MfpAMt has been suggested to interact with DNA gyrase via DNA mimicry (17). The effect of MfpAMt was studied by testing E. coli DNA gyrase, and MfpAMt showed catalytic inhibition (17, 37), but whether it protects gyrase from quinolones was not assessed. Because the structure and functions of the M. tuberculosis gyrase, as well as its interaction with quinolones, differ from those of the E. coli gyrase (2, 3, 20, 26, 28), we suspected that the PRP-topoisomerase interaction exhibits species specificity, i.e., depends on the proteins issued from the same host.
Our objective was to compare the effects of MfpAMt and Qnr on their respective targets, i.e., the effect of MfpAMt on the M. tuberculosis gyrase and the effect of Qnr on the E. coli gyrase, by assessing (i) the catalytic reactions of the enzyme and (ii) the interaction with the DNA gyrase-DNA-fluoroquinolone ternary complex. Among the Qnr proteins, we selected the QnrB4 protein, which is a frequent variant of QnrB and has not yet been purified and studied. We cloned, expressed, and purified the two PRPs, MfpAMt and QnrB4, as recombinant His tag fusion proteins and assessed their functions under the same experimental conditions.
MATERIALS AND METHODS
Cloning, expression, and purification of recombinant PRPs.
The Rv3361c open reading frame, encoding MfpAMt, was amplified from M. tuberculosis H37Rv genomic DNA with the Expand Long Template PCR system (Roche Diagnostics, Meylan, France) and the following primers: Fw-mfpa (5′CGGTTGAAAACATATGCAGCAGTGGGTTGA), Rv-mfpa-29a (5′CCGGCTCACCGATCTCGAGCCCTGCCAAGC), or Rv-mfpa-19b (5′GTCCCGGCTCCTCGAGCTAGCCCCCTGCCA) (NdeI or XhoI sites are underlined). The qnrB4 open reading frame was amplified by PCR from the clinical strain Enterobacter cloacae HM05-184 (qnrB4+) isolated at Henri Mondor Hospital using the primers Fw-qnrB4 (5′CAGGTTAATCCATATGACTCTGGCGTTAGT) and Rv-qnrB4 (5′-ACCGCTCCCTGACTCGAGACCCATGACAGC). Amplified fragments were cloned into the expression vectors pET-29a and pET-19b (Novagen Merck Eurolab, Fontenay-sous-Bois, France) as described previously (3) for the gyrase genes. The recombinant plasmids carrying mfpAMt or qnrB4 were transformed by electroporation into E. coli BL21-CodonPlus(λDE3)-RP (Stratagene). Three different clones were grown separately at 37°C in 4 ml of LB medium with appropriate selective antibiotics, and subcultures were mixed to inoculate 250 ml of LB medium with selective antibiotics. Cells were grown at 37°C until the optical density at 600 nm reached 0.6, and expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation at 18°C for 18 h. The soluble recombinant proteins were purified and eluted as described previously (2). Protein was concentrated with an Amicon Ultra-4 filter (Millipore) and measured with a Nanodrop ND-1000 instrument. Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12.5% acrylamide gels with Coomassie blue staining. Proteins were flash frozen in 30% glycerol, 1 mM dithiothreitol (DTT), and EDTA and stored at −80°C.
Mycobacterium tuberculosis GyrA and GyrB subunit overexpression and purification.
Plasmids expressing M. tuberculosis DNA gyrase subunits were constructed as described previously (3). Plasmids expressing gyrA genes containing mutations were generated by site-directed mutagenesis of gyrA in the pATB expression vector (pET-29a expressing gyrA) (3, 28). Expression and purification of both subunits were performed as described previously for wild-type GyrA and GyrB subunits and the three modified GyrA subunits GyrA A83S, GyrA D87G, and GyrA D87H (E. coli numbering system). Activities of the different recombinant DNA gyrases were controlled by supercoiling, relaxation, decatenation, and DNA cleavage assays.
Topoisomerase assays.
For the DNA supercoiling assay, 0.4 μg relaxed plasmid pBR322 (Roche Diagnostics, Meylan Cedex, France) was mixed in a basic DNA gyrase assay buffer (40 mM Tris-HCl [pH 7.5], 25 mM KCl, 2 mM spermidine, 4 mM DTT, 0.1 mg/ml E. coli tRNA, 0.36 mg/ml bovine serum albumin, 6 mM magnesium acetate, and 1 mM ATP at pH 8.0) enriched with 100 mM potassium glutamate (KGlu) for the tests carried out with 1 U of M. tuberculosis gyrase. For M. tuberculosis, 1 unit was the amount of DNA gyrase that converted 400 ng of relaxed pBR322 to the supercoiled form in 1 h, i.e., 40 ng of GyrA and 110 ng of GyrB. For E. coli, DNA gyrase was purchased from John Innes Enterprises Ltd. Tests were carried with various ratios of purified pentapeptide proteins and concentrations of ciprofloxacin, as indicated in the figure legends. Moxifloxacin and ciprofloxacin were provided by Bayer Pharma (Puteaux, France). Reaction mixtures were incubated at 37°C for 1 h, and reactions were terminated by addition of 50% glycerol containing 0.25% bromophenol blue. The total reaction mixtures were loaded on a 1% agarose gel in 0.5× TBE buffer for electrophoresis (4 h, 50 V). Fluorescence of the bands was quantified with an Alpha Innotech digital camera and software (Bio-Rad). In order to avoid a possible variability between experiments, assays with wild-type and mutant enzymes were carried out and processed in parallel on the same day under identical conditions. Enzyme assays were done at least twice, with reproducible results.
For the DNA relaxation assay, the protocol was similar to that for DNA supercoiling except that 0.4 μg of supercoiled plasmid pBR322 (John Innes Enterprises Ltd., Norwich Research Park, Colney, Norwich, United Kingdom) was mixed in the gyrase assay buffer without ATP and 1 U of M. tuberculosis gyrase as described previously (2).
For the decatenation assay, 450 ng of kinetoplast DNA (kDNA) from Crithidia fasciculata (Topogen, Denver, CO) was mixed in decatenation buffer (40 mM Tris-HCl [pH 7.5], 10 mM NaCl, 6 mM magnesium acetate, 10 mM DTT, 0.5 mg·ml−1 bovine serum albumin, and 1 mM ATP [pH 8.0]) with 600 ng of GyrA and 810 ng of GyrB M. tuberculosis gyrase subunits as described previously (2).
For the assay that measures the quinolone-induced DNA cleavage, supercoiled pBR322 plasmid was mixed in the supercoiling assay buffer supplemented with ciprofloxacin or moxifloxacin (10 μg/ml). M. tuberculosis gyrase, consisting of 300 ng of GyrA and 300 ng of GyrB, was able to cleave 25% of 400 ng of supercoiled pBR322 in the presence of 30 μM ciprofloxacin in 1 h at 37°C. After the addition of 3 μl of 2% sodium dodecyl sulfate and 3 μl of 1-mg/ml proteinase K in order to release DNA breaks, incubation was continued for 30 min at 37°C.
RESULTS
Purification of recombinant MfpAMt and QnrB4 proteins.
To verify that the His tag did not hamper the interaction of MfpAMt with gyrase, we constructed two vectors expressing MfpAMt fusion proteins with a C-terminal His6 tag (pET-29a) or an N-terminal His10 tag (pET-19b). The QnrB4 protein was produced only with the C-terminal His6 tag, since Tran et al. obtained similar results with recombinant QnrA1 with a C-terminal or an N-terminal His tag (42, 44). Various conditions were tested for the induction phase, and the best results were obtained after an 18-h induction at 18°C (data not shown). MfpAMt-His6 and His10-MfpAMt were obtained at 0.2 mg/ml (0.4-mg yield per liter of culture) in the soluble fraction of the cell extract with a purity of >90% and at the expected sizes, i.e., 21 kDa for MfpAMt-His6 and 25 kDa for His10-MfpAMt, which correspond to MfpAMt plus the histidine residues and the amino acids added due to the insertion into the pET vectors. The QnrB4-His6 fusion protein was homogeneous as a 25-kDa soluble protein and was obtained at high quantities, i.e., 7 mg/ml after concentration for a yield of 7 mg per liter of culture.
Effect of MfpAMt and QnrB4 on DNA gyrase catalytic reactions.
We assessed the effect of MfpAMt on M. tuberculosis DNA gyrase functions, i.e., supercoiling, relaxation, double-stranded DNA cleavage, and decatenation (2). Overall, MfpAMt inhibited the catalytic activity of M. tuberculosis DNA gyrase. Although dimerization of MfpAMt is thought to occur via the C-terminal α helix (17), no difference was observed between the effects of MfpAMt-His6 and His10-MfpAMt on inhibition of gyrase.
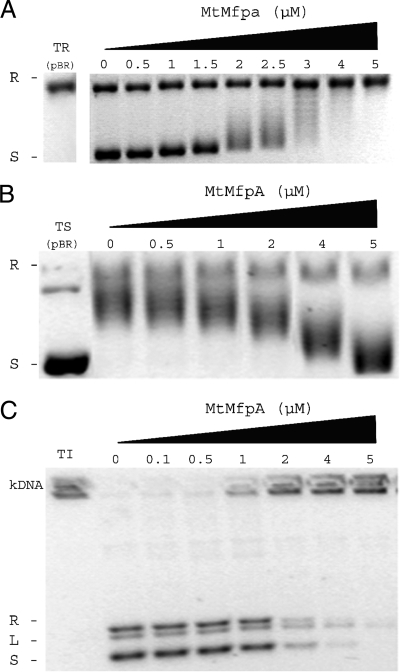
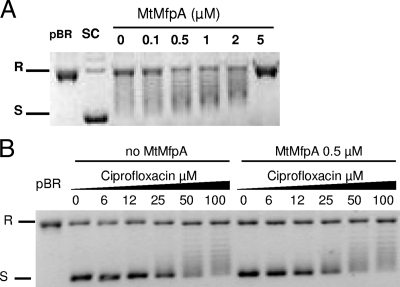
MfpAMt inhibited the activity of the M. tuberculosis DNA gyrase in a concentration-dependent manner. In the supercoiling assay (Fig. 1A), the concentration of MfpAMt required to inhibit 50% of M. tuberculosis gyrase supercoiling activity (IC50) was determined to be 1.75 μM. In the presence of 4 μM of MfpAMt, gyrase supercoiling activity was abolished and pBR322 remained completely relaxed. The inhibitory effect of MfpAMt on M. tuberculosis gyrase relaxation activity was also observed, with an IC50 of 2 μM (Fig. 1B). Since M. tuberculosis DNA gyrase is a better decatenase than E. coli gyrase (2), we also determined whether MfpAMt could interfere with decatenation. MfpAMt inhibited the decatenation activity of M. tuberculosis DNA gyrase in a concentration-dependent manner with an IC50 of 2 μM. When 5 μM of MfpAMt was used in the decatenation assay, the decatenation activity of M. tuberculosis gyrase was suppressed and kDNA remained completely interlocked and did not migrate in the agarose gel (Fig. 1C).
FIG. 1.
MfpAMt inhibits the catalytic activity of M. tuberculosis DNA gyrase in topoisomerase assays. (A) Concentration-dependent inhibitory effect of MfpAMt on supercoiling activity of M. tuberculosis DNA gyrase. Supercoiling assays were performed using relaxed pBR322 as the substrate (TR) and final concentrations of MfpAMt (μM) as indicated. R, relaxed pBR322; S, supercoiled pBR322. (B) Concentration-dependent inhibitory effect of MfpAMt on ATP-independent relaxation of supercoiled DNA. Supercoiled pBR322 (TS) was the substrate. (C) Concentration-dependent inhibitory effect of MfpAMt on decatenation activity of M. tuberculosis DNA gyrase. Interlinked kDNA (kDNA) was used as the substrate (TI). Decatenated minicircles were visualized in three forms: relaxed (R), linearized (L), and supercoiled (S).
Double-stranded DNA breakage is normally transient but can be trapped by quinolones or Ca2+, resulting in a stable gyrase-DNA complex in a conformation in which DNA is cleaved (5). To test the effect of MfpAMt on M. tuberculosis gyrase cleavage activity, we performed assays in the presence of a high concentration of ciprofloxacin or moxifloxacin. In the presence of 0.1 μM, 1 μM, or 2 μM of MfpAMt, no significant effect on DNA cleavage activity was noted, but a decrease in quinolone-promoted cleavage by M. tuberculosis gyrase was observed in the presence of 4 μM MfpAMt (data not shown). This can be explained by the inhibition of gyrase catalysis, which occurred at 4 μM of MfpAMt, as shown above.
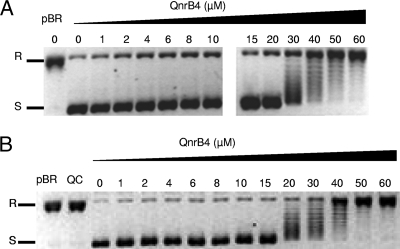
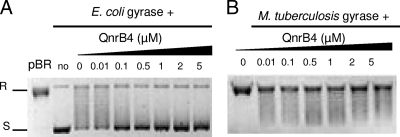
The direct effect of QnrB4 on E. coli DNA gyrase was tested in parallel. In contrast to MfpAMt, QnrB4 exhibited no effect on E. coli gyrase supercoiling activity, unless concentrations were over 30 μM (Fig. 2A).
FIG. 2.
The direct effect of QnrB4 on E. coli DNA gyrase (A) and M. tuberculosis gyrase (B) supercoiling is minimal and is observed only at high concentrations. Supercoiling assays were carried out with 400 ng of relaxed pBR322 as the substrate (pBR) and 1 U of E. coli DNA gyrase or 1 U of M. tuberculosis DNA gyrase with increasing concentrations of QnrB4-His6 (μM) as indicated. R, relaxed pBR322; S, supercoiled pBR322. QC, QnrB4 with pBR322 without gyrase.
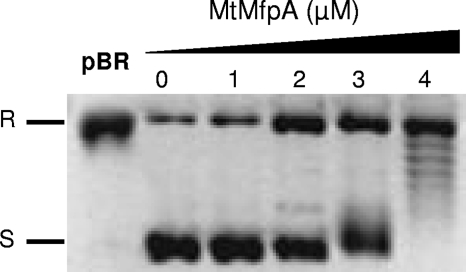
We then tested the specificity of the two PRPs by testing them on a gyrase from a different species. QnrB4 did not display any inhibition of M. tuberculosis gyrase supercoiling activity until concentration was elevated to 20 μM (Fig. 2B). In contrast, MfpAMt inhibited DNA supercoiling mediated by the E. coli gyrase, with an IC50 of 3 μM (Fig. 3).
FIG. 3.
MfpAMt inhibits E. coli DNA gyrase supercoiling activity with an IC50 of 3 μM. Supercoiling assays were performed with 400 ng of relaxed pBR322 as the substrate (pBR) and 1 U of E. coli DNA gyrase with increasing concentrations of MfpAMt (μM) as indicated. R, Relaxed pBR322; S, supercoiled pBR322.
Interaction between MfpAMt and altered M. tuberculosis DNA gyrases.
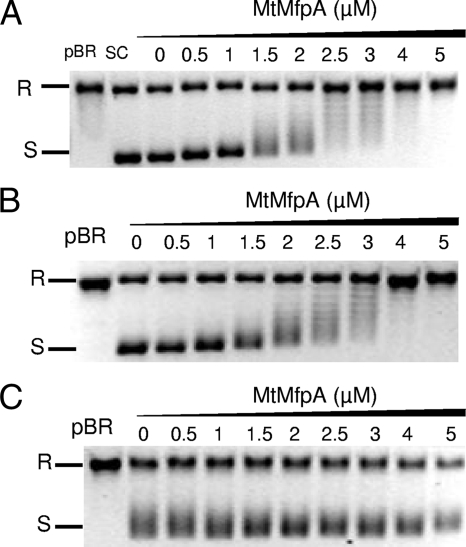
To investigate the site of interaction between MfpAMt and the M. tuberculosis DNA gyrase, we performed supercoiling assays with gyrases composed of modified GyrA subunits and the wild-type GyrB, referred to as AmodifiedBwt. Three alterations were introduced in the GyrA subunits, A83S, D87H, and D87G. The inhibitory effect of MfpAMt was completely abolished with the altered gyrases AD87GBwt (Fig. 4) and AD87H Bwt (data not shown). In contrast, we still observed a direct effect with the AA83SBwt gyrase, at a concentration similar to that for the wild-type gyrase (1.8 μM versus 2 μM, respectively).
FIG. 4.
The MfpAMt effect on DNA gyrase catalysis is abolished with M. tuberculosis DNA gyrase harboring the D87G but not the A83S substitution of GyrA. (A) Supercoiling assays were performed with wild-type M. tuberculosis gyrase and relaxed pBR322 as a substrate. R, relaxed pBR322; S, supercoiled pBR322. Reactions were carried out without MfpAMt (lane 0) and with increasing concentrations of MfpAMt (final concentrations in μM are indicated above the gel). MfpAMt inhibited gyrase supercoiling in a concentration-dependent manner. (B) Same experiment with an altered M. tuberculosis DNA gyrase composed of GyrA A83S and wild-type GyrB subunit, which efficiently supercoiled pBR322 (lane 0). MfpAMt inhibited altered gyrase supercoiling in a concentration-dependent manner, as for the wild-type gyrase. (C) Same experiment with an altered M. tuberculosis DNA gyrase composed of GyrA D87G and the wild-type GyrB subunit, which efficiently supercoiled pBR322 (lane 0). No modification of this supercoiling activity in the presence of MfpAMt (from 0.5 to 5 μM) was observed.
Interaction of MfpAMt and QnrB4 with the gyrase-DNA-quinolone ternary complex.
To measure the effects of MfpAMt on the gyrase-DNA-quinolone ternary complex, we conducted experiments using (i) fixed concentrations of quinolones and an increasing quantity of MfpAMt and (ii) increasing concentrations of ciprofloxacin and moxifloxacin in the presence of a fixed quantity of MfpAMt.
We first tested increasing concentrations of MfpAMt (0.01 μM to 5 μM) with fixed concentrations of ciprofloxacin or moxifloxacin under the standard conditions for the M. tuberculosis gyrase supercoiling assay. At ciprofloxacin and moxifloxacin concentrations corresponding to twofold and fourfold the IC50, i.e., 50 and 100 μM ciprofloxacin and 25 and 50 μM moxifloxacin, respectively, no modification of the quinolone inhibition effect, i.e., no increase in the supercoiled DNA band, was observed (Fig. 5A). Moreover, when using concentrations of MfpAMt of above 1.5 μM, the direct effect of the PRP on the catalytic activity was cumulative with the fluoroquinolone inhibitory effect. Second, we ascertained whether MfpAMt could increase the IC50s of fluoroquinolones, which is predicted in the case of quinolone resistance mediated by gyrase protection. We measured the IC50s of ciprofloxacin and moxifloxacin in the absence and presence of a fixed concentration of MfpAMt; the experiments were conducted simultaneously under the same conditions. In the absence of MfpAMt, the ciprofloxacin and moxifloxacin required IC50s for the M. tuberculosis DNA gyrase supercoiling activity were 24 μM and 11 μM, respectively. Different concentrations of MfpAMt (0.01 μM, 0.1 μM, 0.5 μM, and 1 μM) were tested. The IC50s of ciprofloxacin and moxifloxacin were not modified in the presence of MfpAMt, despite the concentration: IC50s of 24 and 29 μM for ciprofloxacin and 11 μM and 14 μM for moxifloxacin was measured in the absence and presence of 0.5 μM of MfpAMt, respectively (Fig. 5B).
FIG. 5.
MfpAMt does not modify ciprofloxacin inhibition of M. tuberculosis gyrase. (A) Supercoiling assays were performed with relaxed pBR322 as a substrate (pBR). In the absence of fluoroquinolone, M. tuberculosis DNA gyrase efficiently supercoiled pBR322 (SC). In the presence of 50 μM ciprofloxacin, supercoiling activity was inhibited (lane 0). Increasing concentrations of MfpAMt (final concentrations in μM are indicated above the gel) were added to reaction mixtures containing M. tuberculosis DNA gyrase and ciprofloxacin. (B) Determination of the ciprofloxacin IC50 in the absence and presence of MfpAMt. The IC50 was measured by comparing the intensities of the bands corresponding to supercoiled pBR322.
Similar experiments were performed to assess the interaction of QnrB4 with the DNA-E. coli gyrase-quinolone ternary complex. In the presence of six times the IC50 of ciprofloxacin (6 μM), reversal of supercoiling inhibition was observed with concentrations of QnrB4 of as low as 0.1 μM; the effect was dependent on the QnrB4 concentration (Fig. 6A). A protective effect of QnrB4 on E. coli DNA gyrase activity was still observed in the presence of 15 μM ciprofloxacin (data not shown). We also measured the ciprofloxacin IC50 for E. coli DNA gyrase supercoiling in the absence and presence of QnrB4. In the absence of QnrB4, the IC50s of ciprofloxacin and moxifloxacin for E. coli DNA gyrase were 1 μM and 1.1 μM, respectively. As shown in Fig. 7, the ciprofloxacin IC50 increased fivefold in the presence of 0.5 μM QnrB4. Recombinant QnrB4 protein exhibited protection of E. coli gyrase from quinolone inhibition, and in a concentration-dependent manner, in contrast to the results obtained with MfpAMt and M. tuberculosis gyrase.
FIG. 6.
QnrB4 has a protective effect on fluoroquinolone inhibition of E. coli DNA gyrase supercoiling activity but not toward M. tuberculosis gyrase. Supercoiling assays were performed with 400 ng of relaxed pBR322 (pBR) as the substrate and 1 U of E. coli DNA gyrase or 1 U of M. tuberculosis DNA gyrase in the presence of increasing concentrations (μM) of QnrB4. R, relaxed pBR322; S, supercoiled pBR322. (A) In the presence of 6 μM ciprofloxacin, supercoiling activity was inhibited (lane 0); pBR322 remained in a relaxed form (R). When QnrB4 was added (final concentrations in μM are indicated above the gel), the intensity of the band corresponding to the supercoiled form of pBR322 (S) increased (lanes 0.1 to 5), showing that the inhibition of E. coli DNA gyrase supercoiling activity by ciprofloxacin was diminished. (B) Same experiment with QnrB4, M. tuberculosis gyrase, and ciprofloxacin at 50 μM. When QnrB4 was added, the topoisomeres produced by gyrase inhibition by ciprofloxacin remained and no supercoiling form (result of gyrase protection) was observed, even at high concentrations (5 μM) of QnrB4.
FIG. 7.
MfpAMt does not protect against E. coli gyrase inhibition by quinolones even in the presence of KGlu. (A) Comparison of inhibition by ciprofloxacin of pBR322 (pBR) supercoiling by E. coli gyrase in the presence of QnrB4 or MfpAMt at the same concentration (0.5 μM). R, relaxed pBR322; S, supercoiled pBR322. (B) Similar experiments with 100 mM KGlu in the assay buffer.
The only difference between the experiments conducted in parallel with E. coli and M. tuberculosis was the presence of KGlu in the supercoiling assay buffer for the M. tuberculosis gyrase, since KGlu is necessary for the catalytic activities of this gyrase. Since KGlu may modify the gyrase-quinolone interaction (41), we controlled the protective effect of the two PRPs on E. coli gyrase in the presence of 100 mM KGlu. As shown in Fig. 7, the QnrB4 fluoroquinolone-protective effect on E. coli DNA gyrase was not modified in the presence of KGlu. Thus, the difference between the protective effects of the PRPs cannot be explained by the presence of KGlu in the M. tuberculosis assays.
To determine if the difference in PRP behavior was due to the protein specificity or the gyrase with which they interacted, we conducted experiments with one PRP from one species with the gyrase of the other species; i.e., we measured the effect of QnrB4 on the M. tuberculosis gyrase and the effect of MfpAMt on the E. coli gyrase. We showed that MfpAMt does not exhibit a quinolone-protective effect on the E. coli gyrase, as the ciprofloxacin IC50 was not modified in the presence of increasing concentrations of MfpAMt (Fig. 7). More surprisingly, we observed that QnrB4 has no protective effect on M. tuberculosis gyrase, even after a preincubation phase as described previously (1) (Fig. 6B).
DISCUSSION
The pentapeptide repeat family includes a variety of proteins whose common feature is their ability to interact with DNA-binding proteins, such as DNA gyrase (Qnr, MfpAMt, and McbG) or DNA polymerase (OxrA) (45). Genes encoding PRPs were found in the genomes of various bacteria, such as mfpA-like genes in M. tuberculosis and other mycobacteria (M. avium, M. bovis, and M. leprae) (29) or qnr-like genes in environmental bacteria such as Shewanella and Vibrionaceae (10, 31, 40). Plasmid-borne qnr genes detected in quinolone-resistant strains of Enterobacteriaceae are derived from these chromosome-borne genes. The physiological role of the chromosomally encoded PRPs is unknown. Therefore, we investigated their direct effect on gyrase, the protein with which they associate.
We compared the effects of MfpAMt and QnrB4 on their respective DNA gyrases using classical topoisomerase assays (5). The important finding from our work is the strong inhibitory effect of MfpAMt on the catalytic reactions of the type II topoisomerase of M. tuberculosis (ATP-dependent supercoiling, ATP-independent relaxation, and decatenation) and decreased DNA cleavage of double-strand DNA. This inhibitory effect was partially shown by Hegde et al. (17) with E. coli gyrase. In contrast, QnrB4 did not modify E. coli DNA gyrase supercoiling unless concentrations were at least as high as 30 μM (Table 1). The QnrB4 result is in agreement with previously reported results for QnrA1 and QnrB1 (23, 44), for which inhibition of supercoiling was observed only at high concentrations.
TABLE 1.
Effects of MfpAMt and QnrB4 on DNA gyrases
| Type of effect | Type II topoisomerase | Effect of:
|
|
|---|---|---|---|
| MfpAMt | QnrB4 | ||
| Direct effect on catalysis | M. tuberculosis DNA gyrase | Inhibition of supercoiling activity (IC50, 2 μM) | No effect until 20 μM |
| E. coli DNA gyrase | Inhibition of supercoiling activity (IC50, 3 μM) | No effect until 30 μM | |
| Protection from fluoroquinolone inhibition | M. tuberculosis DNA gyrase | No protection, no modification of ciprofloxacin or moxifloxacin IC50 | No protection, no modification of ciprofloxacin IC50 |
| E. coli DNA gyrase | No protection, no modification of ciprofloxacin IC50 | Concentration-dependent protective effect, increase of ciprofloxacin and moxifloxacin IC50s in the presence of QnrB4 | |
Fluoroquinolones are commonly used drugs against E. coli and other enterobacteria and are also one of the rare new groups of antituberculous drugs (14, 17, 37). M. tuberculosis has been shown to withstand fluoroquinolones better than E. coli. The mechanisms of intrinsic quinolone resistance in M. tuberculosis are due to the low permeability of its thick, lipid-containing cell wall (25) and to the low affinity of its gyrase for quinolones (10-fold lower than that of E. coli gyrase) (3). Since mfpA was described as a new fluoroquinolone resistance determinant in M. smegmatis (29), the corresponding protein in M. tuberculosis, MfpAMt, was proposed to be another determinant of intrinsic quinolone resistance. Moreover, its similarity to Qnr proteins led to the hypothesis that it could mediate fluoroquinolone resistance by gyrase protection (43).
Using the methodology employed to demonstrate the protective effect of QnrA1 (42), we compared the effects of MfpAMt and QnrB4 on quinolone activity on its target, DNA gyrase. The assays performed with the M. tuberculosis gyrase showed that MfpAMt does not protect gyrase from fluoroquinolone inhibition (Table 1). On the contrary, the inhibitory effect of MfpAMt was additive with that of fluoroquinolones. These results were very different from those obtained with the control pair, QnrB4 and E. coli DNA gyrase. Similar to previous observations with QnrB1 (23), we observed that a low concentration of QnrB4 protects E. coli DNA gyrase from supercoiling inhibition caused by fluoroquinolones. This concentration-dependent protection was inversely proportional to the fluoroquinolone concentration but was maintained at high quinolone concentrations. For Qnr, the in vitro protection effect on topoisomerase is consistent with the increase in the inhibitory concentrations necessary to achieve E. coli bacteriostasis. mfpA was shown to increase the level of quinolone resistance by eightfold in M. smegmatis (29) and by twofold in M. bovis BCG, a species very similar to M. tuberculosis. The apparent discrepancy between the results obtained with M. smegmatis and M. tuberculosis could be due to several differences between the two species, such as a second topoisomerase in M. smegmatis (topoNM) (24), the presence of multiple other antibiotic resistance determinants (11), and differences in the MfpA and MfpAMt structures (see Fig. S1 in the supplemental material).
In order to better understand the difference between the effects on the two PRPs, MfpAMt and QnrB4, we first controlled the specificity of their effect by crossover experiments. Whereas MfpAMt has an effect on the E. coli gyrase that is similar to its effect on the M. tuberculosis gyrase, QnrB4 did not modify M. tuberculosis gyrase activity and did not show quinolone protection toward this gyrase. This may be related to the specificity of the PRP-gyrase interaction, which may possibly be based on the PRP structure. Although the data obtained thus far from the three PRPs that have been crystallized show that all show the same right-handed quadrilateral beta-helix on which DNA mimicry relies (8), MfpAMt and QnrB4 share only 26% identity. The only residue identified so far to be involved in the PRP-gyrase interaction is C115, as substitution of this residue for tyrosine in the Qnr-like protein (VPA0085) from Vibrio parahaemolyticus conferred an increase in quinolone MICs, suggesting gyrase protection from quinolone (40). The alignment of QnrB4 and VPA0085 with MfpA and MfpAMt (Fig. 8) showed that the cysteine residue is conserved between the two former proteins (and also in QnrA [42] and QnrS[(15] [data not shown]) and is replaced by an arginine in the two latter. There are many other positions in addition to C115 at which amino acids in MfpA and MfpAMt are similar and differ from the amino acid present in QnrB4 and VPA0095 (based on V. parahaemolyticus numbering, positions 62 to 67, 76, 88, 126, 128 to 129, 135, 143 to 144, 153, 157, 160, 164 to 165, 177, 198, and 207). The environment of C- or R-115 is also clearly different between the two groups of proteins. Site-directed mutagenesis of MfpAMt and crystallization of Qnr would be necessary in order to further understand the relationship between PRP structure and its effect. Using computer modeling, Hegde et al. (17) proposed that the MfpAMt protein mimics a 30-bp double-stranded DNA and lies across the entire saddle-shaped active site of DNA gyrase in place of DNA. This corresponds to the head dimer interface as shown by crystallization of the 59-kDa N-terminal domain of E. coli GyrA (30). The proposed region of interaction between MfpAMt, QnrB4, and gyrase should thus include the QRDR domain of GyrA, in which residues S83 and D87 are strongly involved in the quinolone-gyrase interaction (4, 16, 48). We performed experiments with GyrA subunits modified at these two positions and found that the effect of MfpAMt on gyrase catalysis was nearly abolished when the residue at position 87 in GyrA was changed, but not with substitutions at position 83. Position 87 in gyrase, and the corresponding position 84 in ParC of topoisomerase IV, is involved in the topoisomerase-DNA interaction (4, 18). Residue 87 in GyrA is likely a preferential residue for the interaction with MfpAMt. This effect seems to be more likely due to the loss of an acidic amino acid (Asp) than to the bulkiness of the residue, since a reduced effect was observed with either a D87G or a D87H substitution. It was previously shown that the absence of an acidic residue at position 87 greatly enhances the stability of topoisomerase-DNA complexes (4, 18). Therefore, the abolition of the MfpAMt effect on catalysis of M. tuberculosis gyrases with GyrA87 modifications may result from the difficulty in displacement of DNA strands tightly linked to gyrase subunits.
FIG. 8.
Comparison of sequences of four PRPs shows major differences in the region including residue 115, which was demonstrated previously to be involved in gyrase protection for quinolones. An alignment of MfpA (29), MfpAMt (Rv 3361c), QnrB4 (AAZ04784), and VPA0085 is shown. Conserved residues in the four sequences are in black, and those in three sequences are in light gray. C115 is indicated.
Our finding that MfpAMt and Qnr exhibit different effects on DNA gyrase suggests that these factors have different physiological roles in the bacterial cell. These in vitro results should be extended in the future by in vivo experiments with M. tuberculosis.
Supplementary Material
Acknowledgments
This work was supported by grants from Ministère de l'Education Nationale et de la Recherche (grant UPRES EA 1541), the Association Française Raoul Follereau, the Chancellerie de l'Université de Paris, and the Fondation pour la Recherche Médicale.
The authors have no conflict of interest.
Footnotes
Published ahead of print on 5 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arsene, S., and R. Leclercq. 2007. Role of a qnr-like gene in the intrinsic resistance of Enterococcus faecalis to fluoroquinolones. Antimicrob. Agents Chemother. 513254-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry, A., L. M. Fisher, V. Jarlier, and E. Cambau. 2006. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 348158-165. [DOI] [PubMed] [Google Scholar]
- 3.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 481281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrob. Agents Chemother. 451994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett, J. F., T. D. Gootz, P. R. McGuirk, C. A. Farrell, and S. A. Sokolowski. 1989. Use of in vitro topoisomerase II assays for studying quinolone antibacterial agents. Antimicrob. Agents Chemother. 331697-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, A., A. G. Murzin, and S. A. Teichmann. 1998. Structure and distribution of pentapeptide repeats in bacteria. Protein Sci. 71477-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, and A. A. Vernon. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167603-662. [DOI] [PubMed] [Google Scholar]
- 8.Buchko, G. W., S. Ni, H. Robinson, E. A. Welsh, H. B. Pakrasi, and M. A. Kennedy. 2006. Characterization of two potentially universal turn motifs that shape the repeated five-residues fold—crystal structure of a lumenal pentapeptide repeat protein from Cyanothece 51142. Protein Sci. 152579-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cambau, E., C. Lascols, W. Sougakoff, C. Bebear, R. Bonnet, J. D. Cavallo, L. Gutmann, M. C. Ploy, V. Jarlier, C. J. Soussy, and J. Robert. 2006. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002-2005. Clin. Microbiol. Infect. 121013-1020. [DOI] [PubMed] [Google Scholar]
- 10.Cattoir, V., L. Poirel, D. Mazel, C. J. Soussy, and P. Nordmann. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 512650-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 12.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 3395-118. [DOI] [PubMed] [Google Scholar]
- 13.Gay, K., A. Robicsek, J. Strahilevitz, C. H. Park, G. Jacoby, T. J. Barrett, F. Medalla, T. M. Chiller, and D. C. Hooper. 2006. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 43297-304. [DOI] [PubMed] [Google Scholar]
- 14.Ginsburg, A. S., J. H. Grosset, and W. R. Bishai. 2003. Fluoroquinolones, tuberculosis, and resistance. Lancet Infect. Dis. 3432-442. [DOI] [PubMed] [Google Scholar]
- 15.Hata, M., M. Suzuki, M. Matsumoto, M. Takahashi, K. Sato, S. Ibe, and K. Sakae. 2005. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49801-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 191249-1264. [DOI] [PubMed] [Google Scholar]
- 17.Hegde, S. S., M. W. Vetting, S. L. Roderick, L. A. Mitchenall, A. Maxwell, H. E. Takiff, and J. S. Blanchard. 2005. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 3081480-1483. [DOI] [PubMed] [Google Scholar]
- 18.Hiasa, H. 2002. The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interactions. Biochemistry 4111779-11785. [DOI] [PubMed] [Google Scholar]
- 19.Hooper, D. 2003. Mechanisms of quinolone resistance, p. 41-67. In D. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents. ASM Press, Washington, DC.
- 20.Huang, Y. Y., J. Y. Deng, J. Gu, Z. P. Zhang, A. Maxwell, L. J. Bi, Y. Y. Chen, Y. F. Zhou, Z. N. Yu, and X. E. Zhang. 2006. The key DNA-binding residues in the C-terminal domain of Mycobacterium tuberculosis DNA gyrase A subunit (GyrA). Nucleic Acids Res. 345650-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacoby, G., V. Cattoir, D. Hooper, L. Martinez-Martinez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 522297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2)S120-S126. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby, G. A., K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robicsek, and D. C. Hooper. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 501178-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain, P., and V. Nagaraja. 2005. An atypical type II topoisomerase from Mycobacterium smegmatis with positive supercoiling activity. Mol. Microbiol. 581392-1405. [DOI] [PubMed] [Google Scholar]
- 25.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 12311-18. [DOI] [PubMed] [Google Scholar]
- 26.Manjunatha, U. H., M. Dalal, M. Chatterji, D. R. Radha, S. S. Visweswariah, and V. Nagaraja. 2002. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 302144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351797-799. [DOI] [PubMed] [Google Scholar]
- 28.Matrat, S., A. Aubry, C. Mayer, V. Jarlier, and E. Cambau. 2008. Mutagenesis in the α3α4 GyrA helix and in the toprim domain of GyrB refines the contribution of Mycobacterium tuberculosis DNA gyrase to intrinsic resistance to quinolones. Antimicrob. Agents Chemother. 522909-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montero, C., G. Mateu, R. Rodriguez, and H. Takiff. 2001. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 453387-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388903-906. [DOI] [PubMed] [Google Scholar]
- 31.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56463-469. [DOI] [PubMed] [Google Scholar]
- 32.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 503953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks, W. M., A. R. Bottrill, O. A. Pierrat, M. C. Durrant, and A. Maxwell. 2007. The action of the bacterial toxin, microcin B17, on DNA gyrase. Biochimie 89500-507. [DOI] [PubMed] [Google Scholar]
- 34.Perichon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 512464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierrat, O. A., and A. Maxwell. 2005. Evidence for the role of DNA strand passage in the mechanism of action of microcin B17 on DNA gyrase. Biochemistry 444204-4215. [DOI] [PubMed] [Google Scholar]
- 36.Poole, K. 2005. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 5620-51. [DOI] [PubMed] [Google Scholar]
- 37.Putnam, C. D., and J. A. Tainer. 2005. Protein mimicry of DNA and pathway regulation. DNA Repair (Amsterdam) 41410-1420. [DOI] [PubMed] [Google Scholar]
- 38.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6629-640. [DOI] [PubMed] [Google Scholar]
- 39.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 1283-88. [DOI] [PubMed] [Google Scholar]
- 40.Saga, T., M. Kaku, Y. Onodera, S. Yamachika, K. Sato, and H. Takase. 2005. Vibrio parahaemolyticus chromosomal qnr homologue VPA0095: demonstration by transformation with a mutated gene of its potential to reduce quinolone susceptibility in Escherichia coli. Antimicrob. Agents Chemother. 492144-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strahilevitz, J., A. Robicsek, and D. C. Hooper. 2006. Role of the extended α4 domain of Staphylococcus aureus gyrase A protein in determining low sensitivity to quinolones. Antimicrob. Agents Chemother. 50600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 995638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 493050-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vetting, M. W., S. S. Hegde, J. E. Fajardo, A. Fiser, S. L. Roderick, H. E. Takiff, and J. S. Blanchard. 2006. Pentapeptide repeat proteins. Biochemistry 451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 472242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamane, K., J. I. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 513354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yonezawa, M., M. Takahata, N. Banzawa, N. Matsubara, Y. Watanabe, and H. Narita. 1995. Analysis of the NH2-terminal 87th amino acid of Escherichia coli GyrA in quinolone-resistance. Microbiol. Immunol. 39517-520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.