Abstract
The uncharacterized protein family UPF0042 of the Swiss-Prot database is predicted to be a member of the conserved group of bacterium-specific P-loop-containing proteins. Here we show that two of its members, YvcJ from Bacillus subtilis and YhbJ, its homologue from Escherichia coli, indeed bind and hydrolyze nucleotides. The cellular function of yvcJ was then addressed. In contrast to results recently obtained for E. coli, which indicated that yhbJ mutants strongly overproduced glucosamine-6-phosphate synthase (GlmS), comparison of the wild type with the yvcJ mutant of B. subtilis showed that GlmS expression was quite similar in the two strains. However, in mutants defective in yvcJ, the transformation efficiency and the fraction of cells that expressed competence were reduced. Furthermore, our data show that YvcJ positively controls the expression of late competence genes. The overexpression of comK or comS compensates for the decrease in competence of the yvcJ mutant. Our results show that even if YvcJ and YhbJ belong to the same family of P-loop-containing proteins, the deletion of corresponding genes has different consequences in B. subtilis and in E. coli.
Functional annotation of genomes is a major challenge of the postgenomic era, especially for eukaryotes, which have many more genes than prokaryotes. Nevertheless, prokaryotic genomes have revealed an unexpected complexity, since about one-third of their genes code for proteins of unknown functions (13). Among these proteins, some contain a specific signature in their sequence known as the Walker A motif (39). This signature [(G/A)X4GK(T/S)] is often used as a fingerprint to detect new putative ATP- or GTP-binding proteins. Indeed, the 3-dimensional structures show that proteins bearing this motif invariably form a loop that wraps around the polyphosphate moiety of the bound nucleotide, and hence it is referred to as the P-loop motif (35). However, the presence of this motif is not enough to clarify the physiological role of these proteins. Furthermore, the sequencing of genomes has revealed that genes encoding P-loop proteins are numerous in both prokaryotic and eukaryotic genomes (20), and many of them have evaded characterization.
In this study, we have investigated a bacterial P-loop-containing protein that belongs to the uncharacterized protein family UPF0042 of the Swiss-Prot database. This protein is conserved in several bacteria whose genomes have been sequenced. This broad conservation would suggest a central role in bacterial physiology. In Bacillus subtilis and Escherichia coli, the paradigmatic gram-positive and gram-negative bacteria, the designations of the corresponding genes are yvcJ and yhbJ, respectively. Nevertheless, neither yvcJ nor yhbJ is an essential gene in these two bacteria. In several proteobacteria, yhbJ is found with genes related to the phosphoenolpyruvate:carbohydrate phosphotransferase system (4), whereas in most gram-positive bacteria, yvcJ is clustered with yvcK. The latter gene is required for the growth of B. subtilis on substrates of the pentose phosphate pathway and Krebs cycle intermediates, and its deletion has been shown to have a pleiotropic effect, since it affects the cell envelope and cell shape (14). A previous study of the yvcIJKL-crh-yvcN operon in B. subtilis showed that the deletion of yvcJ does not affect bacterial growth in any of the carbon sources tested (14). This observation would suggest a broader role of this operon in cell metabolism apart from its role in carbon source utilization.
Recently, it was shown that E. coli YhbJ is involved in glucosamine-6-phosphate synthase (GlmS) expression, by affecting the processing and stability of a small RNA, GlmZ (24, 32). The molecular mechanism that regulates the processing of GlmZ in E. coli is still unknown, but the authors suggest a pleiotropic role for YhbJ in regulating the activities of genes or proteins involved in RNA turnover control. In the gram-positive bacterium B. subtilis, the genetic mechanism for the control of glmS expression is well characterized (2, 42). Briefly, the 5′ untranslated mRNA of the glmS gene contains a riboswitch, a highly structured domain sensitive to glucosamine-6-phosphate, which acts as a ribozyme. Indeed, direct activation of this ribozyme in the presence of glucosamine-6-phosphate leads to the processing of the glmS transcript by a site-specific self-cleavage, thereby repressing the expression of glmS. The participation of YvcJ in this mechanism has never been mentioned.
In this paper, we have biochemically characterized YvcJ and its homologue YhbJ, and we have investigated the cellular function of YvcJ in B. subtilis.
MATERIALS AND METHODS
Plasmids, bacterial strains, and general growth conditions.
The strains used in this study are listed in Table 1. DNA manipulations were performed by standard procedures. Luria-Bertani (LB) broth was routinely used for bacterial growth. CSK-citrate contains 70 mM K2HPO4, 30 mM KH2PO4, 25 mM (NH4)2SO4, 0.5 mM MgSO4, 10 μM MnSO4, 22 mg/liter ferric ammonium citrate, 20 mg/liter l-Trp, 8 g/liter potassium glutamate, 1 mM MgCl2, and 0.5% (wt/vol) citrate. Competence experiments were carried out in MD medium, containing 10.7 g/liter K2HPO4, 6 g/liter KH2PO4, 1 g/liter trisodium citrate · 5H2O, 2% (wt/vol) glucose, 50 mg/liter l-Trp, 11 mg/liter ferric ammonium citrate, 2.5 g/liter potassium aspartate, and 3 mM MgSO4. When necessary, the media were supplemented with the appropriate antibiotics (ampicillin at 100 μg/ml for E. coli; chloramphenicol at 5 μg/ml, spectinomycin at 150 μg/ml, and kanamycin at 10 μg/ml for B. subtilis). E. coli and B. subtilis cells were transformed with DNA by standard procedures (25, 34). Sequencing of PCR-derived DNA fragments in the final plasmid constructs was carried out by Genome Express (Meylan, France).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| SG91 | trpC2 ΔyvcJ::cat | This work |
| SG106 | trpC2 ΔyvcJ::cat amyE::yvcI kan | This work |
| SG107 | trpC2 ΔyvcJ::cat amyE::yvcIJ kan | This work |
| BD3836 | hisB2 leu-8 metB5 amyE::P hs-comK spec | 27 |
| SG119 | trpC2 amyE::P hs-comK spec | BD3836→168 |
| SG120 | trpC2 ΔyvcJ::cat amyE::P hs-comK spec | SG119→SG91 |
| BD2528 | hisB2 leu-8 metB5 pUB110 comS kan | 15 |
| SG121 | trpC2 pUB110 comS kan | BD2528→168 |
| SG122 | trpC2 ΔyvcJ::cat pUB110 comS kan | SG121→SG91 |
| SG131 | trpC2 ΔyvcJ::tet | This work |
| BD3196 | hisB2 leu-8 metB5 Δrok::kan | 1 |
| BD2711 | hisB2 leu-8 metB5 comK-gfp cat | 37 |
| SG147 | hisB2 leu-8 metB5 Δrok::kan comK-gfp cat | BD2711→BD3196 |
| SG152 | hisB2 leu-8 metB5 ΔyvcJ::tet Δrok::kan PcomK-gfp cat | SG131→SG147 |
| BD1243 | hisAl leuA8 metB5 comA124::Tn9171acZ | 33 |
Deletion of the yvcJ gene.
For the deletion of the B. subtilis yvcJ gene, its flanking sequences were amplified by PCR with two pairs of primers (see the list of primers in Table S1 in the supplemental material): BG1 (yvcI [−20 to −3]) and BG49 (yvcI [+477 to +459]), containing a BamHI site, and EF1 (yvcK [−83 to −61]) and BG56 (yvcK [+954 to +936]), containing an EcoRI site. These two PCR products were digested by BamHI or EcoRI. In parallel, a chloramphenicol or tetracycline resistance cassette without promoter and transcriptional terminators was amplified from pAC5 (29) or pDG1515 (23), respectively, using primers EF2 (cat [−19 to +6]) and EF3 (cat [+651 to +630]) or 5′-tet and 3′-tet, and was then digested at the BamHI and EcoRI sites within the primers. After ligation of the three fragments, the resulting DNA was reamplified using primers BG1 and BG56 before transformation of B. subtilis strain 168 by a double-crossover event and selection for either chloramphenicol or tetracycline resistance. In the resulting strain SG91 or SG131, the cat gene or the tet gene, which replaces the yvcJ gene, is expressed from the promoter located in front of yvcI, like the other genes of the operon, yvcK, yvcL, crh, and yvcN, located downstream (14).
Strains for complementation.
Two PCR fragments containing the promoter region with either the yvcI gene or the yvcIJ genes were amplified with two pairs of primers, BG38 (promoter region [−100 to −81])-BG49 and BG38-BG67 (yvcJ [+888 to +870]), and were digested at the BamHI and EcoRI sites within the primers. These fragments were then ligated into the BamHI-EcoRI sites of pAC7 (41), so as to place these fragments between the arms of amyE, and were transformed into SG91 to create strains SG106 and SG107.
Plasmid construction for protein purification.
Fragments containing the yvcJ gene or the yhbJ gene were amplified by PCR using chromosomal DNA of B. subtilis strain 168 or E. coli strain K-12 and two specific primers for each amplification; one containing a BamHI or EcoRI site and another containing an XhoI site. The BamHI-XhoI fragment containing the yvcJ gene or the EcoRI-XhoI fragment containing the yhbJ gene was cloned into the expression vector pET21a(+) (Novagen). A modified YvcJ or YhbJ protein, carrying a T7 tag fused to the N terminus and a polyhistidine tag fused to the C terminus, was expressed from the resulting plasmid pJL1 or pJL2 after transformation in the E. coli strain C41(DE3) (30). Then YvcJ or YhbJ was affinity purified on nickel-nitrilotriacetic acid resin (Qiagen) as previously described (12).
Limited proteolysis.
Six micrograms of YvcJ was mixed with 40 mM NaCl,-10 mM Tris-HCl (pH 8.0) in the absence or presence of 1 mM ATP or GTP. After addition of 0.6 μg of endoproteinase Glu-C (Promega), the reaction mixture was incubated for 0, 5, 15, or 30 min at 37°C. Digestion was stopped by addition of an equal volume of electrophoresis loading buffer to the assay mixtures and by heating for 5 min at 100°C before the samples were subjected to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Fluorescence measurements.
All experiments were performed at 25 ± 0.1°C using a SAFAS flx-Xenius 5117 spectrofluorimeter. All spectra were corrected for buffer fluorescence. Fluorescence measurements were routinely carried out after dilution of YvcJ or YhbJ (final concentration, 0.5 μM) and equilibration for 5 min in 2 ml of a buffer containing 25 mM HEPES-KOH (pH 8) and 1 mM MgCl2.
Increasing concentrations of N-methylanthraniloyl (Mant) derivatives (Molecular Probes) were then added, and the emission fluorescence was scanned in the range of 300 to 500 nm, upon excitation at 282 nm. Fluorescent resonance energy transfer (FRET) between tryptophan residues of purified protein and bound Mant nucleotide derivatives was monitored by the appearance of a fluorescence emission peak between 400 and 500 nm, characteristic of bound nucleotide analogues. Peak integration was carried out for each ligand concentration, and the observed FRET was used for the calculation of ligand affinity. Each measure was determined in triplicate, and the curve fitting of the data was performed by using Graphit (version 4.0) software as previously described (11).
Enzymatic activity measurements.
Enzymatic activities were measured at 37°C. For nucleoside triphosphatase (NTPase) activity, the buffer contained 25 mM morpholinepropanesulfonic acid (MOPS)-HCl (pH 7.5), 5 mM MgCl2, 0.5 mM EDTA, 100 mM NaCl, 2 mM dithiothreitol (DTT), and 5 mM ATP or GTP, in the presence of an auxiliary regenerating system coupled to the oxidation of 0.3 mM NADH (4 mM phosphoenolpyruvate, 30 μg of pyruvate kinase, and 16 μg lactate dehydrogenase). The addition of 30 μg of purified protein (YvcJ or YhbJ) in a total volume of 1 ml initiated the reaction, and then NADH oxidation was recorded at 340 nm for 5 min. For p-nitrophenyl phosphatase (pNPPase) activity, the buffer contained 50 mM Tris-HCl (pH 7.4), 1 mM DTT, and 5 mM pNPP in a total volume of 100 μl. The addition of 2 μg of YvcJ or YhbJ initiated the reaction, and pNPP hydrolysis was recorded at 405 nm for 10 min. The kinetic parameters (Km and Vmax) were determined by double-reciprocal plots.
mRNA isolation.
Bacteria were cultivated in competence medium until the transition phase was reached. Then 1 ml of the cell suspension was harvested, and RNA was prepared using the standard protocol with the High Pure RNA isolation kit (Roche, Basel, Switzerland) except that two DNase treatments were performed to avoid any DNA contamination. All the isolations were performed at least in triplicate from independent cultures.
q-RT-PCR.
One microgram of RNA was reverse transcribed using the standard protocol with Superscript II reverse transcriptase (Invitrogen) and 100 ng of random primers. The resulting cDNA was diluted (1/16), and 5 μl was used for quantitative reverse transcription-PCR (q-RT-PCR). This step was performed on a Mastercycler ep realplex instrument (Eppendorf) by using the SYBR Premix Ex Taq (Perfect Real Time) PCR kit (Takara Bio Group, Japan) according to the manufacturer's instructions in a final volume of 20 μl. Specific primers are described in Table S1 in the supplemental material. Melting curves were analyzed to control for the specificity of the PCRs. Data from three independent experiments were analyzed and normalized with the software supplied with the Mastercycler. The relative units were calculated from a standard curve plotting four different dilutions (1/80, 1/400, 1/2,000, and 1/10,000) against the PCR cycle number at which the measured fluorescence intensity reached the threshold (CT), specified so that it is significantly above the noise band of the baseline (10 times the standard deviation).
Global transcriptional analysis.
Experiments were done using DNA microarrays from Eurogentec (Seraing, Belgium). The Pronto Plus system (Promega, Madison, WI) was used, by following the protocol described by the manufacturer, to label cDNA by direct incorporation of Cy3- and Cy5-dCTP. Hybridization was performed at 42°C for 16 h with DIG Easy hybridization solution (Roche, Basel, Switzerland) and herring sperm DNA (10 mg/ml; 1 μl added to 30 μl hybridization solution). After hybridization, the glass array was washed using 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS for 5 min, 0.2× SSC for 2 min, and 0.1× SSC for 2 min, dried by centrifugation (1,000 rpm for 1 min), and then scanned with a Packard BioChip ScanArray, model 5000 (Perkin-Elmer), at a resolution of 10 μm per pixel. Image and data analyses were performed with GenePix Pro (version 6.0) and Acuity (version 4.0) software (Axon Instruments, Inc.) using locally weighted scatterplot smoothing (LOWESS) normalization (3). Data from spots with diameters of <90 μm and with signal intensities lower than twice the background were removed. Microarray analysis was done for three independent experiments.
Measurement of transformation frequencies.
Strains were grown in MD medium at 37°C with shaking. Cells were transformed 2 h after the onset of stationary phase (T2), as described in reference 25, with chromosomal DNA carrying either a spectinomycin or a kanamycin marker, and were then plated on LB medium in the presence and absence of antibiotic. The transformation frequency was expressed as the ratio between the number of transformants per milliliter and the number of cells per milliliter.
Phosphorylation assays.
Bacteria were grown at 37°C in 400 ml MD medium and were collected at the onset of stationary phase. They were then harvested by low-speed centrifugation, resuspended in 4 ml of lysis buffer containing 100 mM Tris-HCl (pH 7.4), 10.5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 5 mg/ml lysozyme, and 10 U/ml of benzonase (Merck), and disrupted by sonication. After centrifugation, the supernatant was dialyzed against 100 mM Tris-HCl (pH 7.4) and 140 μM β-mercaptoethanol. Phosphorylation assays were carried out in a total volume of 20 μl. Three micrograms of YvcJ was incubated for 15 min at 37°C with 100 μg of crude extract in 50 mM Tris·HCl (pH 7.4), 5 mM MgCl2, 1 mM DTT, and 50 μM [γ-33P]ATP (2 μCi). The phosphorylation reaction was quenched by addition of an equal volume of SDS sample buffer to the reaction mixtures before SDS-PAGE analysis. Gels were then dried and exposed to autoradiography.
Microarray data accession number.
All DNA microarray data, including the detailed protocol, the slide images, and the raw data, obtained in this study are available online at Array Express (http://www.ebi.ac.uk/arrayexpress; accession number: E-MEXP-1471).
RESULTS
ATP and GTP bind to B. subtilis YvcJ and induce conformational changes.
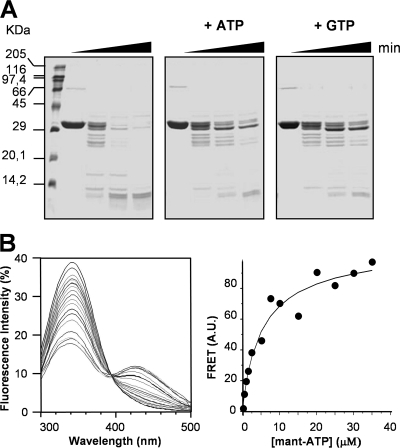
The presence of Walker A and B motifs in the N-terminal part of YvcJ (Swiss-Prot accession no. O06973) suggests that it is able to bind and hydrolyze ATP or GTP (Fig. 1) (39). In order to analyze its ability to bind nucleotides, the protein was overexpressed, purified, and assessed for its sensitivity to limited protease digestion. As shown in Fig. 2A, in the absence of nucleotides, YvcJ is sensitive to the endoproteinase Glu-C. Indeed, after 30 min of digestion, the band at around 30 kDa, corresponding to YvcJ, is degraded, and only bands below 14 kDa are still detectable. In contrast, in the presence of ATP or GTP, YvcJ is more resistant to proteolysis, since bands around 30 kDa were still visible after 30 min of exposure to the endoproteinase Glu-C. This indicates direct binding of both ATP and GTP to YvcJ, and this binding probably induces conformational changes in YvcJ that result in protection from endoproteinase Glu-C cleavage.
FIG. 1.
Partial sequence alignment of YvcJ homologues in various bacteria, including Bacillus anthracis, Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria gonorrhoeae, as well as B. subtilis and E. coli. The putative Walker A and B motifs are indicated by the two shaded boxes. Walker A is normally quoted in the literature by the consensus sequence (A/G)X4GK(T/S), which is centered at a loop between a β-strand and an α-helix (35, 39). The Walker B motif, much less conserved, can be represented by the consensus sequence DX2G (38), located at the end of a hydrophobic β-sheet.
FIG. 2.
YvcJ is a nucleotide-binding protein. (A) Coomassie blue-stained SDS-PAGE gel showing a partial proteolysis profile of YvcJ. YvcJ was incubated with endoproteinase Glu-C (Promega) in the absence or presence of ATP or GTP for 0, 5, 15, or 30 min at 37°C. The digestion profiles were assessed by 15% SDS-PAGE. (B) Effect of Mant-ATP on the fluorescence of YvcJ. Increasing concentrations of Mant-ATP (from 0 μM to 40 μM) were added to a 2-ml assay medium containing 25 mM HEPES-KOH (pH 8.0), 1 mM MgCl2, and 0.5 μM YvcJ, and the fluorescence intensity was recorded after each addition. The FRET, taken as the increase in fluorescence between 400 and 500 nm, was plotted against the concentration of Mant-ATP. A.U., arbitrary units.
Since YvcJ contains two tryptophans in its sequence (Trp-89 and Trp-223), we investigated the nucleotide binding by fluorescence measurements. The binding of the fluorescent nucleotide analogues Mant-ATP and Mant-GTP was measured. The addition of increasing concentrations of Mant nucleotides to YvcJ progressively quenched the Trp fluorescence emission peak (centered at around 340 nm) and gave rise concomitantly to a new peak of fluorescence (centered at around 430 nm). This new peak corresponds to a FRET between Trp residues and the Mant moiety (Fig. 2B). Fitted KD (equilibrium dissociation constant) values were obtained from the plot of the FRET intensity against the Mant-ATP or Mant-GTP concentrations; they indicate that YvcJ possesses a better affinity for Mant-GTP than for Mant-ATP (apparent KD values, 0.46 ± 0.04 μM and 3 ± 0.7 μM, respectively).
YvcJ has GTPase and ATPase activities.
Since YvcJ binds nucleotides and possesses a Walker B motif often found in GTPases (38), we tested if YvcJ exhibited an intrinsic ATPase or GTPase activity. The NTPase activities were studied by a coupled enzymatic assay following NADH oxidation at 340 nm. Our results indicate that YvcJ is able to hydrolyze 14.1 mol of ATP · min−1 · mol of enzyme−1 and 28.3 mol of GTP · min−1 · mol of enzyme−1 in the presence of magnesium and, to a lesser extent, manganese (data not shown). Thus, GTP is hydrolyzed with a higher efficiency than ATP. In order to test if YvcJ is able to hydrolyze other substrates apart from nucleotides, we tested pNPP, which is often used to measure protein phosphatase activity but has also been found to be hydrolyzed by the Na+/K+-ATPase (22). Our results indicate that YvcJ is able to hydrolyze 20.9 mol of pNPP · min−1 · mol of enzyme−1. To check if pNPPase and NTPase activities are interconnected, we carried out competition experiments. For this purpose, we measured pNPPase activities in a range of 2 to 10 mM pNPP in the presence of a constant concentration (4 mM) of ATP. We observed that the Vmax of the enzyme did not change, but the Km for pNPP increased (from 14.7 mM in the absence of ATP to 84.5 mM in the presence of 4 mM ATP), suggesting competitive inhibition between pNPP and ATP for YvcJ.
YhbJ from E. coli is also a nucleotide-binding protein with NTPase activity.
YvcJ and YhbJ (Swiss-Prot accession no. P33995) possess 38% identity. Since YhbJ also contains two tryptophans (Trp-190 and Trp-228) in its sequence, we tested its ability to bind to nucleotides by fluorescence measurements as described previously. The binding of Mant-ATP and Mant-GTP was observed and measured. The KD values obtained with YhbJ are similar to those obtained with YvcJ and indicate that YhbJ also has a better affinity for Mant-GTP than for Mant-ATP (apparent KD values, 0.65 ± 0.03 μM and 1.45 ± 0.54 μM, respectively). We also investigated the enzymatic properties of YhbJ. We observed that YhbJ also displays NTPase and pNPPase activities with Km values of the same magnitude but with a maximum velocity about 10-fold lower than that of YvcJ. Indeed, YhbJ was able to hydrolyze 1.75 mol of pNPP · min−1 · mol of enzyme−1, with a Km of 8.65 mM.
Deletion of yvcJ affects competence efficiency.
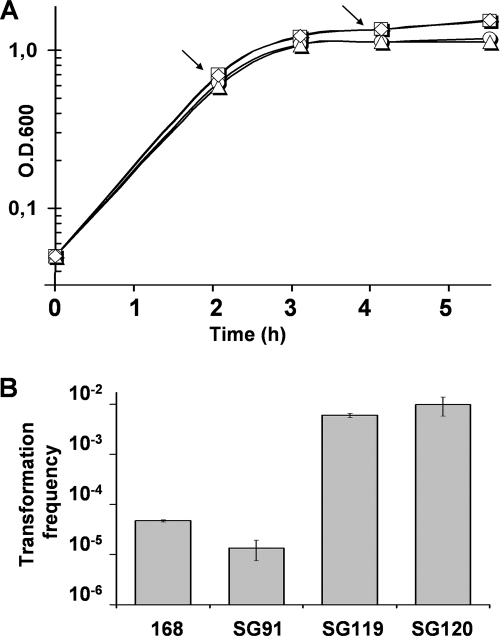
In order to examine the role of YvcJ in B. subtilis, we first constructed a nonpolar mutant strain, SG91 (yvcJ::cat), in which yvcJ was replaced by a cat cassette, since the yvcK gene, located just downstream of yvcJ, was crucial for growth on some poor carbon sources (14). We then analyzed the crude extract from yvcJ mutant cells after growth on LB medium by SDS-PAGE and staining with Coomassie blue. In contrast to recent results obtained with E. coli (strong overexpression of GlmS in a yhbJ mutant strain [24]), no obvious accumulation of proteins was detected (data not shown). We also monitored growth in other media, but no difference in growth curves was observed between the wild-type and mutant strains (Fig. 3A) (reference 14 and data not shown). We then undertook a phenotype screening. In particular, we tested if motility, biofilm formation, the secretion of degradative enzymes, competence, and sporulation were affected in the yvcJ mutant strain compared to the wild-type strain (data not shown). No obvious differences were detected between the two strains except for their competence efficiencies. Indeed, we observed that the transformation frequency of the mutant strain was about eightfold lower than that of the wild-type strain (0.6 × 10−5 for SG91 versus 4.6 × 10−5 for the wild-type strain). To determine if the competence phenotype observed for strain SG91 was due only to the disruption of the yvcJ gene, we carried out complementation experiments by expressing yvcJ from the amyE locus under the control of its own promoter. When the frequency of transformation of SG106 (yvcJ::cat amyE::yvcI) was compared to that of the isogenic strain SG107 (yvcJ::cat amyE::yvcIJ), strain SG106 showed no complementation of the yvcJ mutation by yvcI. Strain SG107 grew more slowly than the wild-type strain, but the expression of the yvcIJ genes from their own promoter at the amyE locus almost fully restored (about 90%) the wild-type efficiency of competence. In contrast, the frequency of transformation of strain SG106, in which only the yvcI gene is expressed at the amyE locus, was the same as that of the SG91 mutant strain. This result indicates that the competence phenotype observed in strain SG91 is due to the disruption of the yvcJ gene.
FIG. 3.
Growth curves (A) and transformation frequencies (B) of wild-type and yvcJ mutant strains in which ComK was overexpressed. Wild-type strain 168 (squares) and mutant strains SG91 (diamonds), SG119 (circles), and SG120 (triangles) were grown on MD medium. comK expression was induced by 1 mM IPTG at an optical density at 600 nm of 0.7 (first arrow), and cells were transformed 2 h afterwards (second arrow) by using 0.5 μg of a chromosomal DNA carrying a kanamycin marker. Transformation frequencies were determined by selection for Kmr. The transformation frequency corresponds to the ratio between the number of transformants per milliliter and the number of cells per milliliter.
Transcriptome analysis of a yvcJ mutant.
In B. subtilis, competence develops postexponentially and only in some media (9). The effect of the deletion of the yvcJ gene on whole-genome expression was assayed by microarray analysis of DNA from cells grown on MD medium and collected at the beginning of the stationary phase. cDNAs were generated from total RNA and were then hybridized to DNA arrays as described in Materials and Methods. Comparison of the gene expression profiles of the mutant strains versus the wild-type strain yields a total number of about 84 genes with ≥2-fold changes in transcription levels (about 76 downregulated and 8 upregulated genes), many of which have unknown functions and a few of which are implicated in the competence pathway (Table 2). In particular, we observe that the transcription level of comK, the gene encoding the main regulator of competence that activates the transcription of more than 100 genes (19), and of its regulon was repressed in the yvcJ mutant (Table 2). In contrast, the expression of the genes encoding proteins known to regulate comK expression either was not modified or was only slightly modified. We can also note that glmS expression was barely enhanced (1.4-fold) in the yvcJ mutant strain (Table 2), whereas a chromosomal transcriptional fusion of lacZ to glmS had a β-galactosidase activity 28 times higher in the E. coli homologue of the yvcJ mutant, the yhbJ mutant, than in the wild-type strain (24). Apart from the genes involved in the competence pathway, we could not obtain any clue as to the role of YvcJ; that is why we compared the expression profile of the yvcJ mutant strain with the wild-type strain under other growth conditions. We thus carried out transcriptome analyses under different conditions, such as growth of cells on CK-citrate or on LB medium and collection in mid-log phase, but they did not yield any new information on the cellular role of YvcJ (data not shown). We then focused our attention on genes involved in the competence pathway. The expression-profiling data were confirmed by q-RT-PCR performed with primers corresponding to genes involved in competence development (see Table S2 in the supplemental material). The estimated variations of comK expression are quite weak. This is because our result is obtained from the whole population, in which only about 10 to 20% of cells of a culture produce ComK and develop competence, even when all conditions are optimal (8).
TABLE 2.
Relative expression levels of glmS and of genes involved in the competence pathway in a yvcJ mutant strain from transcriptome analysis
| Gene | Expression level | Description |
|---|---|---|
| glmS | 1.39 | Glucosamine-6-phosphate synthase |
| Genes missing in the microarray | ||
| addA | ATP-dependent DNase | |
| comEC | Late competence operon required for DNA binding and uptake | |
| comP | Two-component sensor histidine kinase involved in early competence | |
| Genes with unchanged expression | ||
| comA | 0.84 | Two-component response regulator of late competence genes and surfactin production |
| comX | 1.00 | Competence pheromone precursor |
| comQ | 1.16 | Transcriptional regulator of late competence operon and surfactin expression |
| comS | 0.71 | Assembly link between regulatory components of the competence pathway |
| srfAA | 0.85 | Surfactin synthetase/competence |
| pnpA | 0.85 | Polynucleotide phosphorylase (PNPase) |
| ylbF | 0.73 | Unknown; positively controls ComK at a posttranscriptional level |
| mecA | 1.27 | Negative regulator of competence |
| clpX | 1.13 | ATP-dependent Clp protease |
| clpC | 0.84 | Class III stress response-related ATPase |
| clpP | 0.80 | ATP-dependent Clp protease proteolytic subunit |
| ypbH | 1.18 | Unknown; similar to negative regulator of competence; MecA homologue |
| codY | 0.88 | Transcriptional pleiotropic repressor |
| degU | 0.92 | Two-component response regulator involved in degradative enzyme and competence |
| abrB | 0.81 | Transcriptional pleiotropic regulator of transition state genes |
| rapC | 1.01 | Response regulator, aspartate phosphatase |
| addB | 0.97 | ATP-dependent DNase |
| rok | 1.29 | Repressor of comK |
| recA | 0.67 | Multifunctional protein involved in homologous recombination and DNA repair |
| spx | 1.70 | Negative effector of competence |
| spo0A | 1.35 | Two-component response regulator central for the initiation of sporulation |
| Genes repressed at least twofold in the yvcJ mutant strain | ||
| comK | 0.42 | Competence transcription factor |
| comGA | 0.23 | Late competence gene |
| comGB | 0.24 | DNA transport machinery |
| comGC | 0.27 | Exogenous DNA binding |
| comGD | 0.26 | DNA transport machinery |
| comGE | 0.27 | DNA transport machinery |
| comGF | 0.29 | DNA transport machinery |
| comGG | 0.29 | DNA transport machinery |
| comER | 0.29 | Nonessential gene for competence |
| comEA | 0.20 | Exogenous DNA-binding protein |
| comEB | 0.39 | Nonessential gene for competence |
| comFA | 0.21 | Late competence protein required for DNA uptake |
| comFB | 0.26 | Late competence gene |
| comFC | 0.28 | Late competence gene |
| ywpH | 0.26 | Unknown; similar to single-strand DNA-binding protein |
| nin | 0.34 | Inhibitor of the DNA-degrading activity of NucA |
| comC | 0.41 | Late competence protein |
| yvyF | 0.36 | Unknown; similar to flagellar protein |
| nucA | 0.33 | Membrane-associated nuclease |
ComS or ComK overexpression bypasses yvcJ control of competence.
The effect of ComK overexpression on competence in wild-type or yvcJ backgrounds was tested by measuring the transformation levels in the presence or absence of overexpression of comK. We did not observe any difference in growth (Fig. 3A). However, in both strains SG119 and SG120, strong overexpression of ComK induced very high transformation efficiencies of >150-fold (Fig. 3B). This result clearly shows that ComK overexpression rescues the mutant phenotype of yvcJ inactivation for competence. Then we tested if constitutive expression of comS was able to bypass the mutant yvcJ phenotype for competence. It has been shown previously that ComK, targeted by MecA, is degraded by the ClpC-ClpP complex (36). This proteolysis is inhibited by ComS, an antiadaptor protein that binds to MecA, thus preventing the degradation of the competence transcription factor, ComK (31). As expected, when ComS was overexpressed, it rescued the yvcJ mutant phenotype for competence (Table 3) and brought the transformation efficiency back to the level of the wild-type strain, 168, or strain SG122. We also compared the expression of comS, comK, and comGA genes in these genetic backgrounds by q-RT-PCR. As shown in Table 3, the overexpression of ComS bypasses yvcJ for comK expression. This result confirms that the decrease in competence efficiency is correlated with lower expression of comK and consequently of the late competence genes.
TABLE 3.
ComS overexpression bypasses yvcJ deletion for competence and gene expressiona
| Strain | Relevant genotype | Transformation frequency (106) | Relative gene expression determined by q-RT-PCR
|
||
|---|---|---|---|---|---|
| comS | comK | comGA | |||
| 168 | Wild type | 46.0 ± 7.1 | 1 | 1 | 1 |
| SG91 | yvcJ | 6.6 ± 1.6 | 0.7 ± 0.3 | 0.60 ± 0.18 | 0.60 ± 0.21 |
| SG121 | pcomS | 64.5 ± 10.6 | 3.1 ± 0.4 | 1.06 ± 0.03 | 1.27 ± 0.06 |
| SG122 | yvcJ pcomS | 31.5 ± 7.8 | 4.9 ± 1.8 | 1.60 ± 0.39 | 1.65 ± 0.44 |
Cells of the wild-type strain and of yvcJ mutant strains in which ComS was overexpressed were transformed with 250 ng of a chromosomal DNA containing a spectinomycin cassette. Transformation frequencies were determined as described in Materials and Methods. The relative expression of the comS, comK, and comGA genes in the wild-type strain and in mutant strains in which ComS was overexpressed was determined by q-RT-PCR. All the values are averages of data from at least three independent experiments.
Deletion of yvcJ results in a decrease in the number of cells that express competence.
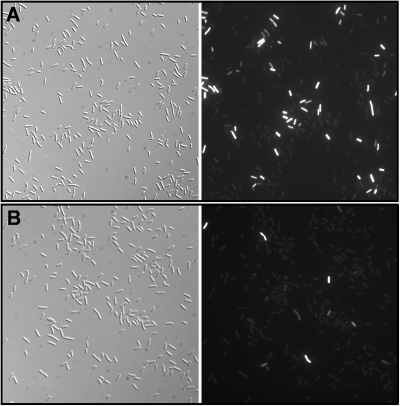
ComK expression shows a bistable pattern and occurs only in about 10% of the cells in a stationary-phase culture (27). Thus, a yvcJ mutation may result either in inhibition of comK expression within the competent cells and/or in a reduction in the number of cells that express competence. To investigate the second hypothesis, we used strain BD2711 (17), which contains a green fluorescent protein (GFP)-tagged ComK fusion protein expressed from its native promoter and in which the yvcJ gene is deleted. To enhance GFP expression, a rok mutation was also introduced to increase the fraction of cells that expressed competence (21). At 90 min after the end of exponential growth, the observation of comK-gfp expression by fluorescence microscopy indicated that strain SG152, in which yvcJ was deleted, showed a significantly lower fraction of fluorescent cells than strain SG147, containing the native yvcJ gene (Fig. 4): only 5% of cells from strain SG152 were fluorescent versus 16% for strain SG147 when about 2,000 cells from each strain were observed.
FIG. 4.
Microscopy of cells harboring comK-gfp in strains SG147 (A) and SG152 (with yvcJ deleted) (B). Shown are images of cells from the cultures of strains SG147 and SG152 90 min after the transition between the exponential and the stationary-growth phase. (Left) Phase-contrast microscopy; (right) fluorescence microscopy.
YvcJ is involved in the phosphorylation of a cellular component.
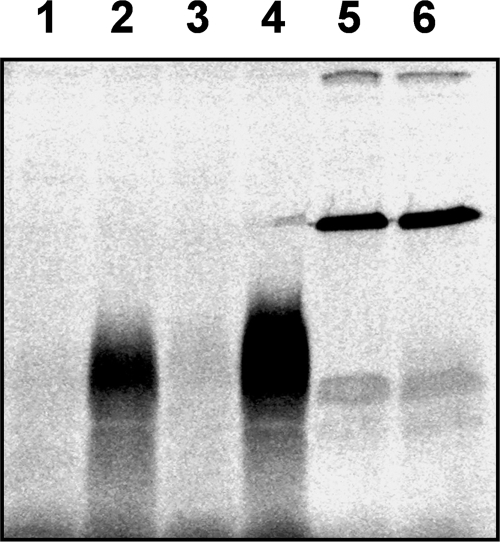
We tried to determine how YvcJ affects the competence pathway. The large structural family of the P-loop-containing proteins is composed of NTPases but also of kinases. Therefore, we tested if YvcJ also possessed a kinase activity. We observed that when a crude extract of the B. subtilis wild-type strain, 168, was incubated with purified YvcJ and [γ-33P]ATP, a phosphorylated band of around 20 kDa could be detected (Fig. 5, lane 2), whereas no phosphorylation of the 20-kDa component could be observed in the absence of purified YvcJ (Fig. 5, lane 1). We tried to identify this substrate by various approaches, including 1- or 2 dimensional SDS gels and mass spectrometry analysis, but the signal was unstable and was thus impossible to isolate and identify by these biochemical techniques. We then tested if this radioactive signal was involved in the competence pathway by using two strains: BD1243, with a mutation in the comA gene and weakened competence, and BD3836, overexpressing comK and highly competent. All strains were grown on MD medium, and crude extracts were prepared and tested in phosphorylation assays. We observed that the intensity of the YvcJ-dependent radioactive signal increased over that for the wild type when a crude extract from strain BD1243 was incubated with purified YvcJ and [γ-33P]ATP (Fig. 5, lane 4). In contrast, no YvcJ-dependent phosphorylation was detected in a crude extract from strain BD3836 (Fig. 5, lane 6). These results suggest that YvcJ may act on the competence pathway via the phosphorylation of a cellular component.
FIG. 5.
ATP-dependent phosphorylation in the presence of YvcJ. Wild-type strain 168 (lanes 1 and 2) and strains BD1243 (lanes 3 and 4) and BD3836 (lanes 5 and 6) were grown on MD medium, and cells were collected at the onset of the stationary phase. For strain BD3836, comK overexpression was induced by addition of 1 mM IPTG to the growth medium. Crude extracts were prepared and then phosphorylated using [γ-33P]ATP in the absence (lanes 1, 3, and 5) or the presence (lanes 2, 4, and 6) of purified YvcJ. Phosphorylation assays were analyzed by 15% SDS-PAGE. Gels were then dried and exposed to autoradiography.
DISCUSSION
In this work we provide evidence that two members of the UPF0042 P-loop-containing proteins, the YvcJ protein from B. subtilis and its homologue in E. coli, YhbJ, are both able to bind and hydrolyze ATP as well as GTP. Furthermore, the nucleotide binding induces a change of conformation in both proteins. In addition, YvcJ and YhbJ have fairly similar affinities for Mant-ATP and Mant-GTP, and both of them have better affinity for GTP. Interestingly, under the experimental conditions described here, YvcJ possesses a maximum velocity of hydrolysis 10-fold higher than that of YhbJ, implying that our in vitro conditions probably were not optimal for measuring nucleotide and pNPP hydrolysis by YhbJ and suggesting that some cofactor would be necessary to achieve a better enzymatic velocity.
Conserved P-loop ATPases and GTPases of unknown function constitute a significant group of proteins in bacteria (5). Among these, most studies have been carried out on essential proteins that seem to play a role in ribosome biogenesis. The YvcJ and YhbJ proteins do not belong to this group of conserved GTPases, since they are dispensable for the cell. For E. coli, it was shown that YhbJ was involved in the control of GlmS expression by affecting the processing and stability of a small RNA, GlmZ (24). In B. subtilis, we observed that YvcJ was not involved in GlmS expression but that the deletion of yvcJ decreased the efficiency of competence and weakly repressed the expression of comK and that of its downstream genes, thereby decreasing the proportion of cells that express competence.
yvcJ is a highly conserved gene (4), found in naturally competent bacteria but also in several noncompetent bacteria, and its role in the competence pathway, a phenomenon resulting from several cellular events, could be indirect. Indeed, the expression of comK is highly regulated, and it depends on the presence of several proteins and regulators (19). For example, the srfA operon, containing comS, is also involved in the synthesis of the lipopeptide surfactant surfactin, which is required for swarming motility (6). Furthermore, two phosphorelay proteins, Spo0A, involved in sporulation, and DegU, involved in degradative enzyme secretion, have been shown to regulate comK expression (16, 18). In the yvcJ mutant strain, motility, biofilm formation, secretion of degradative enzymes, and sporulation seem not to be affected, and no significant change in the expression of srfA, spo0A, or degU was detected by microarrays or q-RT-PCR. Our epistatic study showed that either ComK or ComS overexpression bypasses yvcJ deletion. Our transcriptome data indicate that comS transcription and the expression of the genes encoding proteins known to regulate comK expression either were not modified or were only slightly modified, highlighting the fact that only comK transcription is significantly repressed in the yvcJ mutant. Furthermore, our observation of the comK-gfp fusion indicates that yvcJ deletion is responsible for a threefold decrease in the number of cells that express competence. All these data suggest that the specific decrease in comK expression, due to the yvcJ deletion, is responsible for the decrease in the number of cells that express competence, thus causing a decrease in competence efficiency.
In order to obtain some clues about the mechanism by which YvcJ affects the expression of competence, we tested its potential kinase activity. We observed that when bacteria were grown on MD medium, YvcJ was involved in the phosphorylation of a 20-kDa cellular component. Despite our efforts, we could not characterize this labile radioactive signal, which, however, seems correlated with the competence pathway. Indeed, in a comA mutant strain, affected in competence, the intensity of the YvcJ-dependent radioactive signal is higher than that for the wild-type strain. In contrast, no YvcJ-dependent phosphorylation was detected in a crude extract from a highly competent strain overexpressing comK. We also observed that under these conditions, the deletion of the yvcJ gene has no effect on competence efficiency (see the transformation frequencies of strains SG119 and SG120 [Fig. 3B]). These results suggest that YvcJ affects the expression of competence by phosphorylation of a cellular component.
In E. coli, YhbJ is proposed to have a pleiotropic effect by regulating the activities of genes or proteins involved in RNA turnover control (24). Hence, the yvcJ gene was expected to have a similar pleiotropic function in B. subtilis. However, our global transcriptional studies of cells grown under various conditions and our phenotypic screening yielded a role for YvcJ only in the competence of cells. Yet it has previously been reported that disruption of RNA stability affects competence (10, 28). In particular, PnpA, a polynucleotide phosphorylase involved in RNA stability (10), was shown to be necessary for competence and for the expression of late competence genes in B. subtilis (26). It may modify and stabilize the srfA transcript, since it stimulates the synthesis of surfactin synthetase and ComS (26). But while the whole-genome transcript analysis of a pnpA mutant strain showed high overexpression of trp operon genes when the strain was grown in the presence of tryptophan (7), the expression levels of two trp genes highly induced in a pnpA mutant, trpB and trpE, were not affected in the yvcJ mutant strain (data not shown). Furthermore, the yvcJ strain is able to swarm like the wild-type strain and does not have a cold-sensitive growth phenotype (data not shown), in contrast to the pnpA mutant (40). All these observations suggest that, in the yvcJ mutant strain, the activity of PnpA is not affected.
In conclusion, the deletion of the yvcJ and yhbJ genes, encoding two members of the P-loop protein family UPF0042, has different consequences in B. subtilis and E. coli. However, from our data, we cannot exclude the possibility that YvcJ, like its E. coli counterpart, can be involved, directly or indirectly, via a phosphorylation event, in the stability of some transcripts and thus can modulate the synthesis of ComS or ComK. All these hypotheses are now being tested in the laboratory to elucidate the molecular mechanism involving YvcJ and how this P-loop-containing protein is connected to the competence pathway.
Supplementary Material
Acknowledgments
This research was supported by the CNRS, the University of Aix-Marseille II, and the French FRM (Fondation pour la Recherche Médicale). J.L. was supported by an MENRT fellowship from the French government.
We thank François Denizot, Frédéric Barras, and Vincent Méjean for helpful discussions, Adrien Ducret for help in microscopy, and Yann Denis from the IBSM transcriptome platform. We also thank Dave Dubnau and his laboratory for the kind gift of strains and for advice.
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albano, M., W. K. Smits, L. T. Ho, B. Kraigher, I. Mandic-Mulec, O. P. Kuipers, and D. Dubnau. 2005. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J. Bacteriol. 1872010-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrick, J. E., K. A. Corbino, W. C. Winkler, A. Nahvi, M. Mandal, J. Collins, M. Lee, A. Roth, N. Sudarsan, I. Jona, J. K. Wickiser, and R. R. Breaker. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 1016421-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, J. A., S. Hautaniemi, A. K. Jarvinen, H. Edgren, S. K. Mitra, and J. Astola. 2004. Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics 5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boël, G., I. Mijakovic, A. Maze, S. Poncet, M. K. Taha, M. Larribe, E. Darbon, A. Khemiri, A. Galinier, and J. Deutscher. 2003. Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria. J. Mol. Microbiol. Biotechnol. 5206-215. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. D. 2005. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 83738-746. [DOI] [PubMed] [Google Scholar]
- 6.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8821-831. [DOI] [PubMed] [Google Scholar]
- 7.Deikus, G., P. Babitzke, and D. H. Bechhofer. 2004. Recycling of a regulatory protein by degradation of the RNA to which it binds. Proc. Natl. Acad. Sci. USA 1012747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53217-244. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D. 1991. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55395-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farr, G. A., I. A. Oussenko, and D. H. Bechhofer. 1999. Protection against 3′-to-5′ RNA decay in Bacillus subtilis. J. Bacteriol. 1817323-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forouhar, F., M. Abashidze, H. Xu, L. L. Grochowski, J. Seetharaman, M. Hussain, A. Kuzin, Y. Chen, W. Zhou, R. Xiao, T. B. Acton, G. T. Montelione, A. Galinier, R. H. White, and L. Tong. 2008. Molecular insights into the biosynthesis of the F420 coenzyme. J. Biol. Chem. 28311832-11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stülke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 948439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., and E. V. Koonin. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 325452-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görke, B., E. Foulquier, and A. Galinier. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 1513777-3791. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, J., A. Luttinger, and D. Dubnau. 1996. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 21763-775. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, J., M. Roggiani, and D. Dubnau. 1995. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 1773601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haijema, B. J., J. Hahn, J. Haynes, and D. Dubnau. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 4052-64. [DOI] [PubMed] [Google Scholar]
- 18.Hamoen, L. W., A. F. Van Werkhoven, G. Venema, and D. Dubnau. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 979246-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 1499-17. [DOI] [PubMed] [Google Scholar]
- 20.Hegyi, H., J. Lin, D. Greenbaum, and M. Gerstein. 2002. Structural genomics analysis: characteristics of atypical, common, and horizontally transferred folds. Proteins 47126-141. [DOI] [PubMed] [Google Scholar]
- 21.Hoa, T. T., P. Tortosa, M. Albano, and D. Dubnau. 2002. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 4315-26. [DOI] [PubMed] [Google Scholar]
- 22.Homareda, H., and M. Ushimaru. 2005. Stimulation of p-nitrophenylphosphatase activity of Na+/K+-ATPase by NaCl with oligomycin or ATP. FEBS J. 272673-684. [DOI] [PubMed] [Google Scholar]
- 23.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 1834317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalamorz, F., B. Reichenbach, W. Marz, B. Rak, and B. Görke. 2007. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 651518-1533. [DOI] [PubMed] [Google Scholar]
- 25.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 1772403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luttinger, A., J. Hahn, and D. Dubnau. 1996. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19343-356. [DOI] [PubMed] [Google Scholar]
- 27.Maamar, H., and D. Dubnau. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mäder, U., L. Zig, J. Kretschmer, G. Homuth, and H. Putzer. 2008. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 70183-196. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Verstraete, I., M. Debarbouille, A. Klier, and G. Rapoport. 1992. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J. Mol. Biol. 22685-99. [DOI] [PubMed] [Google Scholar]
- 30.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260289-298. [DOI] [PubMed] [Google Scholar]
- 31.Prepiak, P., and D. Dubnau. 2007. A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol. Cell 26639-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reichenbach, B., A. Maes, F. Kalamorz, E. Hajnsdorf, and B. Görke. 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 362570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roggiani, M., J. Hahn, and D. Dubnau. 1990. Suppression of early competence mutations in Bacillus subtilis by mec mutations. J. Bacteriol. 1724056-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15430-434. [DOI] [PubMed] [Google Scholar]
- 36.Schlothauer, T., A. Mogk, D. A. Dougan, B. Bukau, and K. Turgay. 2003. MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. USA 1002306-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 176730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Via, A., F. Ferre, B. Brannetti, A. Valencia, and M. Helmer-Citterich. 2000. Three-dimensional view of the surface motif associated with the P-loop structure: cis and trans cases of convergent evolution. J. Mol. Biol. 303455-465. [DOI] [PubMed] [Google Scholar]
- 39.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, W., and D. H. Bechhofer. 1996. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 1782375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinrauch, Y., T. Msadek, F. Kunst, and D. Dubnau. 1991. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J. Bacteriol. 1735685-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkler, W. C., A. Nahvi, A. Roth, J. A. Collins, and R. R. Breaker. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428281-286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.