Abstract
The organization of chromatin has a major impact on cellular activities, such as gene expression. For bacteria, it was suggested that the spatial organization of the genetic material correlates with transcriptional levels, implying a specific architecture of the chromosome within the cytoplasm. Accordingly, recent technological advances have emphasized the organization of the genetic material within nucleoid structures. Gemmata obscuriglobus, a member of the phylum Planctomycetes, exhibits a distinctive nucleoid structure in which chromatin is encapsulated within a discrete membrane-bound compartment. Here, we show that this soil and freshwater bacterium tolerates high doses of UV and ionizing radiation. Cryoelectron tomography of frozen hydrated sections and electron microscopy of freeze-substituted cells have indicated a more highly ordered condensed-chromatin organization in actively dividing and stationary-phase G. obscuriglobus cells. These three-dimensional analyses revealed a complex network of double membranes that engulf the condensed DNA. Bioinformatics analysis has revealed the existence of a putative component involved in nonhomologous DNA end joining that presumably plays a role in maintaining chromatin integrity within the bacterium. Thus, our observations further support the notion that packed chromatin organization enhances radiation tolerance.
It is generally accepted that the prokaryotic cytoplasm maintains a defined order in which proteins and DNA are spatially localized so as to accurately orchestrate cellular processes (13, 22). Bacterial chromatin is generally organized into a subcellular nucleoid structure, while in the phylum Planctomycetes, chromatin is bounded by a membrane to present a boundary analogous to the defining feature of the eukaryotic cell, the nucleus (20, 55). Cellular compartmentalization within members of the phylum Planctomycetes thus provides novel insight into the cellular compartmentalization of bacteria and their nucleoid structures. The Planctomycetes are regarded as a rapidly evolving (56) or early-branching (4) group within the domain Bacteria and consist of budding, peptidoglycan-lacking organisms (16). Within this phylum, Gemmata obscuriglobus exhibits a double-membrane-bounded nucleoid structure, as revealed by electron microscopy studies, termed the nuclear body (20). Given the similarities of this compartment to the double-membrane nuclear envelope of eukaryotic cells, it has been suggested that G. obscuriglobus corresponds to the missing link between prokaryotes and eukaryotes (20, 35, 42). Nonetheless, compartmentalization within G. obscuriglobus and the consequences of this phenomenon remain enigmatic.
A fundamental property of the organization of the genetic material is reflected in its influence on the ability of organisms to tolerate radiation. Indeed, the ability of some microorganisms to tolerate DNA lesions caused by high doses of radiation correlates with their ability to maintain the integrity of their genomic material, as exemplified by studies of highly radiation-resistant bacteria, such as Deinococcus radiodurans and spores of Bacillus species (7, 18). Such studies suggest that two primary factors promote radiation resistance: enzymatic repair pathways and physical protection of the genomic material (39). In bacteria, two main enzymatic pathways are thought to provide protection against severe DNA damage, i.e., homologous recombination and nonhomologous end joining (NHEJ) (10, 53). Homologous recombination is promoted by binding of the RecA protein to single-stranded DNA, forming a presynaptic filament. The filament acts as a sequence-specific DNA-binding entity, capable of scanning and binding intact homologous double-stranded DNA sites. The resulting ternary complex promotes DNA strand exchange and heteroduplex extension. This mechanism is supported by numerous studies showing the involvement of the ternary complex in the repair of double-strand breaks (DSBs) (reviewed in references 30-32 and 47), in addition to other DNA repair mechanisms, such as replication-dependent recombination (reviewed in references 6 and 8).
NHEJ is a crucial mechanism for recognizing and repairing DNA breaks that was considered, until recently, exclusive to eukaryotes (9, 54). In eukaryotes, the NHEJ mechanism participates in the meiosis recombination process and in the VDJ locus rearrangement that occurs in B cells, as well in DSB repair processes (10, 53). It was recently shown that a unique family of ATP-dependent DNA ligases (ADDLs) is present in several bacterial species, including Mycobacterium tuberculosis, Bacillus subtilis, and Agrobacterium tumefaciens, in addition to the typical NAD-dependent ligases (9, 54, 57). In some cases, these ligases were found to be recruited by the Ku-like protein, a bacterial homologue of the eukaryotic NHEJ system Ku proteins (2, 10). These proteins are targeted to the edges of DNA strands, keeping DNA ends in close proximity until they are ultimately ligated (43, 52).
In addition to enzyme-based mechanisms, an alternate mode of radiation resistance is based on physical protection through the packed conformation of the genetic material. This passive form of protection is usually enlisted in states in which high-energy compounds are depleted, such as during starvation and lowered metabolic activity, causing a virtually complete inactivation of DNA repair enzymes, as in the case of dormant bacterial spores (17, 50). A link between condensed chromatin and radiation resistance was previously suggested to promote bacterial chromatin integrity under stress conditions and in unusual cases, such as D. radiodurans (12, 34, 37). From those studies, it was postulated that the tightly packed DNA organization of D. radiodurans chromatin, whether organized in a toroidal or two-dimensional crystal-like conformation (11, 34), renders NHEJ repair activity feasible by enforcing spatial proximity of the DNA fragments. Therefore, NHEJ is thermodynamically favored in the case of DSB repair (19, 34).
Here, we show that cultured G. obscuriglobus cells exposed to UV radiation exhibit a survival rate that is approximately 40 times higher than that of Escherichia coli. G. obscuriglobus can also tolerate large doses of ionizing radiation (IR), gamma rays, indicating an efficient DNA DSB repair mechanism. We have applied a variety of electron microscopy and tomography approaches suitable for biological specimens to characterize the three-dimensional (3D) organization of G. obscuriglobus internal membranes and chromatin. These analyses imply that the DNA in this organism is highly condensed. Genomic analyses have indicated the involvement of direct ligation of DSBs rather than homologous-recombination-based repair. Our observations thus support a link between highly condensed DNA structures and radiation tolerance in bacteria through direct ligation of DNA DSBs.
MATERIALS AND METHODS
Culture.
Lyophilized G. obscuriglobus DSM 5831 was acquired from the German Collection of Microorganisms and Cell Cultures. Cultures were generated by inoculation of M1 medium (48) and incubation at 30°C for 5 or 10 days, as discussed in Results. E. coli strains BL21 and AB1157 were cultured in Luria-Bertani (LB) medium at 37°C.
Specimen preparation. (i) High-pressure freezing, cryosubstitution, and sectioning.
G. obscuriglobus was grown for 4 days in M1 medium. The culture was harvested, concentrated, and drawn into cellulose capillary tubes with an inside diameter of 200 μm (28), which were cut into short segments. The cut segments were placed into 100-μm-deep aluminum sample carriers, and the excess empty volume was filled with hexadecene (Sigma, St. Louis, MO). The samples were frozen in an HPM 010 high-pressure freezing machine (Bal-Tec, Liechtenstein) under a pressure of 2.1 × 108 Pa for 250 ms. Following cryofixation, the samples were freeze-substituted (Leica EM AFS) with anhydrous acetone containing 1% OsO4 (Electron Microscopy Supplies, West Chester, PA) for 3 days at −90°C and then warmed up to −30°C over 24 h, kept for 1 h on ice, and brought to room temperature. Samples were then infiltrated for 7 days at room temperature in a series of increasing concentrations of Epon (Electron Microscopy Supplies) in acetone (10%, 20%, 30%…100% Epon), followed by four exchanges of pure resin. The Epon block was sectioned (70- to 80-nm-thick sections) using a Leica UCT microtome and a Diatome Ultra 45° diamond knife. The sections were mounted on 200- or 300-mesh carbon-coated electron microscopy grids. The sections were stained with 1% uranyl acetate and lead citrate and examined on a Jeol 1230 electron microscope operating at 120 kV.
(ii) DNA-specific staining.
Intracellular DNA localization was performed using the DNA-specific stain osmium ammine-SO2 (41). Grid-mounted thin sections of Epon-embedded bacteria were floated on 5 N HCl for 30 min at room temperature, washed with distilled water, and treated with osmium ammine-B (Polysciences) in 8 N acetic acid and 40 mM sodium metabisulfite for 1 h at 37°C. The sections were then thoroughly rinsed with distilled water, dried, and studied without additional staining.
(iii) Fully hydrated (vitreous) cryosectioning.
G. obscuriglobus cultures grown to logarithmic and stationary phases were harvested by centrifugation at 4,000 × g for 5 min and resuspended at a 1:1 ratio in fresh culture medium containing 40% (wt/vol) dextran (Mr, 100,000 to 200,000; product D-4876; Sigma, St. Louis, MO). The suspension was drawn into copper capillary tubes with an inside diameter of 250 μm. The suspension in the capillary tube was vitrified using an EMPact hyperbaric freezer (Leica Microsystems, Vienna, Austria), as described by Studer et al. (50a). After the sides were trimmed from the copper tube, sections with a nominal thickness setting of 50 nm, 85 nm, 100 nm, or 150 nm were cut using a Leica UC6 microtome and cryo 25° or 35° diamond knives (Diatome, Biel, Switzerland). An ionizer was positioned close to the knife's edge to facilitate gliding of the sections. Ribbons of sections were transferred to bare 200-mesh copper grids (Polysciences, Warrington, PA) and attached by pressing them between two polished ceramic plates. The loaded grids were stored in liquid nitrogen until tomography was performed.
(iv) Cryoelectron tomography of vitrified sections and image processing.
Sample-containing grids were cryotransferred into a Tecnai Polara electron microscope operating at 300 kV and equipped with a Gatan postcolumn GIF energy filter. Tomographic tilt series were acquired at 2° intervals over an angular range of −64° to +64° at an underfocus of −15 μm (24). The projection image was recorded on a 2K charge-coupled-device camera. The tilt series were aligned by cross-correlation, and the tomographic volume was reconstructed by a weighted back-projection algorithm as implemented in the EM software package (26).
For 3D visualization purposes, the tomogram was processed using an anisotropic-diffusion denoising algorithm to increase the S/N ratio (15). Image segmentation and isosurface rendering were performed with the Amira 3.1 software package.
Radiation procedures. (i) UVC radiation.
A 5-day-old G. obscuriglobus culture was harvested and filtered through Whatman Nuclepore 5-μm filters in order to eliminate cell aggregates. The filtered culture was placed on ice until irradiation was performed. Radiation tolerance experiments were performed by exposing the culture to a series of increased irradiation durations. Five-milliliter aliquots from the same batch were placed in sterile petri dishes for each exposure. The bacteria were irradiated under continuous agitation at 100 rpm in a fixed diameter via a polychromatic UV source (Philips; Holland UVC 15W G15T8; maximum, 254 nm). An unexposed control replica tube with the same culture was kept on the side.
After each irradiation, the exposed and unexposed cultures were serially diluted (by a factor of 10) in cold fresh medium, and the appropriate dilutions from each series were plated on M1 agar plates. The plates were incubated at 30°C for 18 days until colonies became visible for counting (Suntex [Taiwan] colony counter 570).
A logarithmic-phase E. coli BL21(DE3) culture (optical density, ∼0.6) was irradiated under the same conditions as G. obscuriglobus as an additional control. Exposed and unexposed E. coli cultures were also diluted in a similar manner and plated on LB agar plates. The plates were incubated overnight at 37°C, and the colonies were counted. For each irradiation dose, the so-called “relative survival value” was calculated by dividing the numbers of culture CFU from irradiated cultures by the corresponding control culture CFU and multiplying by 100%. A radiation tolerance (relative survival as a function of the radiation dose) curve was produced using Origin 6.1 software (OriginLab Corporation) for both G. obscuriglobus and E. coli based on the colony-counting data.
(ii) IR.
The same culture conditions described above for UVC radiation experiments were used. Tubes were irradiated by a 60Co radioactive source for approximately 50 min. The estimated dose was 1,000 to 1,200 Gy. A set of replica tubes were left unexposed as a control. After irradiation, the exposed and unexposed cultures were serially diluted in cold, fresh medium, and the appropriate dilutions from the series were plated onto M1 agar plates. The plates were incubated at 30°C for 18 days, at which time colonies were visible for counting. A logarithmic-phase E. coli BL21(DE3) culture (optical density, ∼0.46) was irradiated in the same manner as G. obscuriglobus to serve as a control. Exposed and unexposed E. coli cells were also diluted in a similar manner and plated onto LB agar plates. The plates were incubated overnight at 37°C, after which the colonies were counted.
Bioinformatics analysis.
A partial genome sequence of G. obscuriglobus based on shotgun analysis was provided by The Institute for Genomic Research. Sequences of the proteins of interest were identified using tBLASTn. Homologous sequences showing high fidelity rates (low E values and high percentages of amino acid similarity) were recorded, and the open reading frames (ORFs) were searched for using the NCBI ORFinder applet. When an ORF was determined in the suspicious region, its translational frame was compared to the frame of the homologous sequence achieved by the BLAST search. Multiple alignments were performed using the ClustalW applet, aligning the homologous predicted protein of G. obscuriglobus with accession numbers gi 15608078 and 2633694.
RESULTS
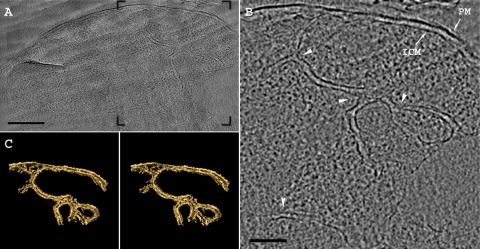
To acquire deeper insight into the organization of the double-membrane system of G. obscuriglobus, we examined actively growing 4-day-old cells using cryoelectron tomography of vitrified sections (CETOVIS) (25). Such analysis allowed us to define the 3D organization of the internal membranes in a fully hydrated, close-to-native state (1). A complex double-membrane system emanating from the intracytoplasmic membrane was elucidated (Fig. 1). A rendered view of such a network, shown in stereo (Fig. 1C), highlights the complexity of the membrane network, implying that the bacterial nucleoid is not completely sealed by the double-membrane system. The internal membrane system is fully continuous with the intracytoplasmic membrane, presumably formed by membrane invaginations. This observation is in agreement with earlier studies showing connections between the compartments, confirmed here by means of electron microscopy imaging of sections from freeze-substituted and polymer-embedded preparations (20) (Fig. 2A and B).
FIG. 1.
Cryoelectron tomography of a freeze-hydrated section of G. obscuriglobus. (A) A 15-nm-thick tomographic slice from the reconstructed volume shows the intracytoplasmic meshwork of double membranes. (B) The region framed in panel A shows the plasma membrane (PM) and the morphology of the intracytoplasmic membrane (ICM) system. The membrane curvature and junctions are indicated by arrowheads. (C) Stereo image of the membranes found within the tomogram shown in panel B. The surface-rendering view shows that the internal membranes in G. obscuriglobus are continuous with the ICM. Scale bars, 250 nm (A) and 125 nm (B).
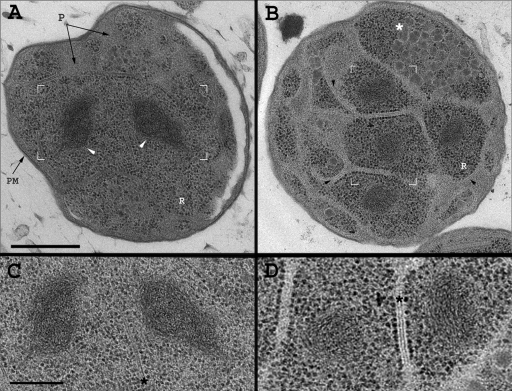
FIG. 2.
Electron microscopy analysis of G. obscuriglobus reveals a densely packed chromatin structure. (A and B) Transmission electron micrographs of G. obscuriglobus, high-pressure frozen and freeze-substituted (see Materials and Methods), exhibiting the condensed chromatin (white arrowheads). Typical G. obscuriglobus cell characteristics, i.e., the plasma membrane (PM), intracytoplasmic membrane (black arrowheads), peryphoplasm (P), riboplasm (R), and glycogen storage (white asterisk), are indicated. (C and D) Enlarged views of the regions framed white in panels A and B, respectively. The region framed in panel B is rotated 90° in panel D. The dense chromatin regions reveal a cholesteric, liquid crystal-like morphology. The double-membrane structure, separate between densely packed DNA regions, is evident (black asterisks). Scale bars, 500 nm (A and B) and 250 nm (C and D).
G. obscuriglobus presents a condensed-chromatin organization.
In many bacterial species, the chromatin usually appears in electron micrographs as an amorphous, ribosome-free space distributed irregularly throughout the cytoplasm. Such dispersed chromatin distribution reflects a relatively small extent of chromatin condensation, consistent with a state of high metabolic activity and ongoing DNA transcription (27, 46).
Using DNA-specific staining, we found that the chromatin of G. obscuriglobus is condensed in specific and distinct positions within the cell (see Fig. S1 in the supplemental material). Figures 2A and B show sections through the midplane of the bacterium. In such a view, all canonical components of the cell are detected, including the plasma membrane, an intracytoplasmic membrane, an internal double-membrane system, glycogen storage particles (51), and ribosomes, distributed in both compartments (35). Large, electron-dense, ribosome-free regions are also present, revealing a “nested-arches” structure (Fig. 2) typical of liquid crystalline DNA organization (33, 45), as was indeed confirmed by DNA-specific staining (see Fig. S1 in the supplemental material). Our results further support the notion that G. obscuriglobus exhibits multiple nucleoid domains enclosed by a double-membrane system. The heterogeneity in the multiple chromatin domains presumably stems from the specific growth state of the bacteria and the sectioning plane. These results thus encouraged us to further look for a plausible physiological function of highly condensed DNA.
G. obscuriglobus is highly resistant to UVC radiation.
Accumulating experimental evidence (34, 44, 58) implies that a condensed-chromatin organization usually corresponds to enhanced tolerance for DNA-damaging factors. To assess a possible link between chromatin condensation and UV radiation tolerance in G. obscuriglobus, we exposed the bacteria to various doses of UV light.
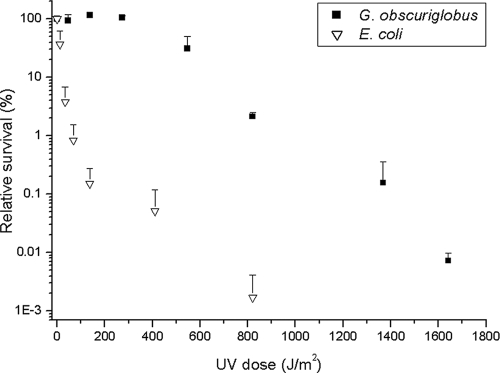
We irradiated 5-day-old cultures with elevated UVC doses, with a constant flux of 2.28 J/m2 s. Survival rates were estimated by quantifying CFU. Since G. obscuriglobus tends to develop clumps, the culture was filtered prior to exposure and plating, thereby allowing measurement of the behavior of individual cells. The relative radiation tolerance of a given microorganism is defined by the D37 and D10 values, reflecting the exposure dosage that causes the population to decrease to a level of 37% or 10% of the initial density, respectively.
Figure 3 shows semilogarithmically scaled results comparing the colony-forming abilities of actively growing G. obscuriglobus and E. coli cells. The results indicate that the UV D37 of G. obscuriglobus is 490.75 J/m2 and the D10 is 675.8 J/m2. Similar analysis of E. coli revealed a D37 value of 10.73 J/m2 and a D10 value of 24.33 J/m2, comparable to previously published data (21). During this experiment, E. coli BL21 was irradiated and used as a reference point; however, we verified that wild-type E. coli (AB1157) behaved similarly (see Fig. S2 in the supplemental material).
FIG. 3.
G. obscuriglobus tolerates high levels of UVC irradiation. Actively growing cultures of G. obscuriglobus and E. coli were irradiated with increasing doses of UV radiation using a germicidal UVC lamp. The corresponding D10 values were calculated as described previously (21) and were found to be 675.8 J/m2 and 24.33 J/m2 for G. obscuriglobus and E. coli, respectively. Statistical analysis of a covariance test revealed a significant “species by exposure time” interaction (F1,49 = 24.19; P < 0.001), indicating that the UVC radiation sensitivity of E. coli was significantly higher than that of G. obscuriglobus. Bars indicate standard errors.
G. obscuriglobus can tolerate high doses of IR.
Since the nature of DNA damage is diverse, including single-strand breaks and formation of thymidine dimers and DSBs, and since the repair pathways involved in each damage type are different, we examined the effect of typical DNA DSB lesions caused by IR on the capability of G. obscuriglobus to reproduce. Actively dividing and stationary cells, corresponding to 5- and 10-day-old cultures, respectively, were exposed to IR from a 60Co source. Table 1 shows that G. obscuriglobus is capable of reproducing following exposure to doses ranging from 500 to 1,000 Gy.
TABLE 1.
G. obscuriglobus can tolerate high doses of IRa
| Dose (Gy) | Bacterial culture | Age (days) | No. of CFU (106)
|
Relative survival (%) | |
|---|---|---|---|---|---|
| Before irradiation | After irradiation | ||||
| 500 | G. obscuriglobus | 5 | 1.83 | 1.53 | 83.6 |
| 10 | 21.1 | 20.4 | 97 | ||
| E. coli | 100 | <0.002 | <0.0002 | ||
| 1,000 | G. obscuriglobus | 5 | 7.2 | 2.9 | 40.27 |
| 10 | 7.6 | 4.67 | 61.4 | ||
| E. coli | 63.3 | <0.001 | <0.00015 | ||
Five- and 10-day-old cultures of G. obscuriglobus and logarithmic-phase E. coli cultures (optical density, 0.4 to 0.6) were harvested and irradiated with 500- and 1,000-Gy doses emitted from a radioactive 60Co source. The CFU were examined, and relative survival values were calculated by dividing the irradiated culture CFU by the untreated control culture CFU.
Variations in survival rates were observed between the 5- and 10-day-old G. obscuriglobus cultures, consistent with radiation tolerances found in other radio-resistant bacteria (29). At a dose of 500 Gy, the relative survival rate for the 5-day-old culture was 83.6%, while in the 10-day-old culture, the survival rate was 97%. The relative survival rates of 1,000-Gy-irradiated cultures were 40.3% for the 5-day-old culture and 61.4% for the 10-day-old culture. Cells from a stationary-phase culture were more resistant to radiation, in agreement with previous studies of other systems, such as D. radiodurans (49). The measured relative survival rates show that G. obscuriglobus viability deteriorated by less than an order of magnitude in response to such IR doses. Thus, it can be postulated that the IR D37 value for actively growing cultures of G. obscuriglobus is slightly higher than 1,000 Gy. This estimated D37 value is 15 times higher than the value for wild-type strains of E. coli (38).
Genomic analysis reveals high homology among the ATP-dependent DNA ligases (ADDL) of G. obscuriglobus, M. tuberculosis, and B. subtilis.
G. obscuriglobus cells show extensive tolerance for UVC radiation and IR. Recently, it was suggested that the maintenance of a tightly packed genome in the bacterium D. radiodurans and other members of the same genus is responsible for the enhanced repair capabilities of these species (58). A tight alignment of DNA strands would restrict damaged DNA fragments from diffusing following exposure of bacteria to IR, UVC, or DSB-inducing chemicals (34). In such a scenario, error-free joining of DNA ends generated at a break site through an NHEJ-like repair mechanism would be enhanced. Since the repair of double-stranded DNA breaks may occur through either homologous recombination or NHEJ, we explored the G. obscuriglobus genome for genes participating in either repair system.
This bioinformatics-based approach revealed that the G. obscuriglobus genome contains gene sequences homologous to those for RecA, RecB, and RecD. No homologue of the γ subunit of the RecBCD enzyme, RecC, was detected. Similar searches for genes involved in the NHEJ mechanism revealed sequences presenting domain homology to members of a family of ADDL, proteins that possess functional ligation activity in M. tuberculosis and B. subtilis (23, 54) (Fig. 4). The predicted G. obscuriglobus protein shows ∼30% amino acid identity and 44% similarity to the M. tuberculosis DNA repair ligase protein (LigMt) and is predicted to contain an ADDL domain. In contrast, Ku-like proteins, which participate in the conjugation of adjacent DNA strand edges (10), were not found.
FIG. 4.
Genomic analysis of G. obscuriglobus shows high homology to known ADDL of other microorganisms. The sequences of the ADDL protein of M. tuberculosis (LigDMt) and the putative B. subtilis ADDL protein, YkoU, were aligned with the homologous G. obscuriglobus ORF (LigGo) using the ClustalW multiple-alignment procedure. The five nucleotidyltransferase sequence elements conserved in the DNA ligases, designated motifs I, III, IIIa, IV, and V, are shown. The asterisks indicate identical or conserved residues in all sequences in the alignment, the colons indicate conserved substitutions, and the periods indicate semiconserved substitutions.
The predicted G. obscuriglobus DNA repair ligase protein has an apparent molecular mass of 58.4 kDa, whereas the M. tuberculosis LigDMt homologue protein has a mass of 83.6 kDa. This mass difference is significant, as it enhances the accessibility of the smaller ADDL to the DNA breaks within the condensed DNA environment.
DISCUSSION
Compartmentalization within prokaryotic cells is not a common phenomenon. Nevertheless, G. obscuriglobus has evolved an impressive 3D network comprising a double-membrane system. Despite this double-membrane network surrounding the genomic material, ribosome-like particles have been detected in both the nuclear body and riboplasm compartments (35). In agreement with our data and the nuclear nature of eukaryotic cells, the double-membrane network of G. obscuriglobus cells emanates from the intracytoplasmic membrane to form unsealed compartments. At rather large defocus values, the application of cryoelectron tomography to freeze-hydrated sections emphasizes the membrane structure and therefore allows easy characterization of the double-membrane system in a close to life state (Fig. 1). Complementary approaches, such as freeze-substitution (14), enable detailed insight into the G. obscuriglobus cellular organization (Fig. 2). Unfortunately, membranes in bacteria stain poorly, making it a challenging task to draw accurate conclusions about their organization and the nature of their cellular compartmentalization.
The diverse habitats of G. obscuriglobus may have induced the bacterium to evolve a versatile mechanism allowing it to cope with different physiological, environmental, and stress conditions. In this study, we have evaluated the capability of the organism to survive under stress conditions by studying resistance to UVC and IR. Our irradiation experiments revealed the impressive capability of the bacteria to cope with potent sources of DNA damage. Specifically, we showed that G. obscuriglobus may tolerate large doses of UVC radiation, similar to the doses endured by members of the genus Deinococcus. Though the functional connection between enhanced tolerance for UV-induced damage and condensed DNA structures has yet to be demonstrated, several studies (34, 38-40, 58) imply such a connection.
A link between condensed chromatin and radiation resistance was demonstrated for IR within bacteria (18, 58). In keeping with this correlation, G. obscuriglobus cells are reported here to tolerate impressive dosages of IR and to exhibit a condensed-chromatin organization (Table 1 and Fig. 2), resembling other radio-resistant bacteria, such as members of the genus Deinococcus.
Genomic analysis has indicated that G. obscuriglobus does not contain a RecC homologue. The absence of this subunit is intriguing, since RecBC plays an essential role in homologous recombination. Surprisingly, D. radiodurans has also been shown to lack the RecBC complex (36). These findings emphasize the ability of some bacteria to tolerate high radiation levels even in the absence of key components of the homologous-recombination machinery. Moreover, the alternative yet common mechanism for homologous recombination involving products of the addAB gene cassette (5) is also absent in G. obscuriglobus. Though homologous recombination is considered to be the main mechanism by which bacteria deal with DSBs, it is not clear how G. obscuriglobus could conceivably pursue homologous DNA repair without the complete set of required genes. Moreover, our data, like those of previous studies (20), emphasize the internal membrane network that encloses the DNA and provides a physical barrier, a spatial arrangement that would limit the efficiency of homologous-recombination-based DNA repair to a single compartment. Therefore, it is plausible that an alternative pathway has evolved in G. obscuriglobus to cope with DNA damage.
The highly radio-resistant bacterium D. radiodurans has four internal compartments, each of which contains an individual copy of the genome during stationary phase, thus reducing the probability of these redundant genomes interacting and utilizing a homologous-recombination repair process. It was suggested, therefore, that these bacteria utilize NHEJ as a central mechanism to resist high doses of radiation, in agreement with the condensed nature of the D. radiodurans chromosome (11, 34). Consequently, the comparable membrane barriers within G. obscuriglobus may reduce the feasibility of homologous recombination with the heavily condensed chromatin arrangement, making the NHEJ pathway the mechanism of choice for repairing DNA lesions. Indeed, we have shown the putative existence of an ADDL protein, a key player in direct DSB repair, involved in the DNA repair mechanism of several bacteria exhibiting condensed chromatin (3, 23, 54). The molecular masses of ADDL show wide variability. While the G. obscuriglobus ADDL has an apparent molecular mass of 58.2 kDa, the M. tuberculosis homologue is an 83-kDa protein. The reduced mass of the G. obscuriglobus DNA ligase may be an advantage in the vicinity of condensed chromatin, where the proteins involved in DNA repair are required to access buried damaged DNA strands. In light of the fact that Ku components are absent in D. radiodurans, their absence in G. obscuriglobus is intriguing. It has been suggested that tight DNA packaging, which results in keeping DNA ends in close proximity, renders the activities of Ku-like proteins redundant (12, 38).
In summary, we have shown here the application of several electron microscopy techniques to study the bacterium G. obscuriglobus. In doing so, we revealed the existence of a complex double-membrane system emanating from the internal cell membrane of the bacterium. Although we have acquired novel insight into the intracellular organization of these bacteria, the function of the double-membrane system and the manner by which G. obscuriglobus organizes its genomic material remain unclear.
Supplementary Material
Acknowledgments
We thank Yona Lichtenfeld for her kind contribution to the procedure of plastic-embedded sectioning and grid staining. We thank Eyal Shimoni from the Weizmann Institute for his helpful assistance in the high-pressure freezing and cryosubstitution procedures.
This work was supported by the Israel Science Foundation (grant 794/06 to O.M.).
Footnotes
Published ahead of print on 12 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Al-Amoudi, A., L. P. Norlen, and J. Dubochet. 2004. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 148131-135. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 111365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasius, M., R. Buob, I. V. Shevelev, and U. Hubscher. 2007. Enzymes involved in DNA ligation and end-healing in the radioresistant bacterium Deinococcus radiodurans. BMC Mol. Biol. 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochier, C., and H. Philippe. 2002. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature 417244. [DOI] [PubMed] [Google Scholar]
- 5.Chedin, F., P. Noirot, V. Biaudet, and S. D. Ehrlich. 1998. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol. Microbiol. 291369-1377. [DOI] [PubMed] [Google Scholar]
- 6.Courcelle, J., and P. C. Hanawalt. 2003. RecA-dependent recovery of arrested DNA replication forks. Annu. Rev. Genet. 37611-646. [DOI] [PubMed] [Google Scholar]
- 7.Cox, M. M., and J. R. Battista. 2005. Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol. 3882-892. [DOI] [PubMed] [Google Scholar]
- 8.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 40437-41. [DOI] [PubMed] [Google Scholar]
- 9.Della, M., P. L. Palmbos, H. M. Tseng, L. M. Tonkin, J. M. Daley, L. M. Topper, R. S. Pitcher, A. E. Tomkinson, T. E. Wilson, and A. J. Doherty. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306683-685. [DOI] [PubMed] [Google Scholar]
- 10.Doherty, A. J., S. P. Jackson, and G. R. Weller. 2001. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 500186-188. [DOI] [PubMed] [Google Scholar]
- 11.Eltsov, M., and J. Dubochet. 2005. Fine structure of the Deinococcus radiodurans nucleoid revealed by cryoelectron microscopy of vitreous sections. J. Bacteriol. 1878047-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Englander, J., E. Klein, V. Brumfeld, A. K. Sharma, A. J. Doherty, and A. Minsky. 2004. DNA toroids: framework for DNA repair in Deinococcus radiodurans and in germinating bacterial spores. J. Bacteriol. 1865973-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Errington, J. 2003. Dynamic proteins and a cytoskeleton in bacteria. Nat. Cell Biol. 5175-178. [DOI] [PubMed] [Google Scholar]
- 14.Feder, N., and R. L. Sidman. 1958. Methods and principles of fixation by freeze-substitution. J. Biophys. Biochem. Cytol. 4593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frangakis, A. S., and R. Hegerl. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135239-250. [DOI] [PubMed] [Google Scholar]
- 16.Franzmann, P. D., and V. B. Skerman. 1984. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie van Leeuwenhoek 50261-268. [DOI] [PubMed] [Google Scholar]
- 17.Frenkiel-Krispin, D., S. Levin-Zaidman, E. Shimoni, S. G. Wolf, E. J. Wachtel, T. Arad, S. E. Finkel, R. Kolter, and A. Minsky. 2001. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 201184-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frenkiel-Krispin, D., and A. Minsky. 2006. Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J. Struct. Biol. 156311-319. [DOI] [PubMed] [Google Scholar]
- 19.Frenkiel-Krispin, D., R. Sack, J. Englander, E. Shimoni, M. Eisenstein, E. Bullitt, R. Horowitz-Scherer, C. S. Hayes, P. Setlow, A. Minsky, and S. G. Wolf. 2004. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J. Bacteriol. 1863525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuerst, J. A., and R. I. Webb. 1991. Membrane-bounded nucleoid in the eubacterium Gemmatata obscuriglobus. Proc. Natl. Acad. Sci. USA 888184-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gascon, J., A. Oubina, A. Perez-Lezaun, and J. Urmeneta. 1995. Sensitivity of selected bacterial species to UV radiation. Curr. Microbiol. 30177-182. [DOI] [PubMed] [Google Scholar]
- 22.Gitai, Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120577-586. [DOI] [PubMed] [Google Scholar]
- 23.Gong, C., A. Martins, P. Bongiorno, M. Glickman, and S. Shuman. 2004. Biochemical and genetic analysis of the four DNA ligases of Mycobacteria. J. Biol. Chem. 27920594-20606. [DOI] [PubMed] [Google Scholar]
- 24.Grimm, R., H. Singh, R. Rachel, D. Typke, W. Zillig, and W. Baumeister. 1998. Electron tomography of ice-embedded prokaryotic cells. Biophys. J. 741031-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruska, M., O. Medalia, W. Baumeister, and A. Leis. 2007. Electron tomography of vitreous sections from cultured mammalian cells. J. Struct. Biol. 161384-392. [DOI] [PubMed] [Google Scholar]
- 26.Hegerl, R. 1996. The EM program package: a platform for image processing in biological electron microscopy. J. Struct. Biol. 11630-34. [DOI] [PubMed] [Google Scholar]
- 27.Hobot, J. A., W. Villiger, J. Escaig, M. Maeder, A. Ryter, and E. Kellenberger. 1985. Shape and fine structure of nucleoids observed on sections of ultrarapidly frozen and cryosubstituted bacteria. J. Bacteriol. 162960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hohenberg, H., K. Mannweiler, and M. Muller. 1994. High-pressure freezing of cell suspensions in cellulose capillary tubes. J. Microsc. 17534-43. [DOI] [PubMed] [Google Scholar]
- 29.Keller, L. C., and R. B. Maxcy. 1984. Effect of physiological age on radiation resistance of some bacteria that are highly radiation resistant. Appl. Environ. Microbiol. 47915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalczykowski, S. C. 1991. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 20539-575. [DOI] [PubMed] [Google Scholar]
- 31.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leforestier, A., and F. Livolant. 1993. Supramolecular ordering of DNA in the cholesteric liquid crystalline phase: an ultrastructural study. Biophys. J. 6556-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299254-256. [DOI] [PubMed] [Google Scholar]
- 35.Lindsay, M. R., R. I. Webb, M. Strous, M. S. Jetten, M. K. Butler, R. J. Forde, and J. A. Fuerst. 2001. Cell compartmentalisation in Planctomycetes: novel types of structural organisation for the bacterial cell. Arch. Microbiol. 175413-429. [DOI] [PubMed] [Google Scholar]
- 36.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 6544-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minsky, A. 2003. Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50367-376. [DOI] [PubMed] [Google Scholar]
- 39.Minsky, A., E. Shimoni, and D. Frenkiel-Krispin. 2002. Stress, order and survival. Nat. Rev. Mol. Cell Biol. 350-60. [DOI] [PubMed] [Google Scholar]
- 40.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 1864192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olins, A. L., B. A. Moyer, S. H. Kim, and D. P. Allison. 1989. Synthesis of a more stable osmium ammine electron-dense DNA stain. J. Histochem. Cytochem. 37395-398. [DOI] [PubMed] [Google Scholar]
- 42.Pennisi, E. 2004. Evolutionary biology: the birth of the nucleus. Science 305766-768. [DOI] [PubMed] [Google Scholar]
- 43.Pitcher, R. S., T. E. Wilson, and A. J. Doherty. 2005. New insights into NHEJ repair processes in prokaryotes. Cell Cycle 4675-678. [DOI] [PubMed] [Google Scholar]
- 44.Popham, D. L., S. Sengupta, and P. Setlow. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 613633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich, Z., S. Levin-Zaidman, S. B. Gutman, T. Arad, and A. Minsky. 1994. Supercoiling-regulated liquid-crystalline packaging of topologically-constrained, nucleosome-free DNA molecules. Biochemistry 3314177-14184. [DOI] [PubMed] [Google Scholar]
- 46.Robinow, C., and E. Kellenberger. 1994. The bacterial nucleoid revisited. Microbiol. Rev. 58211-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roca, A. I., and M. M. Cox. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 56129-223. [DOI] [PubMed] [Google Scholar]
- 48.Schlesner, H. 1994. The development of media suitable for the microorganisms morphologically resembling Planctomyces spp., Pirellula spp., and other Planctomycetales from various aquatic habitats using dilute media. Syst. Appl. Microbiol. 17135-145. [Google Scholar]
- 49.Serianni, R. W., and A. K. Bruce. 1968. Radioresistance of Micrococcus radiodurans during the growth cycle. Radiat. Res. 36193-207. [PubMed] [Google Scholar]
- 50.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 4929-54. [DOI] [PubMed] [Google Scholar]
- 50a.Studer, D., W. Graber, A. Al-Amoudi, and P. Eggli. 2001. A new approach for cryofixation by high-pressure freezing. J. Microsc. 203285-294. [DOI] [PubMed] [Google Scholar]
- 51.van Niftrik, L., W. J. Geerts, E. G. van Donselaar, B. M. Humbel, R. I. Webb, J. A. Fuerst, A. J. Verkleij, M. S. Jetten, and M. Strous. 2008. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190708-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weller, G. R., V. L. Brandt, and D. B. Roth. 2004. Doing more with less in bacterial DNA repair. Nat. Struct. Mol. Biol. 111158-1159. [DOI] [PubMed] [Google Scholar]
- 53.Weller, G. R., and A. J. Doherty. 2001. A family of DNA repair ligases in bacteria? FEBS Lett. 505340-342. [DOI] [PubMed] [Google Scholar]
- 54.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. d'Adda di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 2971686-1689. [DOI] [PubMed] [Google Scholar]
- 55.Woebken, D., H. Teeling, P. Wecker, A. Dumitriu, I. Kostadinov, E. F. Delong, R. Amann, and F. O. Glockner. 2007. Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. Isme J. 1419-435. [DOI] [PubMed] [Google Scholar]
- 56.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, H., and S. Shuman. 2007. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 353631-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmerman, J. M., and J. R. Battista. 2005. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.