Abstract
MbeC is a 13-kDa ColE1-encoded protein required for efficient mobilization of ColE1, a plasmid widely used in cloning vector technology. MbeC protein was purified and used for in vitro DNA binding, which showed that it binds specifically double-stranded DNA (dsDNA) containing the ColE1 oriT. Amino acid sequence comparison and secondary structure prediction imply that MbeC is related to the ribbon-helix-helix (RHH) protein family. Alignment with RHH members pointed to a conserved arginine (R13 in MbeC) that was mutated to alanine. The mutant MbeC(R13A) was unable to bind either single-stranded DNA or dsDNA. Limited proteolysis fragmented MbeC in two stable folding domains: the N-terminal domain, which contains the RHH motif, and the C-terminal domain, which comprises a signature shared by nicking accessory proteins. The results indicate that MbeC plays a similar role in conjugation as TraY and TrwA of plasmids F and R388, respectively. Thus, it appears that an extended, possibly universal mechanism of DNA conjugative processing exists, in which oriT-processing is carried out by relaxases assisted by homologous nicking accessory proteins. This mechanism seems to be shared by all major conjugative systems analyzed thus far.
Conjugation is one of the mechanisms through which horizontal gene transfer occurs in bacteria. The appearance of multiresistant bacteria brought horizontal gene transfer to the attention of scientists worldwide, since the dissemination of antibiotic resistance genes by conjugative plasmids poses serious health problems. ColE1 is mobilized by a wide range of conjugative plasmids and is the prototype of a family that comprises mobilizable plasmids found in gram-negative and gram-positive bacteria (14). Its mobilization region contains five genes (mbeA, mbeB, mbeC, mbeD, and mbeE), with two of them (mbeB and mbeD) entirely overlapping mbeA (Fig. 1A). Although the first report on ColE1 mobilization dates back almost 40 years (10), little is known regarding its mobilization properties. Other than the fact that mbeA encodes the ColE1 relaxase (38), only few experimental data are available about the role of the other four mob gene products. It was proposed that MbeB and MbeC are constituents of the ColE1 relaxosome (23); mbeD codes for ColE1 “entry exclusion” protein (40), whereas mbeE is not essential for plasmid mobilization (4). Although mbeA and mbeC genes are present in all ColE1 family plasmids, mbeB, mbeD, and mbeE are not conserved. This observation led to the suggestion of a common ancestral mobilization region containing only mbeA and mbeC (14). Along this line of reasoning, mbeC should play a pivotal role in ColE1 conjugative mobilization.
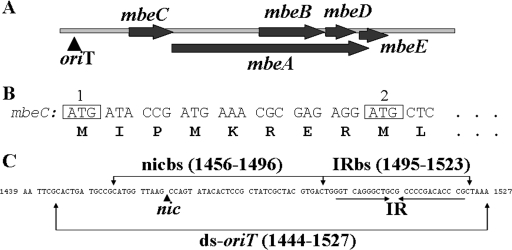
FIG. 1.
(A) mobilization region of ColE1 plasmid. (B) Representation of the two AUG codons of mbeC. (C) oriT region of ColE1 plasmid. The characteristic IR is illustrated by arrows below the nucleotide sequence. The position complementary to the nic cleavage site is indicated with a vertical arrow, since the sequence shown is the strand complementary to the one that is nicked. The different fragments used in the EMSA (Fig. 3) are also shown.
Conjugative DNA processing involves two main groups of proteins: relaxases and nicking accessory proteins. Although the need for nicking accessory proteins has been described in most conjugative and mobilizable plasmids, no sequence relatedness was appreciated for these genes as a class, despite their functional similarity. The best-characterized accessory proteins are the MOBF proteins TraY of plasmid F (TraY_F) and TrwA of plasmid R388 (TrwA_R388). TraY_F enhances the activity of TraI_F relaxase (19, 29). It binds double-stranded DNA (dsDNA) in a way that bends F plasmid oriT, possibly creating a single-stranded DNA (ssDNA) conformation suitable for relaxase binding (24, 39). TraY_F is monomeric in solution (28, 33) and contains two tandem nonidentical ribbon-helix-helix (RHH) motifs, relating it to the RHH proteins Arc and Mnt (3). Similarly, TrwA_R388 enhances the TrwC_R388 relaxase function, as well as participating in the control of the trwABC operon transcription (26), and also belongs to the RHH family (27). Besides MOBF accessory proteins, the MOBP proteins NikA_R64, TraJ_RP4, and MobC_pC221 and the MOBQ protein MobC_RSF1010 have been characterized biochemically. NikA_R64 binds the inverted repeat (IR) proximal to its nic site and induces DNA bending (15). Likewise, TraJ_RP4 binds within RP4 oriT (43) and enhances nic cleavage performed by TraI_RP4 relaxase (30). MobC_pC221 is essential for relaxosome formation and plasmid mobilization (35) and enhances the MobA_pC221 relaxase activity by binding to two sites close to nic (7). The MOBQ protein MobC_RSF1010 assists in oriT strand opening, extending the separation on the DNA strands around nic and thereby increasing the efficiency of cleavage by the relaxase (42).
We demonstrate here that MbeC_ColE1 is a MOBHEN accessory protein, possibly belonging to the RHH family of DNA-binding proteins. Sequence comparisons revealed that MOBF and MOBP accessory proteins share with MOBHEN proteins the RHH DNA-binding domain. Thus, our analysis of MbeC_ColE1 provides a link between the different families of accessory proteins and suggests the existence of a potentially universal mechanism for accessory proteins in conjugative DNA processing.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used were Escherichia coli DH5α [F− endA1 hsdR17 supE44 thi-1 recA1 Δ(argF-lacZYA) u160 φ80dlacZΔM15 gyrA96] (17) for standard plasmid maintenance, BL21(DE3) (F− ompT rB− mB−) (36) for protein overproduction, and HMS174 (recA1 hsdR Rifr) (6) as the recipient in conjugal mobilization experiments. For the maintenance and/or selection of plasmids, growth media were supplemented with antibiotics at the following concentrations: ampicillin (Am), 100 μg/ml; kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 25 μg/ml; tetracycline (Tc), 5 μg/ml; and rifampin (Rif), 100 μg/ml.
Plasmid constructions and genetic techniques.
Plasmids used in the present study are listed in Table 1. All PCR-generated fragments of ColE1 were synthesized by using plasmid pSU4601 DNA as a template. The identity of constructed plasmids was checked by DNA sequencing. Bacterial transformation of BL21(DE3) was carried as previously described (9). DH5α competent cells were prepared (18) and transformed by electroporation (13). Mating experiments were performed as formerly reported (38).
TABLE 1.
Plasmids
| Plasmid | Antibiotic selection | Descriptiona | Size (bp) | Source, reference, or construction strategy |
|---|---|---|---|---|
| pCR2.1 | Km, Am | Cloning vector | 3,900 | Invitrogen |
| pCR-blunt | Km | Cloning vector | 3,500 | Invitrogen |
| pET29c(+) | Km | Expression vector | 5,372 | Novagen |
| pUC19 | Am | Cloning vector | 2,686 | Bioline |
| pSU18 | Cm | Cloning vector | 2,300 | 2 |
| pSU4601 | Km | ColE1::kan | 7,930 | 5 |
| R64drd-11 | Tc, Sm | R64 derepressed for transfer | 56,700 | 20 |
| pUIV201 | Am | pUC19::oriT(ColE1) | 2,886 | 38 |
| pUIV206 | Km | pET29c::mbeA | 6,974 | 38 |
| pUIV230 | Cm, Mob+ | pSU18::mob(ColE1) | 4,700 | 38 |
| pUIV235 | Km, Am | pCR2.1::mbeC* | 4,239 | This study; the 339-bp PCR fragment using the primers mobC-plus and mobC-minus cloned at pCR2.1 |
| pUIV236 | Km | pET29c::mbeC* | 5,591 | This study; the 317-bp NdeI-BamHI fragment from pUIV235 cloned at the corresponding sites of pET29c |
| pUIV238 | Km, Am | pCR2.1::mbeC† | 4,268 | This study; the 368-bp PCR fragment using the primers mobC-P1-plus and mobC-minus cloned at pCR2.1 |
| pUIV239 | Km | pET29c::mbeC† | 5,615 | This study; the 341-bp NdeI-BamHI fragment from pUIV238 cloned at the corresponding sites of pET29c |
| pUIV241 | Km, Am | pCR2.1::89-bp oriT(ColE1) | 4,016 | This study; the 116-bp PCR fragment using the primers plus-oriT and minus-oriT cloned at pCR2.1 |
| pUIV244 | Km | pCR-blunt::(1389-1855)ColE1 | 3,967 | This study; the 467-bp PCR fragment using the primers plus-ΔmbeC and minus-ΔmbeC cloned at pCR-blunt |
| pUIV245 | Cm | pSU18::89-bp oriT(ColE1) | 2,338 | This study; the 89-bp EcoRI-HindIII fragment from pUIV241 cloned at the corresponding sites of pSU18 |
| pUIV247 | Km | pET29c::mob(ColE1)ΔmbeC | 7,267 | This study; the 446-bp NdeI-XbaI fragment from pUIV244 cloned at the corresponding sites of pUIV206 |
| pUIV248 | Cm | pSU18::mob(ColE1)ΔmbeC | 4,310 | This study; the 2,037-bp XbaI-EcoRI fragment from pUIV247 cloned at the corresponding sites of pSU18 |
| pUIV261 | Km | pCR-blunt::mbeC(R13A) | 3,866 | This study; the 366-bp PCR fragment using the primers mbeC-mutR and mobC-minus cloned at pCR-blunt |
| pUIV262 | Km | pET29c::mbeC(R13A) | 5,615 | This study; the 341-bp NdeI-BamHI fragment from pUIV261 cloned at the corresponding sites of pET29c |
| pUIV264 | Km | pCR-blunt::mob(ColE1)ΔoriT | 5,441 | This study; the 1,941-bp PCR fragment using the primers mbeC-P1-plus and minus-ab cloned at pCR-blunt |
| pUIV265 | Cm | pSU18::mob(ColE1)ΔoriT | 4,201 | This study; the 1,941-bp BamHI-EcoRI fragment from pUIV264 cloned at the corresponding sites of pSU18 |
| pUIV266 | Km | pET29c::mob(ColE1)ΔoriT | 7,215 | This study; the 1,941-bp NdeI-BamHI fragment from pUIV264 cloned at the corresponding sites of pET29c |
| pUIV267 | Km | pCR-blunt::oriT(ColE1)ΔIR | 3,549 | This study; hybridizing the ΔIR-plus and ΔIR-minus primers and cloning the dsDNA in pCR-blunt |
*, mbeC cloned from the second ATG (coordinates 1867 to 1869); †, mbeC cloned from the first ATG (coordinates 1843 to 1845).
Oligonucleotides and labeling.
Oligonucleotides used in the present study are listed in Table 2. They were labeled at 5′ end by using [γ-32P]ATP (300 Ci/mmol) and polynucleotide kinase (32). Unbound [γ-32P]ATP was eliminated from the mixture by using MicroSpin G-25 (GE Healthcare) column purification.
TABLE 2.
Oligonucleotides
| Oligonucleotidea | Oligonucleotide sequence (5′→3′)b | Locationc |
|---|---|---|
| mobC-plus | CGC GAG CAT ATG CTC ACA ATA CGG | 1858-1881 |
| mobC-minus | TGA AAT TTA ACT AGG ATC CCG CGC C | 2196-2172 |
| mbeC-P1-plus | CTG TTC ATG GGC ATA TGA TAC CGA TGA | 1829-1855 |
| plus-oriT1 | TAC TTT TCA TAG AAT TCG CAC TGA TGC | 1427-1453 |
| minus-oriT1 | GGC GCG TCA GAA GCT TTT AGC | 1542-1522 |
| plus-ΔmbeC | CTT ACG CAT CTA GAC GGC ATT TCA CAC | 1389-1412 |
| minus-ΔmbeC | TCA TCG GTA TCA TAT GCC CAT GAA CAG | 1855-1829 |
| mbeC-mutR | GTT CAT GGG CAT ATG ATA CCG ATG AAA CGC GAG AGG ATG CTC ACA ATA GCG GTT ACT G | 1831-1888 |
| oligo-plus* | GCA CTG ATG CCG CAT GGT TAA GCC AGT ATA CAC TCC GCT ATC GCT ACG TGA CTG GGT CAG GGC TGC GCC CCG ACA CCC GCT AAA | 1444-1527 |
| oligo-minus* | TTT AGC GGG TGT CGG GGC GCA GCC CTG ACC CAG TCA CGT AGC GAT AGC GGA GTG TAT ACT GG/C TTA ACC ATG CGG CAT CAG TGC | 1527-1444 |
| minus-ab | CCA CTG GAT CCA CTG AAG CTG C | 3769-3748 |
| ΔIR-plus* | GAA TTC TTA AGC CAG TAT ACA CTC CGC TAT CGC TAC GTG ACT AAG CTT | 1454-1502 |
| ΔIR-minus* | AAG CTT AGT CAC GTA GCG ATA GCG GAG TGT ATA CTG G/CT TAA GAA TTC | 1502-1454 |
| nic-plus | CAT GGT TAA GCC AGT ATA CAC TCC GCT ATC GCT ACG TGA CT | 1456-1496 |
| nic-minus | AGT CAC GTA GCG ATA GCG GAG TGT ATA CTG G/CT TAA CCA TG | 1496-1456 |
| IR-plus | CTG GGT CAG GGC TGC GCC CCG ACA CCC GC | 1495-1523 |
| IR-minus | GCG GGT GTC GGG GCG CAG CCC TGA CCC AG | 1523-1495 |
*, The slash shows the position of MbeA-dependent cleavage.
Oligonucleotides used for site-directed mutagenesis and mutant codons are in boldface. Nucleotides introducing a restriction site are underlined.
The numbers refer to coordinates in the ColE1 DNA sequence (GenBank accession no. NC_001371). Base position 1 corresponds to the first T in the single EcoRI site.
Protein sequence analysis.
Protein sequences were aligned by using CLUSTAL W (22). Phylogenetic and molecular evolutionary analyses were conducted by using MEGA version 3.1 (21). The tree was inferred by using the neighbor-joining method and tested by bootstrap (1,000 replicates). The cutoff bootstrap value for condensed tree was set in 50%. Protein secondary structures were predicted by using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) (25), Jpred3 (http://www.compbio.dundee.ac.uk/jpred) (11), and GOR4 (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) (12).
Purification of MbeC-His6 and derivative mutant.
BL21(DE3) cells containing the appropriate plasmid (pUIV239 or pUIV262) were incubated in 2 liters of LB broth containing kanamycin using a micro-DCU fermentation system (B. Biotech International). Cultures were induced at an A600 of 0.6 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 4 h of incubation, cells were pelleted, resuspended in buffer A (100 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 5 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride), and lysed by sonication. No lysozyme was used, since it has a molecular weight similar to that of the MbeC-His6 and MbeC(R13A)-His6 monomer when visualized after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Cellular debris was eliminated after centrifugation at 45,000 × g for 30 min at 4°C. Supernatant was collected, adjusted to buffer B (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1 mM EDTA, 20 mM imidazole), and applied to a HisTrap SP HP column (GE Healthcare). Proteins were eluted in a 20 to 500 mM imidazole gradient of buffer B. Protein samples were collected and dialyzed overnight against buffer C (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 200 mM NaCl) and further purified by HiTrap SP HP column (GE Healthcare) using a 0.2 to 1 M NaCl gradient of buffer C. Finally, proteins were purified by gel filtration through a Superdex 75 HR 10/30 (Pharmacia) using buffer D (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA). The yield was 10 mg of protein per liter of culture for both MbeC-His6 and MbeC(R13A)-His6 purified proteins.
Limited trypsin digestion.
MbeC-His6 protein at a concentration of 0.25 mg/ml (9 μM) in buffer D (18 μM) were incubated with different amounts of trypsin for 15 min at room temperature. The reaction was stopped by adding 200 μM phenylmethylsulfonyl fluoride, and samples were stored at −80°C. Cleavage was verified by SDS-PAGE and Coomassie brilliant blue staining. Molecular masses of intact and protease-digested MbeC-His6 were determined by mass spectrometry. Liquid chromatography-mass spectrometry data were acquired by using a Q-Tof micro-mass spectrometer (Waters) interfaced with a CapLC capillary chromatograph (Waters). Then, 5 μl of each sample was loaded onto a Symmetry 300 C18 NanoEase Trap precolumn (Waters) and washed with 0.1% formic acid for 5 min at a flow rate of 20 μl/min. The precolumn was connected to an Atlantis dC18 NanoEase column (75 μm by 150 mm; Waters) equilibrated in 5% acetonitrile and 0.1% FA. A flow splitter was used to decrease the flow rate to 0.2 μl/min, and peptides were eluted with a 30-min linear gradient of 10 to 60% acetonitrile directly onto a NanoEase Emitter (Waters). Obtained spectra were manually analyzed by using MassLynx 4.1 software (Waters).
DNA electrophoresis mobility shift assay (EMSA).
Increasing amounts of MbeC-His6 were incubated with 10 nM radiolabeled ssDNA or dsDNA in buffer E (20 mM Tris-HCl [pH 7.5], 50 mM NaCl) and in the presence of 1 nM bovine serum albumin (BSA). Reactions (20 μl) were carried out for 30 min at 37°C. Samples were loaded onto a 5% native polyacrylamide Bio-Rad minigel and run for 60 min at 100 V in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.2]). Gels were vacuum dried for 2 h at 80°C and quantified with a Molecular Imager FX System (Bio-Rad). The radiolabeled dsDNA was formed by mixing equal concentration of 5′-end 32P-labeled oligonucleotides with their unlabeled complementary and incubated at 95°C for 15 min. The incubator was then turned off, and the mixture was left in the incubator and allowed to cool slowly to room temperature. The dsDNAs used were ds-oriT (84 bp, formed by using oligo-plus and oligo-minus), nickbs (41 bp, formed by using nic-plus and nic-minus), and IRbs (29 bp, formed by using IR-plus and IR-minus) (Fig. 1C). The ssDNAs used were the oligonucleotides oligo-plus and oligo-minus (Table 2). For the competition experiments, MbeC-DNA complexes were formed as described above. After the formation of the complexes, either 38 nM (1 μg) unlabeled salmon sperm DNA (nonspecific competitor) or various amounts of unlabeled ds-oriT (specific competitor) was added, and the samples were incubated for 30 more min at 37°C. Samples were subjected to electrophoresis and analyzed as described above.
RESULTS
Purification of MbeC protein.
For the mbeC gene, two putative AUG start codons were proposed (Fig. 1B) (8). In order to determine the actual start codon, we constructed two clones: pUIV239 (mbeC cloned in pET29c using the first AUG codon) and pUIV236 (mbeC cloned in the pET29c using the second AUG codon) (Fig. 1B). Both plasmids were tested for their ability to complement pUIV248, a ΔmbeC ColE1-derived plasmid. BL21(DE3) cells carrying (pUIV248 plus pUIV239) or (pUIV248 plus pUIV236) were used as donors, DH5α/R64drd-11 was used as a helper, and HMS174 was used as the recipient. Mating experiments revealed that pUIV239 complemented pUIV248 10,000 times better than pUIV236 (Table 3). This result indicates that the functional protein is expressed from the first AUG codon. Therefore, E. coli strain BL21(DE3)/pUIV239 was used to overproduce MbeC-His6. SDS-PAGE of crude extracts of overproducing cells showed a major band, with electrophoretic mobility corresponding to a molecular mass of ∼15 kDa (close to the 14.9 kDa predicted by sequence). MbeC-His6 was purified by a three-step procedure, using affinity (HisTrap SPHP column), ion-exchange (HiTrap SPHP column), and a final gel filtration chromatography step (Superdex 75 HR 10/30 column) (Fig. 2A). As shown by gel filtration chromatography, MbeC-His6 behaved as a dimer of ∼30 kDa in these solution conditions.
TABLE 3.
Complementation analysis of the ΔmbeC plasmid pUIV248a
| Complementing plasmid | Mobilization frequency (SD) |
|---|---|
| pET29c (negative control) | <10−7 |
| pUIV248 (ΔmbeC, negative control) | <10−7 |
| pUIV239 (pET29c::mbeC cloned from the first ATG) | 3 × 10−2 (1 × 10−2) |
| pUIV236 (pET29c::mbeC cloned from the second ATG) | 3 × 10−6 (1 × 10−6) |
| pUS4601 (ColE1::Km, positive control) | 4 × 10−2 (2 × 10−2) |
| pUIV262 (R13A mutant) | 1 × 10−5 (0.5 × 10−5) |
Derivatives of E. coli strain BL21(DE3) carrying pUIV248 and each of the plasmids shown in the first column were separately used as donors in triparental matings using DH5α/R64drd-11 as the helper and HMS174 as the final recipient. After filter mating, bacteria were plated on selective media containing Rif to counterselect donors and Cm to select for pUIV248 mobilization. Transfer frequencies are expressed as the number of transconjugants per recipient cell and are the average of at least three separate experiments.
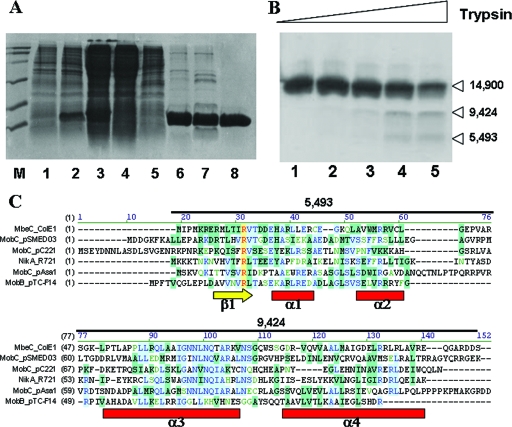
FIG. 2.
(A) Purification of MbeC-His6. Fractions from the different purification steps were analyzed by SDS-12% PAGE. Lane M, molecular weight marker; lane 1, BL21(DE3)/pUIV239 before induction; lane 2, BL21(DE3)/pUIV239 after 4 h induction; lane 3, clear lysate; lane 4, flowthrough from affinity chromatography; lane 5, wash from affinity chromatography; lane 6, 6 μg after affinity chromatography; lane 7, 5 μg of sample after ion-exchange chromatography; lane 8, 3 μg of sample after gel filtration chromatography. (B) Limited trypsin digestion of MbeC-His6. The results are visualized by SDS-15% PAGE. Lane 1, MbeC-His6 without trypsin. Lanes 2, 3, 4, and 5 show results for 0.5, 1.5, 5, and 15 μM trypsin, respectively. Molecular masses (in daltons) of the obtained fragments as determined by MALDI-TOF mass spectrometry are indicated on the right. (C) CLUSTAL W alignment of full-length ColE1 MbeC with other representative MOBHEN and MOBP accessory proteins. Secondary structure prediction (Jpred, PSIPRED, and GOR4) is shown below the alignment, and the length and mass of trypsin-produced fragments is shown above the alignment.
Determination of a minimal ColE1 oriT.
To determine the minimal length of ColE1 oriT, we constructed pUIV245 by cloning a 89-bp segment of ColE1 DNA in pSU18. The 89 bp (coordinates 1439 to 1527 of ColE1, accession no. NC_001371) contain the nic site and the adjacent IR of ColE1 (Fig. 1C). We tested mobilization of pUIV245 and compared it with the mobilization capacity of pUIV201, known to be functional from previous work (38). The results showed that pUIV245 has about the same mobilization frequency as pUIV201 (Table 4), indicating that 89-bp oriT is sufficient for ColE1 mobilization. In order to further reduce the size of the oriT, plasmid pUIV267 was constructed by cloning 41 bp of ColE1-oriT in pCR-blunt. The 41 bp (coordinates 1456 to 1496) correspond to the region including the nic site but not the IR (Fig. 1C). As above, the mobilization of the pUIV267 was tested. Interestingly, results showed that pUIV267 was only about 100 times less efficient than pUIV245, indicating that there is, to some degree, conjugal mobilization even with a 41-bp oriT containing the nic site but not the IR.
TABLE 4.
Complementation analysis of the ΔoriT plasmids pUIV265 and pUIV266a
| Complementing plasmid | Mobilization frequency (SD) |
|---|---|
| Using pUIV265 | |
| pUC19 (negative control) | <10−7 |
| pUIV201 [pUC19::150 bp (ColE1-oriT)] | 5 × 10−2 (1 × 10−2) |
| pUIV247 (pCR-blunt::ΔIR) | 5 × 10−4 (2 × 10−4) |
| pUS4601 (ColE1::Km, positive control) | 4 × 10−2 (2 × 10−2) |
| Using pUIV266 | |
| pSU18 (negative control) | <10−7 |
| pUIV245 [pSU18::89 bp (ColE1-oriT)] | 2 × 10−2 (1 × 10−2) |
| pUIV230 [pSU18::mob(ColE1), positive control] | 2 × 10−2 (1 × 10−2) |
Derivatives of E. coli strain BL21(DE3) carrying pUIV265 and each of the plasmids shown under “using pUIV265” or pUIV266 and each of the plasmids shown under “using pUIV266” were separately used as donors in triparental matings using DH5α/R64drd-11 as the helper and HMS174 as the final recipient. After filter mating, bacteria were plated on selective media containing Rif to counterselect donors and Cm to select for pUIV265 or pUIV266 mobilization. Transfer frequencies are expressed as the number of transconjugants per recipient cell and are the average of at least three separate experiments.
MbeC binds specifically ColE1 oriT but does not recognize the IR.
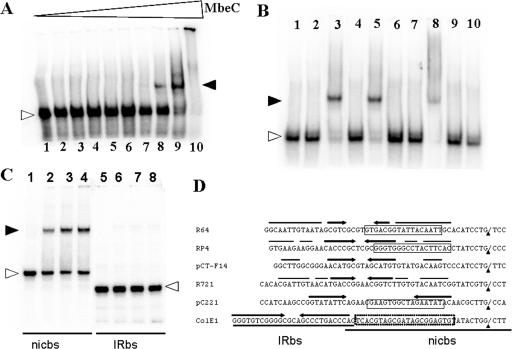
The in vitro binding of MbeC-His6 to ssDNA and dsDNA was tested. The dsDNA and ssDNA were prepared as described in Materials and Methods. Experiments revealed that MbeC-His6 binds specifically to ds-oriT (Fig. 3A). Binding specificity was tested by adding into the reactions 38 nM unlabeled salmon sperm DNA as a nonspecific competitor or various concentrations of unlabeled ds-oriT as a specific competitor (Fig. 3B). When unlabeled salmon sperm DNA was added, the MbeC-DNA complex did not dissociate, indicating that MbeC-His6 binds specifically to ColE1 oriT. This finding is strengthened by the fact that when increasing amounts of unlabeled ds-oriT were added to the binding reactions, the MbeC-DNA complex dissociated and finally disappeared (Fig. 3B). No MbeC-DNA complexes were formed with ssDNA, using either oligo-plus or oligo-minus (data not shown). Given that IR elimination only reduced mobilization 100 times (Table 4), we thought that MbeC binding site could be located within the oriT 1456-1496 segment not containing the IR (Fig. 1C). In order to confirm this possibility, EMSA experiments were performed using the nicbs (nic site fragment) and IRbs (IR-containing fragment) separately (Fig. 1C). As shown in Fig. 3C, MbeC is able to bind the nic site containing fragment but not the IR containing fragment, thus confirming our hypothesis.
FIG. 3.
(A) Gel shift of ds-oriT by MbeC-His6 in the presence of 1 nM BSA. Lane 1, no MbeC-His6 and no BSA; lane 2, no MbeC-His6. Lanes 3, 4, 5, 6, 7, 8, 9, and 10 show results for 10 nM, 30 nM, 0.1 μM, 0.2 μM, 0.4 μM, 0.6 μM, 1 μM, and 3 μM MbeC-His6, respectively. (B) Gel shift of ds-oriT by 1 μM MbeC-His6 in the presence of 1 nM BSA, after the addition of various amounts of specific and nonspecific dsDNA. Lane 1, 10 nM ds-oriT; lane 2, 10 nM ds-oriT and 1 nM BSA; lane 3, 10 nM ds-oriT and 1 μM MbeC-His6, no unlabeled salmon sperm DNA; lane 4, 10 nM ds-oriT and 38 nM unlabeled salmon sperm DNA; lane 5, 10 nM ds-oriT, 38 nM unlabeled salmon sperm DNA, 1 μM MbeC-His6. Lanes 6, 7, 8, 9, and 10 show results for 10, 50, 5, 100, and 200 nM unlabeled ds-oriT, respectively, in 10 nM ds-oriT and 1 μM MbeC-His6. (C) Gel shift of IRbs and nicbs by MbeC-His6. Lane 1, nicbs without MbeC-His6; lanes 2, 3, and 4, nicbs and 0.6, 0.8, and 1 μM MbeC-His6, respectively; lane 5, IRbs without MbeC-His6; lanes 6, 7, and 8, IRbs and 0.6, 0.8, and 1 μM MbeC-His6, respectively. In all gel shifts, free DNA bands are indicated by open arrowheads, while DNA-MbeC complexes are indicated by filled arrowheads. (D) Comparison of different MOBP oriTs around the nic site. Black arrowheads indicate the position of the nic sites. Binding sites of NikA_R64, TraJ_RP4 and MobC_pC221 are squared, and IRs are indicated by arrows. The putative MbeC binding region is indicated by a dotted-line square. The IRbs and nicbs fragments used for EMSAs in panel C are also shown.
MbeC-like proteins belong to the family of accessory proteins for DNA processing in bacterial conjugation.
A PSI-BLAST search using MbeC_ColE1 as a query, setting a score value of 0.00001, converged in the eighth iteration. A total of 103 homologues were recovered. When the most distantly related hits were used as queries in new rounds of BLAST search in order to find more remote homologues to MbeC, the relaxosomal proteins TraJ_RP4 and NikA_R64 were hit, among many of their homologues. MbeC-like proteins are generally smaller than 200 amino acids and share some overall similarity, allowing the construction of a neighbor-joining condensed tree (Fig. 4). It shows clusters of highly related homologues, while the unreliable relationship between clades is not estimated. MbeC homologues were not constrained to mobilizable plasmids but also included conjugative plasmids. Standard PSI-BLAST analysis initiated from NikA_R64 hit MbeC_ColE1 in the third round. The two sequences share 38% homology across 106 residues.
FIG. 4.
Neighbor-joining condensed tree of aligned MbeC homologues. MbeC-like proteins linked to MOBHEN or MOBP relaxases are enclosed by a rectangle. The NLNQ motif associated with each clade is indicated at the right of the figure. The group of elements lacking this motif is gray shaded. When MbeC-like protein is contained in a conjugative plasmid, the element is underlined. An asterisk is placed after plasmids having a truncated relaxase.
MbeC-homologue NikA structure shows an RHH fold.
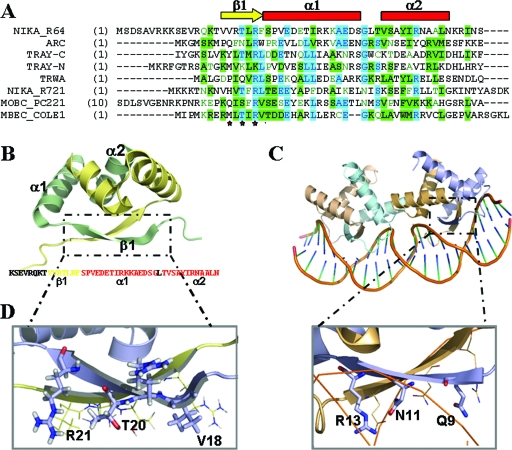
The nuclear magnetic resonance (NMR) structure of R64 plasmid NikA N-terminal fragment (NikA-N, residues 1 to 51) is deposited in the PDB databank under the accession number 2ba3. The final set contains 20 structures with two NikA-N chains. Each NikA-N chain exhibits the RHH fold, consisting of a β-strand formed by residues 17 to 23 (β1), followed by two α-helices formed by residues 24 to 38 (αA) and 42 to 52 (αB) (Fig. 5B). Residues 1 to 16 show a different orientation on each of the 20 models, and thus we consider theses residues as disorder residues in solution. A DALI structural similarity search using chain A of the NikA_R64 NMR structure (residues 15 to 51) found the Arc repressor as the first structural homologue (z-score = 5.7, root mean square deviation over 36 residues = 1.2, and 25% sequence identity). Arc is the paradigm of the RHH-containing Arc/MetJ family of transcriptional repressors. Two Arc dimers contact each side of the operator (Fig. 5C), and the DNA specific contacts are made by using polar amino acids (Gln9, Asn11, and Arg13; Fig. 5D) of their N-terminal β-sheets (31). Sequence alignments showed that NikA_R64 could be contacting DNA via the residues Val18, Thr20, and Arg22 (Fig. 5D). A polar residue could have been expected instead of Val18, but there are some RHH sequences containing a hydrophobic side chain at this position (34). Following the same line of reasoning, the MbeC_ColE1 residues involved in DNA contact could be the polar residues Thr11, Arg13, and Thr15. According to this, the mutation R13A in MbeC should impede or reduce DNA binding.
FIG. 5.
(A) CLUSTALW alignment of the 50 N-terminal amino acids of MbeC with other RHH accessory proteins (TrwA_R388, TraY_F [N-terminal and C-terminal domain], NikA_R721, NikA_R64, and MobC_pC221) and the RHH repressor Arc. The secondary structure of the solved NikA_R64 NMR structure is shown above the alignment (the β-sheet is represented by a yellow arrow, and α-helices are indicated by red squares). (B) Ribbon diagram representation of the NMR structure of NikA_R64 N-terminal domain (PDB accession no. 2ba3). One monomer is shown in blue, and the other is shown in wheat color. The sequence of the solved domain is shown below the structure with a color code secondary structure (yellow β-sheet and red α-helix forming residues). (C) Ribbon diagram representation of the crystal structure of two Arc dimers bound to its operator (PDB accession no. 1par). (D) Comparison of NikA (left) and Arc (right) β-sheet polar residues.
While this manuscript was in preparation, the NMR structure of the R64 plasmid NikA N-terminal 51-residue fragment of NikA was published (41). The characterization of the three-dimensional structure of NikA(1-51) by solution-state NMR has been described, which revealed that NikA is in fact an RHH protein. Residue R22 is oriented toward the outside of the β-sheet and is responsible for binding to the oriT, thus confirming our “in silico” analysis.
Mutation of R13 within the predicted β1 abolished MbeC DNA-binding activity.
In order to test MbeC(R13A)-His6 functionality, the ΔmbeC ColE1-derived nonmobilizable plasmid pUIV248 was used. This plasmid can be rendered mobilizable in the presence of plasmids contributing a functional MbeC protein. Mating experiments showed that complementation by MbeC(R13A)-His6 was 3,000-fold less efficient than complementation by wild-type MbeC. Therefore, the mutant MbeC(R13A) appears to be nonfunctional. For the overproduction of MbeC(R13A)-His6, BL21(DE3)/pUIV262 was used, following the same procedure as for the purification of MbeC-His6. The mutant protein behaved as a dimer in gel filtration like the wild-type MbeC. The purified MbeC(R13A)-His6 was used for in vitro DNA-binding reactions, following the same procedure as described for MbeC-His6. EMSA showed that the mutant MbeC(R13A)-His6 was unable to bind to ssDNA or dsDNA (data not shown).
Identification of two MbeC domains by limited proteolysis.
The predicted tertiary structure of MbeC was probed by limited proteolysis. We observed, after trypsin treatment (Fig. 2B), two discrete bands with lower molecular masses than the full-length MbeC. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometry gave molecular masses corresponding to two C-terminal fragments (5,493 and 9,424 Da), which together add up nicely to the determined molecular mass of the full-length protein (14,900 Da). The fragments perfectly fit the mass of the 46 N-terminal and the 51 C-terminal amino acids of the protein, respectively (Fig. 2C). The existence of this breakpoint possibly signals the interface between two domains as analyzed further in the Discussion.
DISCUSSION
In the present study we analyzed protein MbeC encoded by plasmid ColE1 in an effort to characterize its role in ColE1 mobilization. We first determined the start codon of mbeC by comparing the complementation properties of two potential starts (GenBank accession no. X15873, coordinates 1843 to 1845 and 1867 to 1869; Fig. 1B). Former work proposed the first AUG as the mbeC start codon (8), based on the criterion that a Shine-Dalgarno sequence should be found within 3 to 10 bp upstream. A different work proposed the second AUG as the start codon (4), based on a comparison of mbeC with the related genes mbaC and mbkC of plasmids ColA and ColK, respectively. According to our mobilization results, the conjugation frequency obtained from the MbeC started from the second AUG was 10,000 times lower compared to the MbeC started from the first AUG (Table 3), demonstrating that MbeC starts at the first AUG.
We also localized the ColE1 oriT to a minimal functional DNA segment. According to an earlier study (14), ColE1 oriT should not exceed 100 bp, given the size of the fragment that contains the nic site and the 32-bp upstream IR (Fig. 1C). This assumption was based on the fact that IRs are usually protein-binding sites. In the case of ColE1, the IR was not absolutely required for MbeA_ColE1 binding to DNA, since a shift occurred using an oligonucleotide not containing the IR (38). According to our retardation experiments, the IR is not required for MbeC_ColE1 binding either, since MbeC does not bind to a 29-bp dsDNA fragment containing the IR (Fig. 3C). However, MbeC_ColE1 binds to a 41-bp dsDNA fragment containing the nic site (Fig. 3C). This is in accordance with experiments that showed that ColE1 mobilization frequency only drops 100 times when a 41-bp oriT fragment is used (Table 4). When different oriTs of the MOBP family were compared, we observed that the MobC/NikA binding sites were located in the corresponding coordinates 1456 to 1496 of MbeC_ColE1 oriT (Fig. 3D). Thus, ColE1 oriT, with only 41 bp being sufficient for conjugation, is one of the shortest oriT characterized thus far. The fact that the 89-bp oriT containing the IR was mobilized 100 times better than the minimal 41-bp oriT could be due to the role of this IR in termination of DNA transfer, as proposed for plasmid R64 (16). Other systems, such as R388 and F plasmids, are strongly regulated and thus require longer oriTs (>300 bp). The presence of additional regulatory elements, such as extra IRs and protein binding sites, demonstrates the degree of complication of these systems compared to the simple ColE1-based oriT.
BLAST analysis indicated that MbeC-like proteins show two main regions of homology, which correspond to the N- and C-terminal folding domains (Fig. 2C). The most conserved is the C-terminal domain, which contains an NLNQ motif. The sequence motif [LF]xxx[GS]xNxNQxAxxxN was described for MobC proteins from gram-positive and gram-negative bacteria (1). There are several MbeC homologues that lack this motif, and they are clustered in a single monophyletic group that contains members from conjugative and mobilizable plasmids (Fig. 4). A distinctive feature of the plasmids contained in this group is the organization of their mobilization region, having their mobilization genes located at both sides of oriT. On the other hand, MbeC-like proteins containing the NLNQ motif belong to plasmids whose mobilization genes are placed on the same position with regard to oriT. The NLNQ motif is always present in MOBHEN and in most MOBP accessory proteins, and it could be involved in the interaction with the conjugative helper apparatus, probably with the coupling protein. In fact, TrwA_R388 has been shown to interact via its C-terminal domain with the coupling protein TrwB_R388 (37). This interaction stimulates TrwB ATPase activity, being the energy released from ATP hydrolysis used for DNA pumping. Thus, the interaction of the coupling protein with the C-terminal domain of the accessory protein allows the relaxosome to be recruited by the T4SS for DNA transport. We do not exclude the possibility that the NLNQ motif could interact with the MbeA relaxase or any other proteins, but in any case it seems feasible that this motif plays an important function in conjugation.
The conserved RHH N-terminal domain shows a wide distribution among conjugative accessory proteins. In most MbeC-like proteins, an RHH domain can be recognized at the N terminus by second-structure prediction (Fig. 5A). The RHH motif was not observed up to now probably because of the high conservation of the NxNQ motif in the C-terminal domain and the difficulty in the assignment of an RHH signature, due to the small size of the motif and the similarity to the HTH DNA-binding motif. In a previous study (34), the sequences of characterized and putative RHH proteins were compared and showed that they share alternating hydrophilic and hydrophobic amino acids within the N-terminal β1-strand. In addition, there is a conserved G-X-S/T/N motif in the loop between helix α1 and α2 and some conserved hydrophobic residues in α1 and α2. MbeC predicted β1 strand contains an alternation of hydrophobic (L10, I12, and V14) and hydrophilic (T11 and R13) residues. The MbeC putative loop between helix α1 and α2 contains the sequence GXQ, and there are conserved hydrophobic residues in α1 (L22) and α2 (L31 and L40) (Fig. 5A), which is compatible with the consensus RHH motif (34).
A standard PSI-BLAST analysis initiated from the RHH accessory protein NikA_R64 hit MbeC_ColE1 in the third iteration. Our in silico analysis of the accessory protein NikA and the three-dimensional structure of its N-terminal domain (which was published [41] while the present study was in preparation), as well as secondary structure prediction Jpred3, PSIPRED, and GOR4 on MbeC, gave more credence to the predicted RHH DNA-binding domain of MbeC.
The existence of the N- and C-terminal folding domains found by the BLAST analysis is in accordance with our experimental results of limited proteolysis of MbeC. After trypsin treatment and MALDI-TOF spectrometry, we observed two discrete bands of 5,493 and 9,424 Da. The existence of this breakpoint possibly signals the interface between both domains (Fig. 2C). The N-terminal domain (46 amino acids) contains the predicted RHH domain, while the C-terminal domain (51 amino acids) contains the NxNQ domain.
Comparison with other RHH proteins leads to the prediction that the mutant MbeC(R13A) should be critically affected on DNA binding (Fig. 5A). In fact, the point mutation R13A abrogated MbeC binding to DNA and reduced ColE1 mobilization by 3,000 times (Table 3). A similar effect was reported when the arginine of the DNA binding N-terminal beta sheet (R10) was mutated to alanine for the RHH accessory protein TrwA_R388 (27). This result again confirms the RHH prediction for MbeC and MbeC-like proteins. Moreover, at the mobilization experiments testing the starting codon of MbeC, the MbeC beginning from the second codon, gave conjugation diminution by 10,000 times (Table 4). The predicted N-terminal β-strand of MbeC contains a pattern of alternating hydrophilic and hydrophobic side chains with positive charged Arg or Lys residues at positions 2 or 6 (Fig. 5A) in most of the proteins. MbeC starting from the second AUG looses a predicted polar residue (R6) of the β-sheet (Fig. 5C), which explains the dramatic drop of the mobilization frequency. This finding strengthens even more our hypothesis of a putative RHH N-terminal domain in MbeC.
In conclusion, our study demonstrates that MbeC_ColE1 is a DNA-binding protein possibly belonging to the RHH family. By comparison to other accessory proteins, we propose that MbeC induces DNA bending, helping the MbeA relaxase to melt the DNA around the nic site and cleave the phosphodiester bond. This role has been suggested for most accessory nic-processing proteins such as TrwA_R388, TraJ_RP4, TraY_F, and NikA_R64, all belonging to the RHH family, as confirmed here by sequence comparison. For this reason, despite the differences among the various conjugative systems, we propose that this potentially universal mechanism for DNA processing in bacterial conjugation exists for all accessory proteins.
Acknowledgments
Work in C.D.'s laboratory was cofunded by the EU in the framework of the program Pythagoras I (Operational Program for Education and Initial Vocational Training of the European III Framework Program and the Hellenic Ministry of Education). Work in F.D.L.C.'s laboratory was supported by grants BFU2005-03477/BMC (Spanish Ministry of Education), RD06/0008/1012 (RETICS Research Network, Instituto de Salud Carlos III, Spanish Ministry of Health), and LSHM-CT-2005_019023 (European VI Framework Program).
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Apisiridej, S., A. Leelaporn, C. D. Scaramuzzi, R. A. Skurray, and N. Firth. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 3813-24. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 10275-78. [DOI] [PubMed] [Google Scholar]
- 3.Bowie, J. U., and R. T. Sauer. 1990. TraY proteins of F and related episomes are members of the Arc and Mnt repressor family. J. Mol. Biol. 2115-6. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, A. C., J. A. Archer, and D. J. Sherratt. 1989. Characterization of the ColE1 mobilization region and its protein products. Mol. Gen. Genet. 217488-498. [DOI] [PubMed] [Google Scholar]
- 5.Cabezón, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254400-406. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 752276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caryl, J. A., M. C. Smith, and C. D. Thomas. 2004. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J. Bacteriol. 1863374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, P. T., H. Ohmori, J. Tomizawa, and J. Lebowitz. 1985. Nucleotide sequence and gene organization of ColE1 DNA. J. Biol. Chem. 2608925-8935. [PubMed] [Google Scholar]
- 9.Chung, C. T., and R. H. Miller. 1988. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 163580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clewell, D. B., and D. R. Helinski. 1969. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an operon circular DNA form. Proc. Natl. Acad. Sci. USA 621159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic. Acids. Res. 36(web server issue)W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25147-150. [DOI] [PubMed] [Google Scholar]
- 13.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res. 166127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francia, M. V., A. Varsaki, M. P. Garcillán-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2879-100. [DOI] [PubMed] [Google Scholar]
- 15.Furuya, N., and T. Komano. 1995. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J. Bacteriol. 17746-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuya, N., and T. Komano. 2000. Initiation and termination of DNA transfer during conjugation of IncI1 plasmid R64: roles of two sets of inverted repeat sequences within oriT in termination of R64 transfer. J. Bacteriol. 1823191-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 874645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 19.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 27028381-28386. [PubMed] [Google Scholar]
- 20.Komano, T., N. Funayama, S. R. Kim, and T. Nisioka. 1990. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 1722230-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 22.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 23.Lovett, M. A., and D. R. Helinski. 1975. Relaxation complexes of plasmid DNA and protein. II. Characterization of the proteins associated with the unrelaxed and relaxed complexes of plasmid ColE1. J. Biol. Chem. 2508790-8795. [PubMed] [Google Scholar]
- 24.Luo, Y., Q. Gao, and R. C. Deonier. 1994. Mutational and physical analysis of F plasmid traY protein binding to oriT. Mol. Microbiol. 11459-469. [DOI] [PubMed] [Google Scholar]
- 25.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16404-405. [DOI] [PubMed] [Google Scholar]
- 26.Moncalián, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270188-200. [DOI] [PubMed] [Google Scholar]
- 27.Moncalián, G., and F. de la Cruz. 2004. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim. Biophys. Acta 170115-23. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, W. C., B. S. Morton, E. E. Lahue, and S. W. Matson. 1993. Characterization of the Escherichia coli F factor traY gene product and its binding sites. J. Bacteriol. 1752221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 27028374-28380. [PubMed] [Google Scholar]
- 30.Pansegrau, W., D. Balzer, V. Kruft, R. Lurz, and E. Lanka. 1990. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. USA 876555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raumann, B. E., M. A. Rould, C. O. Pabo, and R. T. Sauer. 1994. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature 367754-757. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Schildbach, J. F., C. R. Robinson, and R. T. Sauer. 1998. Biophysical characterization of the TraY protein of Escherichia coli F factor. J. Biol. Chem. 2731329-1333. [DOI] [PubMed] [Google Scholar]
- 34.Schreiter, E. R., and C. L. Drennan. 2007. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 5710-720. [DOI] [PubMed] [Google Scholar]
- 35.Smith, M. C., and C. D. Thomas. 2004. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J. Bacteriol. 1863363-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 37.Tato, I., I. Matilla., I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezón. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 28225569-25576. [DOI] [PubMed] [Google Scholar]
- 38.Varsaki, A., M. Lucas, A. S. Afendra, C. Drainas, and F. de la Cruz. 2003. Genetic and biochemical characterization of MbeA, the relaxase involved in plasmid ColE1 conjugative mobilization. Mol. Microbiol. 48481-493. [DOI] [PubMed] [Google Scholar]
- 39.Williams, S. L., and J. F. Schildbach. 2007. TraY and integration host factor oriT binding sites and F conjugal transfer: sequence variations, but not altered spacing, are tolerated. J. Bacteriol. 1893813-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada, Y., M. Yamada, and A. Nakazawa. 1995. A ColE1-encoded gene directs entry exclusion of the plasmid. J. Bacteriol. 1776064-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida, H., N. Furuya, Y. Lin, P. Güntert, T. Komano, and M. Kainosho. 2008. Structural basis of the role of the NikA ribbon-helix-helix domain in initiating bacterial conjugation. J. Mol. Biol. 384690-701. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, S., and R. Meyer. 1997. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nic site at the origin of transfer. Mol. Microbiol. 25509-516. [DOI] [PubMed] [Google Scholar]
- 43.Ziegelin, G., J. P. Furste, and E. Lanka. 1989. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J. Biol. Chem. 26411989-11994. [PubMed] [Google Scholar]