Abstract
Regulation of metal ion homeostasis is essential to bacterial cell survival, and in most species it is controlled by metal-dependent transcriptional regulators. In this study, we describe a Corynebacterium diphtheriae ferric uptake regulator-family protein, Zur, that controls expression of genes involved in zinc uptake. By measuring promoter activities and mRNA levels, we demonstrate that Zur represses transcription of three genes (zrg, cmrA, and troA) in zinc-replete conditions. All three of these genes have similarity to genes involved in zinc uptake. Transcription of zrg and cmrA was also shown to be regulated in response to iron and manganese, respectively, by mechanisms that are independent of Zur. We demonstrate that the activity of the zur promoter is slightly decreased under low zinc conditions in a process that is dependent on Zur itself. This regulation of zur transcription is distinctive and has not yet been described for any other zur. An adjacent gene, predicted to encode a metal-dependent transcriptional regulator in the ArsR/SmtB family, is transcribed from a separate promoter whose activity is unaffected by Zur. A C. diphtheriae zur mutant was more sensitive to peroxide stress, which suggests that zur has a role in protecting the bacterium from oxidative damage. Our studies provide the first evidence of a zinc specific transcriptional regulator in C. diphtheriae and give new insights into the intricate regulatory network responsible for regulating metal ion concentrations in this toxigenic human pathogen.
Metal homeostasis in bacteria is mediated by five families of metal-dependent transcriptional regulators: DtxR, Fur, ArsR/SmtB, MerR, and NikR (35). Proteins within each structurally related family respond to the intracellular levels of essential metals such as manganese, iron, and zinc. The DtxR, Fur, and the less-characterized NikR family proteins predominantly regulate genes required for metal uptake, whereas ArsR/SmtB and MerR family proteins regulate metal efflux (35). Control of metal concentrations inside the bacterial cell is essential for survival and plays a key role in bacterial pathogenesis. For instance, a large number of bacterial toxins are expressed under metal-depleted conditions, and this expression is controlled by metal-dependent transcriptional regulators. In Corynebacterium diphtheriae, the etiological agent of the toxin-mediated upper respiratory tract infection diphtheria, DtxR, regulates transcription of the diphtheria toxin gene and multiple other genes involved in iron uptake (18, 38, 40). In addition to DtxR, the C. diphtheriae genome encodes a second DtxR-like protein, MntR, which responds to Mn (37, 38), and it is predicted to encode one Fur, three ArsR/SmtB, four MerR, and zero NikR homologs.
Iron-dependent global gene regulation is controlled by DtxR-like proteins in many gram-positive and acid-fast bacterial species, including C. diphtheriae, and by Fur-like proteins in many gram-negative bacteria. Although DtxR-like and Fur-like proteins share little sequence homology, they both act as transcriptional repressors when complexed with a cognate metal ion. In metal-depleted conditions, apo-Fur and apo-DtxR (metal-free forms) are unable to bind their target promoters, allowing transcription of the downstream genes (22). The Fur homolog in C. diphtheriae is uncharacterized but has similarity to a zinc-dependent regulator found in Mycobacterium tuberculosis, Zur (for zinc uptake regulator) (23).
Zinc is essential for cell survival, serving as a cofactor for more than 300 enzymes such as superoxide dismutase and alcohol dehydrogenase. It also functions as a structural scaffold for RNA polymerase, tRNA synthetases, and approximately 40 additional proteins (10, 11, 31, 41). In fact, Escherichia coli requires as much zinc as it does iron and calcium for cellular processes (31). In addition, zinc protects sulfhydryl groups from free radicals and inhibits free radical formation by competing with redox-active metals such as iron. Alternatively, high concentrations of zinc can be toxic by blocking thiols and binding of other metals to their cognate active sites within enzymes such as cytochrome c oxidase (3, 5, 26).
Unlike iron homeostasis, which has been extensively characterized (1), little research has focused on the uptake and/or storage of other metals, including zinc, by pathogenic bacteria. High-affinity zinc uptake is mediated by several ABC transporters, such as the Ycdhi-yceA and zinc uptake ABC systems (ZnuABC) (16). ZnuA encodes a periplasmic binding protein, ZnuB encodes an integral protein, and ZnuC encodes the ATPase of the transporter. In addition, YciC (CobW) may serve as a low-affinity zinc uptake protein or a metallochaperone (16). The total zinc concentration within bacteria is typically in the millimolar range; however, free unbound intracellular zinc concentrations at femtomolar levels are sufficient to trigger zinc uptake or efflux (31). Zinc uptake is controlled in a zinc-dependent manner by Zur in bacteria such as E. coli, Bacillus subtilis, and M. tuberculosis (14, 23, 34). The importance of zinc homeostasis in bacterial pathogenesis is only beginning to be characterized, but Salmonella enterica serovar Typhimurium mutants lacking Zur or ZnuC have decreased virulence in mice, indicating an essential role for zinc in this model of infection (7).
In the present study, we characterize the only fur-like gene in C. diphtheriae and demonstrate that it is a zinc-responsive regulator and is therefore more accurately described as zur (for zinc uptake regulator). In the presence of zinc, C. diphtheriae Zur represses transcription of three genes predicted to be involved in zinc uptake: cmrA, whose product is predicted to be anchored to the surface of the cell; troA (for transport related operon A), whose product has homology to metal uptake membrane proteins, such as Streptococcus pneumoniae psaA (16) and Treponema pallidum troA (17); and zrg (for zinc-regulated gene), which is similar to yciC (14). Interestingly, the transcription of zur itself is repressed in zinc-depleted conditions by a process that involves Zur. This observation indicates that Zur may have a role in controlling its own expression by a mechanism that is independent of its characterized activity as a zinc-dependent DNA-binding protein. The present study is the first molecular characterization of a zinc-dependent transcriptional regulator in C. diphtheriae, a paradigm for pathogenesis and gene expression in gram-positive and acid-fast organisms.
MATERIALS AND METHODS
Culture media, strains, and growth conditions.
C. diphtheriae strains NCTC13129 (9), the sequenced isolate of the 1990s outbreak in the former Soviet Union, and C7(β) (2), a toxigenic strain used extensively for experimental work since the 1950s, were cultured in PGT, a casein hydrolysate medium (2). Metal ions were removed from the medium by treating with 10g/liter Chelex-100 (Bio-Rad, Hercules, CA) for 2 h, followed by filter sterilization (42). Where indicated, supplementation with specific metals was done at the following concentrations: 10 μM FeCl3, 10 μM MnCl2, 5 μM CuSO4, and 25 μM ZnSO4. To further chelate zinc remaining in the medium following treatment with Chelex-100, a zinc-specific chelator N,N,N′,N′-tetrakis (2-pyridylmethyl)-ethylene diamine (TPEN) was added to a concentration of 20 μM. E. coli TE1 (20) was used for cloning, and E. coli S17-1 (39), an RP4 mobilizing strain, was used as a donor for the conjugative transfer of plasmids into C. diphtheriae. E. coli was cultured in Luria-Bertani broth (LB) (25). Kanamycin, spectinomycin, and nalidixic acid were added at concentrations of 20, 100, and 20 μg/ml, respectively.
Deletion of zur in C. diphtheriae NCTC13129.
To construct a deletion within zur, the plasmid pK19mobsacB, which contains an origin of replication that functions in E. coli, but not C. diphtheriae, was utilized (36). The extreme 5′ and 3′ ends of zur were amplified by using PCR (the primers are listed in Table 1), and the resulting DNA fragments were digested with SalI/ApaI and ApaI/SphI. The digested PCR fragments were ligated to SalI/SphI-digested pK19mobsacB to construct pK19mobsacBΔzur. This plasmid, which includes a deleted zur gene (lacking 369 bp of the 426-bp gene), was transformed into E. coli S17-1 and then mated into NCTC13129 (44). Kanamycin-resistant transconjugants were counterselected for resistance to 10% sucrose, indicating the loss of integrated plasmid vector. A deletion of the zur gene on the chromosome of NCTC13129 was confirmed by PCR and sequencing, and this strain was named NCTC13129Δzur.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′) | Use |
|---|---|---|
| InterArsRFor | CCACGACGCTTACAGAGGTCGACGA | Cloning promoter for arsR-like gene |
| InterArsRev | TGATGATGGTCGTGGAGAAGGATCC | Cloning promoter for arsR-like gene |
| InterZurFor | GTTCTCAAGCAGGCACACATCGTCGAC | Cloning promoter for zur |
| InterZurRev | CCCCAACTTCGGGATCCAACGATC | Cloning promoter for zur |
| ArsRSalI | GTGAGATAAAACGTCGACGGAGGTT | Cloning and deletion of zur |
| Zurdelta1 | CCCCAACTTCGGGCCCGAACGATC | Cloning and deletion of zur |
| Zurdelta2 | CCGAAATCTAGGGCCCCTGCGAAAG | Cloning and deletion of zur |
| ZurSph | CGGCGATTATGTTGGCATGCCC | Cloning and deletion of zur |
| ZrgUpFor | GACGGGAGTCGACTGCGACATTC | Cloning promoter for zrg |
| ZrgUpRev | GCGGAAGATCTGACATGAAGG | Cloning promoter for zrg |
| ArsZurCoFor | AGACATCATTGCGAAAGTCG | Detection of arsR-zur cotranscript |
| ArsZurCoRev | AACGATCAATGGTGCGATTC | Detection of arsR-zur cotranscript |
| KQ-troafor | CGCAATACCATCAATGTTGC | qRT-PCR for troA |
| KQ-troarev | GGTGGTTGCTGGATCGTAGT | qRT-PCR for troA |
| KQ-arsfor | CTCCACGACCATCATCACTG | qRT-PCR for arsR-like gene |
| KQ-arsrev | CGAGTCGAGAGCACTCAAAA | qRT-PCR for arsR-like gene |
| KQ-cmrafor | TGTTGCAAAGTCCAGTGAGC | qRT-PCR for cmrA |
| KQ-cmrarev | ATACCAAAGAACGCCAATGC | qRT-PCR for cmrA |
| KQ-zrgfor | CCAAAGGACACTGCTGGATT | qRT-PCR for zrg |
| KQ-zrgrev | GAGAAGTTGCTGGGCTTGAG | qRT-PCR for zrg |
| KQ-zurfor | TCGTTGGGGTTCTCAAAGAT | qRT-PCR for zur |
| KQ-zurrev | TGATTGCAAGGTTCGGTACA | qRT-PCR for zur |
| KQ-gyrbfor | GGTCTGACCATTACGCTGGT | qRT-PCR for gyrB |
| KQ-gyrbrev | TCTTCTCGCGTTTCTTTGGT | qRT-PCR for gyrB |
Complementation of NCTC13129Δzur.
Full-length wild-type zur, along with its native promoter and ribosomal binding site, was amplified from NCTC13129 by using PCR (see primers in Table 1). The resulting fragment was digested with SacI and ligated into pKPIM (30), which had been identically digested to create pKPIMzur. Purified pKPIMzur was then transformed into S17-1 and transferred to NCTC13129 strains via conjugation. The plasmid pKPIM integrates site specifically at attB2 in NCTC13129. Incorporation of the plasmid into the chromosome via an attP site was confirmed by detecting the recombinant attL2 site created by integration of the plasmid using PCR (30).
Resistance to killing by hydrogen peroxide.
Resistance to killing by hydrogen peroxide, H2O2, was assayed as described previously (28). Briefly, in the zone of inhibition assay, we measured the diameters of the zones of inhibition of bacterial growth when 20 μl of 1 M H2O2 was applied to 0.6-cm-diameter paper disks in the centers of plates containing heart infusion agar and lawns of various strains of C. diphtheriae.
In the percent killing assay, cultures were grown in PGT until the absorbance of the culture measured at 600 nm was between 1 and 2, at which time H2O2 was added to the growth medium at a final concentration of 10 mM, and the cultures were then incubated for an additional 10 min. Viable counts from each culture were then determined by plating dilutions of the culture onto heart infusion agar.
Cloning of promoter regions and β-galactosidase assays.
PCR was used to amplify ∼200 bp of the region upstream of arsR (dip1709), zur (dip1710), and zrg (dip1486) (see primers in Table 1). The resulting fragments were digested with the restriction enzymes BamHI/SalI (arsR and zur) or SalI/BglII (zrg) and then ligated into a similarly digested pSPZ (29), a reporter vector with a promoterless lacZ gene. The upstream regions of cmrA (dip2325) and troA (dip0438) were digested and purified from pcmrA-PO (4) and pdip0438 (M. P. Schmitt, unpublished data), respectively, with the restriction enzymes SphI/XmaI and ligated into identically digested pSPZ. The constructs were transformed via electroporation into C. diphtheriae NCTC13129 and isogenic Δzur strains. For the β-galactosidase assays, strains were inoculated in PGT containing different metal concentrations and incubated with shaking at 37°C overnight. Twenty micromolar TPEN was added to the medium in zinc depleted conditions. β-Galactosidase assays were performed on overnight cultures as previously described (25, 38). pSPZtox (29) and pSPZcmrA were used as positive controls for iron- and zinc-depleted conditions, respectively. The plasmid pSPZmntA was constructed by cloning the mntA promoter containing fragment from pCMmntA (pPO3), and it was used as a control for manganese conditions (37).
Quantitative reverse transcriptase PCR (qRT-PCR).
RNA was isolated, using the RNApro Blue kit (QBioGene), from NCTC13129, NCTC13129Δzur, and C7(β) grown in zinc-replete and -depleted conditions. DNA was eliminated from RNA samples by using New England Biolab RNase-free DNase as described by the manufacturer. cDNA was synthesized from RNA templates by using the SuperScript III RT (Invitrogen). Negative controls contained RNase-free water substituted for RT. The cDNA generated was quantitated by quantitative PCR using a Bio-Rad iQcycler with the Absolute QPCR SYBR green fluorescein mix (Thermo Scientific). For generation of cDNA and quantitative PCR analysis, gene-specific primers were used (Table 1). Standard curves were constructed from serial dilutions of NCTC13129 genomic DNA. The transcript level of gyrB (a gene that is constitutively expressed in the tested conditions [data not shown]) was used as a control for RNA concentration. To determine the relative transcript quantity, the amount of gene-specific transcript was divided by the amount of gyrB transcript.
RESULTS
Characterization of the zur loci.
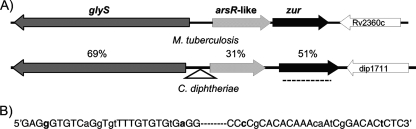
The genome sequence of C. diphtheriae NCTC13129 contains a predicted coding sequence (dip1710) with similarity to genes encoding Fur-like regulators (12). A BLAST search confirmed that dip1710 is the only sequence in the genome with similarity to fur-like genes and that the Zur protein of M. tuberculosis is its closest characterized homolog. Based on its homology to the M. tuberculosis gene and the functional assays described below, we renamed dip1710 as zur. The genomic loci that include zur in M. tuberculosis and C. diphtheriae NCTC13129 are similar in structure (Fig. 1A). Upstream of zur in both organisms is a gene that encodes a putative metal-dependent transcriptional regulator similar to arsR. ArsR family proteins usually repress transcription of genes involved in metal efflux (6). In the M. tuberculosis genome, the zur gene slightly overlaps the 3′ end of the arsR-like gene, and these genes are cotranscribed from a single zinc-dependent promoter located upstream of the arsR-like gene (24). In contrast, there is a 179-bp intergenic region between the arsR-like gene and zur in C. diphtheriae (Fig. 1A). An aminoacyl-tRNA synthetase glyS and a gene with no identified homologs are directly upstream and downstream of arsR-zur, respectively. In the C7(β) strain of C. diphtheriae, which has been studied for many years as a model for gene regulation (2, 27, 28, 38), we identified an insertion sequence (IS element) upstream of the arsR-like gene. DNA sequencing of the region of the C7(β) genome containing the IS element (GenBank no. FJ470294) revealed that the 1,445-bp element includes a transposase whose coding sequence encompasses 1,350 bp. Inverted repeat sequences of 28 bp were found at the ends of the element (Fig. 1B), and its insertion resulted in the direct target site duplication of 8 bp (TTTCGATC). The IS element is located 78 bp upstream of the ATG codon for the arsR-like gene and 240 bp upstream of the GTG start codon of glyS. The effect, if any, of this IS element on expression of the genes at the C7(β) zur locus is unknown. Other than the insertion of the IS element, the gene organization in the zur loci are identical in NCTC13129 and C7(β).
FIG. 1.
Genetic arrangement of the C. diphtheriae and M. tuberculosis zur loci. (A) The dark gray arrow represents glyS, a glycyl-tRNA synthetase beta subunit. The gray striped arrow represents an arsR-like gene, a putative metal-dependent transcriptional regulator. The black arrow represents zur, a zinc-dependent transcriptional regulator. dip1711 and Rv2360c are genes with unknown function. The insertion site of the IS element in C. diphtheriae C7(β) is shown as a triangle, and the region deleted in the Δzur strain is indicated as a dashed line. The percentages of amino acid sequence similarities between genes are indicated between the loci. (B) The sequences of the 28-bp inverted repeats at the ends of the IS element are shown. Bases that are interruptions in the inverted repeat are shown in lowercase, and the rest of the IS element is indicated as a dashed line.
Physiological importance of the Zur protein.
To investigate the function of C. diphtheriae zur, a nonpolar deletion mutation was constructed in the genome of NCTC13129 using the mobilizable vector pK19mobsacBΔzur (see Materials and Methods and Fig. 1), creating NCTC13129Δzur. To complement this strain, we cloned wild-type zur into integration vector pKPIM (30), resulting in pKPIMzur. We then compared the growth curves of NCTC13129 to those of NCTC13129Δzur in the presence or absence of zinc, iron, and manganese to determine whether the absence of Zur affects the growth of C. diphtheriae. Similar growth curves were observed for the NCTC13129 and NCTC13129Δzur regardless of the presence or absence of 10 μM iron, 10 μM manganese, or 25 μM zinc (data not shown). Zinc concentrations of ≥200 μM inhibited growth of both the NCTC13129 and NCTC13129Δzur strains of C. diphtheriae to comparable extents, demonstrating the toxic effect of high zinc concentrations on this organism (data not shown). Also, no differences were observed between NCTC13129 and NCTC13129Δzur in protein profiles or cell morphology, via whole-cell protein lysates and light microscopy, respectively (data not shown). Thus, the absence of Zur had no detectable effect on C. diphtheriae in these experiments.
Since zinc has been shown to protect bacteria from oxidative stress (13), we also tested the ability of NCTC13129 and NCTC13129Δzur to survive challenge with H2O2. H2O2 susceptibility was determined by using two methods: growth inhibition assays and killing assays. Both C7(β) and C7(β)ΔdtxR strains have been assessed for levels of H2O2 stress resistance in previous studies and were used as controls (29). The NCTC13129Δzur pKPIM strain showed a slightly larger zone of inhibition (Table 2) and a higher percentage of killing (Table 2) than both NCTC13129 pKPIM and complemented NCTC13129Δzur pKPIMzur, indicating that it was more sensitive to H2O2 stress than its wild-type parent. Although the P value for the difference between the wild-type and Δzur strain zones of inhibition was >0.05 (i.e., 0.08), a trend of higher peroxide sensitivity is observed in the zur mutant. In the killing assay, the difference between the Δzur strain and the NCTC13129 pKPIM or NCTC13129Δzur pKPIMzur strains had P values of <0.05 (0.01 and 0.034, respectively). Therefore, the absence of Zur in NCTC13129 resulted in a slight increase in sensitivity to H2O2.
TABLE 2.
Peroxide stress susceptibility of C. diphtheriae strains
| Strain | Avg ± SDa
|
|
|---|---|---|
| % of wild-type zone of inhibition | % Killing in exposure assay | |
| C7(β) | 100 | 38 ± 11 |
| C7(β)ΔdtxR | 170 ± 15* | 68 ± 11* |
| NCTC13129/pKPIM | 100 | 24 ± 5 |
| NCTC13129/pKPIMzur | 87 ± 7 | 18 ± 15 |
| NCTC13129Δzur/pKPIM | 110 ± 8 | 50 ± 9* |
| NCTC13129Δzur/pKPIMzur | 91 ± 14 | 30 ± 10 |
Values are given as averages for at least three samples. *, Statistically significant difference from the wild-type parent strain (P < 0.05) as determined by a one-way analysis of variance, followed by the Holm-Sidak method.
Identification of the zur promoter(s).
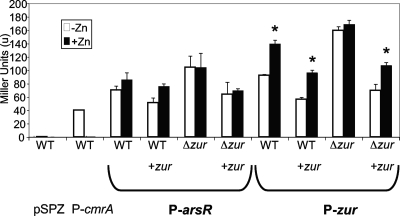
To identify the promoter(s) of zur, we cloned the region immediately upstream of zur into a β-galactosidase promoter reporter vector that replicates in C. diphtheriae, pSPZ (29). Since the promoter of the M. tuberculosis zur gene is located upstream of rv2358/arsR (Fig. 1, (24), the upstream region of the NCTC13129 arsR-like gene was also tested for promoter activity. In contrast to M. tuberculosis, C. diphtheriae NCTC13129 contains promoters directly upstream of both the arsR-like gene and zur (Fig. 2). We also investigated whether there was an arsR/zur cotranscript using RT-PCR with primers that annealed within the intergenic region (Table 1). No arsR-zur cotranscript was observed but transcripts that contained only the arsR-like gene or zur were detected (data not shown).
FIG. 2.
arsR and zur promoter activity. NCTC13129 (indicated as wild type [WT]) and NCTC13129Δzur (indicated as Δzur) strains with either vector control, pKPIM, or complementing wild-type zur, pKPIMzur (shown as +zur), were inoculated into zinc-replete (+Zn) or -depleted (-Zn) conditions and tested for arsR and zur promoter activity. β-Galactosidase activity is expressed in Miller units. The cmrA promoter is a positive control for zinc-depleted conditions. The asterisks denote statistical significance (<0.05) between zinc-replete and -depleted conditions, as determined by a one-way analysis of variance, followed by the Holm-Sidak method.
Regulation of zur transcription.
In many organisms, transcription of metal-dependent transcriptional regulators is regulated in response to the presence of their cognate metal (24). Therefore, we tested the activity of the promoters upstream of the arsR-like gene and zur in the presence or absence of zinc. The promoter upstream of the arsR-like gene showed no significant difference in response to differential zinc conditions or to the presence of Zur (Fig. 2). In contrast, the zur promoter had lower activity in zinc depleted versus replete conditions in both the NCTC13129 and NCTC13129Δzur pKPIMzur with P values <0.05 (<0.001). Strikingly, zur promoter activity remained high in the NCTC13129Δzur background irrespective of the presence or absence of zinc, suggesting that Zur has a role in the regulation in its own expression. This phenomenon was confirmed by the observation that the zinc dependence of the zur promoter was restored in NCTC13129Δzur pKPIMzur (Fig. 2). Both the arsR-like gene promoter and zur promoter activities were unaltered by the presence or absence of either iron or manganese (data not shown). These data indicate that the zur promoter is repressed specifically in zinc-depleted conditions in a Zur-dependent manner.
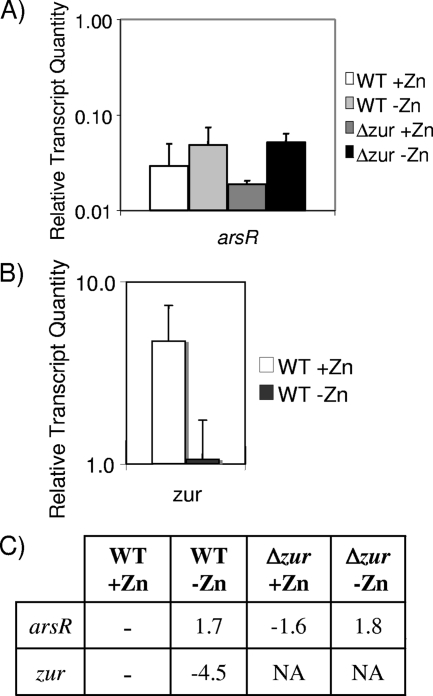
β-Galactosidase assays determine promoter strength without being impacted by mRNA stability, whereas qRT-PCR, although affected by transcript stability, more precisely quantifies the transcripts present at a single time point. Therefore, we used qRT-PCR to further analyze the arsR-like gene and zur transcript levels in NCTC13129 and NCTC13129Δzur under zinc-replete and -depleted conditions. The arsR-like gene transcripts, although barely detectable, remained constant in NCTC13129 and NCTC13129Δzur regardless of the zinc concentration, confirming the promoter fusion data described above (Fig. 3A and C). In contrast, the zur transcript level was 4.5-fold higher in zinc-depleted conditions (Fig. 3B and C), which correlated with the observed promoter activity (Fig. 2). The zur transcript was only assayed in NCTC13129 because the majority of the zur gene was deleted in NCTC13129Δzur, preventing its detection in this assay. With a weakly transcribed constitutive promoter, the accumulation of β-galactosidase may indicate a higher promoter activity compared to transcript levels at a single time point detected in qRT-PCR data (compare arsR in Table 3 and Fig. 2).
FIG. 3.
Detection of arsR and zur transcripts. RNA isolated from NCTC13129 (WT) and NCTC13129Δzur (Δzur) in zinc-replete and -depleted conditions was analyzed for relative transcript levels (normalized to gyrB mRNA levels) of arsR (A) and zur (B). (C) Fold change of these genes from NCTC13129 in zinc-replete conditions.
TABLE 3.
Detection of arsR and zur transcripts
| Gene | Strain | Relative transcript quantity (avg ± SD)a
|
Fold changeb | |
|---|---|---|---|---|
| +Zn | -Zn | |||
| arsR | C7(β) | 0.01 ± 0.00 | 0.01 ± 0.00 | <2 |
| NCTC13129 | 0.03 ± 0.02 | 0.05 ± 0.03 | <2 | |
| zur | C7(β) | 0.58 ± 0.13 | 0.56 ± 0.06 | <2 |
| NCTC13129 | 4.8 ± 2.7 | 1.1 ± 0.67 | 4.5 | |
Normalized with gyrB. Values are averages for at least three samples.
The fold change is calculated as +Zn/-Zn.
To determine whether zinc-dependent regulation of zur occurs in other strains of C. diphtheriae, transcription of zur and the arsR-like gene was assayed in the C7(β) strain by using qRT-PCR. The transcript levels of arsR were lower in C7(β) than in NCTC13129 but remained unchanged in zinc-replete and -depleted conditions (Table 3). Interestingly, not only was the transcript of zur at lower levels in C7(β), the differential regulation observed between zinc-replete and -depleted conditions was not seen in this strain. Thus, the regulation and accumulation of the zur transcript differed in these C. diphtheriae strains.
Transcriptional regulation by Zur.
We performed a BLAST search of the C. diphtheriae genome to identify genes that may be regulated by Zur. Sequence probes that were used for the BLAST search included genes that are known to be regulated by Zur in other bacteria and genes that encode proteins involved in metal transport. This analysis identified a gene we named zrg (dip1486), with homology to cobW/yciC genes from a variety of bacterial species and which encodes a putative low-affinity zinc uptake protein, as well as a gene we named troA (dip0438), with sequence similarity to T. pallidum troA and to S. pneumoniae psaA, that is predicted to encode a component of an ABC-type metal transporter. The cmrA gene, which was previously shown to be zinc regulated (4), is predicted to encode a sortase anchored cell wall protein. The mechanism of the zinc-dependent repression of cmrA has not previously been investigated. We thus sought to determine whether Zur had a role in regulating transcription of these three genes.
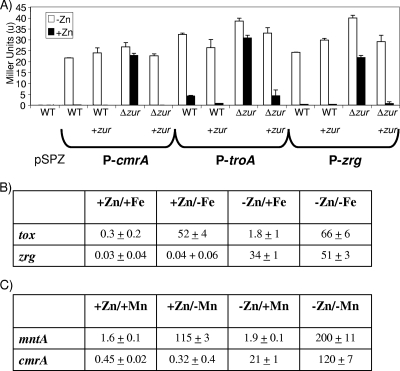
We cloned the upstream regions of zrg, cmrA, and troA, into the promoter reporter vector pSPZ and assayed the activity of each in NCTC13129 and NCTC13129Δzur. All three promoters were repressed in zinc-replete conditions and derepressed in zinc-depleted conditions in NCTC13129 (Fig. 4A). In NCTC13129Δzur, the activity of all three promoters was constitutive in response to zinc. Zinc-dependent promoter activity was restored when zur was provided from pKPIMzur in NCTC13129Δzur. These data indicate that the activities of these three promoters were controlled by Zur in response to changes in zinc concentration.
FIG. 4.
Activity of Zur-regulated promoters. NCTC13129 (indicated as WT) and NCTC13129Δzur (indicated as Δzur) strains with either vector control, pKPIM, or complementing wild-type zur, pKPIMzur (shown as +zur), inoculated into zinc-replete (+Zn) or -depleted (-Zn) conditions were used to test the activity of promoters (zrg, cmrA, and troA) in the reporter plasmid pSPZ. (A) The β-galactosidase activity was determined under differential conditions. The activity of the zrg promoter under iron and zinc stress conditions (B) and activity of the cmrA promoter under manganese and zinc stress conditions (C) are also presented. The promoters of tox, mntA, and cmrA genes (cloned into the reporter vector) were used as positive controls for iron-, manganese-, and zinc-depleted conditions, respectively.
We tested each of the promoters for activity in the presence or absence of iron and manganese. When the activity of each promoter was compared under iron-replete or -depleted conditions in the wild-type NCTC13129 strain, no activity was observed. Identical results were obtained when manganese was substituted for iron (Fig. 4B and C, first two columns). This result was not unexpected since the assay medium contained zinc, which represses the expression of all three promoters. Interestingly, we did observe higher levels of activity from the zrg promoter under iron-depleted conditions in the NCTC13129Δzur strain (23 Miller units in iron-replete medium versus 33 Miller units in iron-depleted medium). Similarly, we observed higher activity from the cmrA promoter in manganese-depleted medium in the NCT13129Δzur strain background (28 Miller units in manganese-replete versus 92 Miller units in manganese-depleted medium). The activity of the promoter for troA was unchanged when the medium was depleted of either iron or manganese in the NCTC13129Δzur background. These observations led us to hypothesize that the effects of iron and manganese on the zrg and cmrA promoters, respectively, might be epistatic to the effects of zinc and Zur.
We next examined the activities of the zrg and cmrA promoters in medium depleted of both iron and zinc or both manganese and zinc. As shown in Fig. 4B (columns 3 and 4), depletion of both iron and zinc resulted in a 1.5 increase in the activity of the zrg promoter compared to the activity in medium depleted only for zinc. As a control we used the well-characterized tox promoter, whose activity is responsive only to iron and not to zinc (38). The activity of the cmrA promoter increased sixfold when the growth medium was depleted of both zinc and manganese compared to activity in medium depleted only of zinc (Fig. 4C, columns 3 and 4). For this assay, we used the mntA promoter as a control for manganese-dependent regulation since it is controlled by MntR in response to manganese (37). Interestingly, the mntA promoter showed an increase in activity when the growth medium was depleted of both manganese and zinc (compared to medium lacking only manganese), indicating that MntR-dependent manganese regulation is epistatic to the effect of zinc.
Our observations imply that the zrg promoter is controlled first by zinc in a process that requires Zur and secondarily by the presence of iron in a process that is independent of Zur. The cmrA promoter is controlled first by zinc and Zur and secondarily by manganese in a Zur-independent mechanism. Clearly, the regulation of the zrg, cmrA, and mntA promoters is more complex than the one-metal, one-regulator models proposed for many metal-dependent promoters.
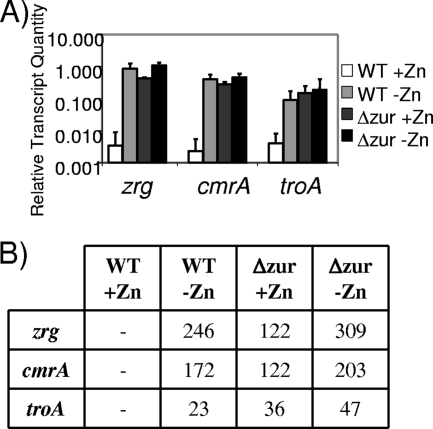
To confirm that the transcript levels correlated with the promoter activity, we used qRT-PCR to assay cmrA, zrg, and troA transcription in NCTC13129 and NCTC13129Δzur under zinc-replete and -depleted conditions. The transcript levels of all three genes were significantly lower in NCTC13129 under zinc-replete conditions than the levels in NCTC13129 in zinc-depleted conditions and in NCTC13129Δzur in all conditions (Fig. 5A). The fold change was calculated by dividing all transcript levels by the corresponding NCTC13129 wild-type strain transcript level under zinc-replete conditions (Fig. 5B). The largest transcript level fold change was observed for zrg with an average of 250. The cmrA transcript had an average of a 170-fold change, and the troA transcript level had an average fold change of 36. Large differences between the fold changes of zrg, cmrA, and troA transcript levels correlate with the differences in transcripts present under induced conditions (i.e., zinc depleted or lacking Zur), rather than a difference in the repression or uninduced state (i.e., zinc replete and wild-type Zur levels). These data confirm that Zur represses the transcription of zrg, cmrA, and troA in zinc-replete conditions.
FIG. 5.
RNA levels of Zur-regulated genes. RNA isolated from NCTC13129 and NCTC13129Δzur under zinc-replete and -depleted conditions was analyzed by using qRT-PCR. (A) Relative level of transcripts (normalized to gyrB mRNA levels) of cmrA, zrg, and troA. (B) Fold change of these genes from NCTC13129 under zinc-replete conditions.
DISCUSSION
To regulate metal homeostasis, bacteria use multiple metal-dependent transcriptional regulators, each controlling different regulons in response to different inducers. We describe here the characterization of a zinc-dependent Fur family homolog in C. diphtheriae. To our knowledge, Zur, is the first zinc-dependent transcriptional regulator described in Corynebacteria. We identified three genes, cmrA, zrg, and troA whose transcription is repressed by Zur. In addition, we demonstrated that the zur gene is transcribed from a promoter that is repressed by Zur when zinc is scarce. Our data also indicate that C. diphtheriae is capable of integrating signals from different metals to exert exquisite control over the transcription of specific genes.
Although C. diphtheriae lacking Zur do not have a growth defect compared to the wild-type, all C. diphtheriae strains exhibited slowed growth rates in medium containing concentrations of zinc greater than 200 μM, thus indicating that zinc toxicity occurs in C. diphtheriae. Due to uncontrolled uptake of zinc, one might expect the C. diphtheriae zur mutant strain to be more sensitive to high concentrations of zinc than its wild-type parent, as is observed for Xanthomonas campestris zur mutants (43). This was not the case. In X. campestris Zur controls the expression of both zinc uptake and efflux; thus, when Zur is absent, X. campestris not only undergoes unregulated uptake of zinc, but it also fails to activate zinc efflux systems (19). Our data suggest that, unlike X. campestris Zur, C. diphtheriae Zur is not required for the activation of zinc efflux. In addition, given the roles of zinc as an antioxidant and as a cofactor for enzymes such as superoxide dismutase, we tested the ability of a Δzur mutant strain to survive challenge with H2O2. The C. diphtheriae NCTC13129Δzur strain demonstrated an increased susceptibility to peroxide stress, suggesting that Zur is required for expression of oxidative stress defenses.
Interestingly, in C. diphtheriae NCTC13129, zur transcript levels are affected by zinc concentrations, but the levels of the adjacent arsR-like gene transcript are not. This distinguishes C. diphtheriae zur from its homolog in M. tuberculosis, where the ArsR-like protein represses the arsR-zur cotranscript in response to zinc-depleted conditions (8, 24). In addition, the repression of zur transcription in C. diphtheriae NCTC13129 in zinc-depleted conditions is dependent on the presence of Zur. These variations highlight important differences and suggest that Zur may play different roles in the physiology of these two species. The regulation of C. diphtheriae zur transcription by zinc and Zur is the first indication of Zur activity in zinc-depleted conditions. However, in other species, Fur proteins have been shown to exhibit uncharacteristic activity, such as functioning as a direct DNA-binding transcriptional activator in Neisseria meningitidis and as a repressor in the absence of metal in Helicobacter pylori (22). DNA binding experiments are currently in progress to determine whether this effect on the zur transcript is a result of direct Zur binding to its promoter.
We observed differences in transcription of zur in C. diphtheriae strains C7(β) and NCTC13129, demonstrated by both the absence of zinc-dependent zur regulation and the lower overall zur transcript levels in C7(β). Using qRT-PCR, the Zur-regulated genes zrg, cmrA, and troA were all shown to be regulated by zinc and Zur in C7(β) and, unlike the finding for the zur transcript levels, the transcript levels of zrg, cmrA, and troA in C7(β) were all extremely similar in NCTC13129 under identical growth conditions (data not shown). It is unlikely that the zinc stress conditions used were significantly different or that transcription of all genes is generally lower in C7(β) compared to NCTC13129. Although the IS element upstream of arsR in C7(β) could result in lower arsR transcription, it is unlikely that this IS element is affecting zur transcription directly, since zur has its own promoter. In addition, we determined the DNA sequence of the region between arsR and zur in C7(β), and it was identical to that in NCTC13129. There remains the possibility that there are other differences between these strains that account for the loss of zinc-dependent regulation of zur transcript in C7(β), including unknown trans-acting factors. C7(β) is a lab strain originally isolated in the 1950s, while NCTC13129 is a recent clinical isolate representative of an outbreak that occurred in the late 1990s (2, 9). The differences in zur regulation and in the sequence of the loci in these two strains highlight the value of assaying recent clinical isolates and the importance of genomic sequencing of multiple strains of a single bacterial species.
In both strains of C. diphtheriae the zrg transcript is strongly repressed in high zinc conditions in a Zur-dependent manner. This is also the case for its homologs in B. subtilis and M. tuberculosis (14, 23, 32). Sequence homology suggests that Zrg is a low-affinity zinc transporter protein, or a metallochaperone, that “passes” zinc ions between transport proteins and enzymes. Our data support the notion that Zrg is a low-affinity zinc transporter protein given that repression of a zinc chaperone under high zinc concentrations would not likely be of value to the cell. The evidence that Zrg is a zinc transporter is supported by observations that the B. subtilis Zrg homolog YciC is important for zinc utilization (14, 15). Similar to zrg, the transcript of cmrA is highly repressed by Zur under zinc-replete conditions in C. diphtheriae. Based on this observation and the prediction that cmrA encodes a sortase anchored cell wall protein, we hypothesize that CmrA is a surface protein associated with an ABC transporter involved in the uptake of zinc.
The final C. diphtheriae gene that we characterized was troA, which is repressed by Zur in zinc-replete conditions. In contrast to the observations with cmrA and zrg, troA promoter activity was observed in zinc-replete conditions, albeit at low levels (Fig. 5A). We also observed that induction of troA transcription occurs in the presence of higher concentrations of zinc than the transcription of either zrg or cmrA (data not shown). These observations suggest that the troA promoter is released from Zur repression when the concentrations of zinc are sufficient to allow binding of Zur at the zrg and cmrA promoters. The troA gene encodes a membrane protein with homology to S. pneumoniae PsaA and is the first gene in a seven gene operon. The genes of this operon are predicted to encode the components of an ABC transporter, a putative surface-anchored sortase protein, and three putative membrane proteins of unknown function. Some of these genes have homology to genes in the ZnuABC metal uptake machinery family. The similar gene cluster in S. pneumoniae is thought to be a manganese transporter, but recent evidence suggests that it may transport zinc (16). In addition to its role in metal ion transport, S. pneumoniae PsaA also contributes to virulence and oxidative stress resistance (21).
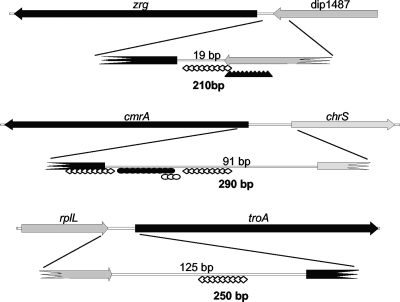
One of the most intriguing findings from our investigation of C. diphtheriae Zur-dependent gene regulation is the identification of two genes whose transcription is affected by more than one metal ion. Zur is required for the zinc-dependent primary regulation of both zrg and cmrA. We demonstrated that the zrg promoter is secondarily regulated by the availability of iron and that the cmrA promoter is secondarily regulated by the presence of manganese. In both cases, the secondary regulation is independent of Zur, suggesting that a second metal-dependent regulator may be involved. There are examples of bacterial promoters regulated by more than one metal-dependent regulator, including the promoter for rv0282, a gene of unknown function in M. tuberculosis, which undergoes dual zinc- and iron-dependent regulation by Zur and IdeR (a DtxR homolog), respectively (23). Similarly, E. coli mntH, a homolog of the eukaryotic natural resistance associate macrophage protein (NRAMP), is regulated by iron and manganese by mechanisms that require Fur and MntR (33). Since zrg and cmrA are secondarily regulated by iron and manganese, respectively, we hypothesize that DtxR, the only iron-dependent transcriptional regulator, and MntR, the only manganese-dependent transcriptional regulator characterized in C. diphtheriae, are involved. The binding sites for DtxR and MntR have been characterized (37, 38), and we have searched the regions upstream of zrg and cmrA for sequences with similarity to these sites. The DNA region with highest similarity (10/19) to the consensus DtxR binding site (TTAGGTTAGGCTAACCTAA) upstream of zrg is 74 bp upstream of the start codon (Fig. 6). The binding site for MntR is less well characterized (37), but there are two possible binding motifs for MntR upstream of cmrA. A five-out-of-seven match to the direct repeat (TGAACAA) found in the MntR binding site is located 19 bp upstream, and a 19-bp sequence with 63% identity to the MntR inverted repeat is located 3 bp upstream of the cmrA start codon (Fig. 6). The roles of MntR and DtxR in controlling transcription of cmrA and zrg are under active investigation.
FIG. 6.
Regulator binding sites in the upstream regions of zrg, cmrA, and troA. The locations of the putative binding sites for Zur (diamonds), DtxR (triangles), and MntR (black and white circles) are indicated for the zrg, cmrA, and troA promoter regions. The black circles indicate the inverted repeat, and the white circles indicate the direct repeat in the MntR binding site The lengths of the DNA fragment used in the promoter assays (Fig. 5) are shown in boldface below each region, and the distance between adjacent genes is shown in normal typeface above each region. The diagram is not drawn to scale.
The binding sites for C. diphtheriae Zur have not been defined, but given its similarity to M. tuberculosis Zur, we searched regions upstream of cmrA, zrg, and troA for sequences similar to the 21 bp at the center of the M. tuberculosis Zur binding site (TATTGAAAATNATTTTCAATA) (23). There are two putative Zur binding sites upstream of cmrA (Fig. 6). The first site overlaps the start codon (running from +19 to −2) and matches the M. tuberculosis consensus site at 16 out of 20 conserved positions. The second site is located 26 bp upstream of the cmrA start codon and is a 65% match to the consensus. We identified a single putative Zur binding site located 5 bp upstream of the zrg start codon that matches the consensus sequence at 13 of 20 positions (Fig. 6). Finally, upstream (70 bases from the start codon) of troA there is a single putative Zur binding site (14 of 20 bases identical). The locations of the putative Zur binding sites upstream of cmrA, zrg, and troA are consistent with the notion that they overlap the promoter sequences in these regions.
In summary, we have characterized zinc-dependent Zur regulation of three genes and the interplay between zinc and other metals in controlling transcription in C. diphtheriae. The positions of the putative binding sites for Zur, DtxR, and MntR in the promoter regions of zrg, cmrA, and troA is suggestive that the regulators act at these promoters. DNA-binding assays are in progress to confirm the roles of each regulator. In addition, we characterized the transcription of zur and observed that, unlike most other fur family genes, zur is repressed under low-zinc conditions in a process requiring Zur (in strain NCTC13129). Finally, we observed increased sensitivity of the C. diphtheriae zur mutant strain to H2O2 stress and, although the exact mechanism of this sensitivity is unknown, this indicates a role for Zur both in metal homeostasis and in protection against host defenses that utilize oxygen radicals to kill pathogenic bacteria.
Acknowledgments
We thank Mark Strauch, Mark Oram, and Kelley Hovis for their careful reading and helpful comments of the manuscript and Dean Dessem for assistance with statistical analysis.
This study was supported by research grant NIH/NIAID K22 AI60882 to D.M.O.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 2.Barksdale, L., and M. Pappenheimer, Jr. 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard, S. J., M. N. Hughes, and R. K. Poole. 1995. Inhibition of the cytochrome bd-terminated NADH oxidase system in Escherichia coli K-12 by divalent metal cations. FEMS Microbiol. Lett. 131205-210. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, L. A., N. D. King, C. A. Kunkle, and M. P. Schmitt. 2005. Analysis of a heme-dependent signal transduction system in Corynebacterium diphtheriae: deletion of the chrAS genes results in heme sensitivity and diminished heme-dependent activation of the hmuO promoter. Infect. Immun. 737406-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, T. M., and W. J. Bettger. 1990. The physiological role of zinc as an antioxidant. Free Radic Biol. Med. 8281-291. [DOI] [PubMed] [Google Scholar]
- 6.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27131-143. [DOI] [PubMed] [Google Scholar]
- 7.Campoy, S., M. Jara, N. Busquets, A. M. Perez De Rozas, I. Badiola, and J. Barbe. 2002. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 704721-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canneva, F., M. Branzoni, G. Riccardi, R. Provvedi, and A. Milano. 2005. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. J. Bacteriol. 1875837-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 316516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, J. E. 1998. Zinc enzymes. Curr. Opin. Chem. Biol. 2222-234. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, K. L., J. L. Farrant, P. R. Langford, and J. S. Kroll. 2003. Bacterial [Cu,Zn]-cofactored superoxide dismutase protects opsonized, encapsulated Neisseria meningitidis from phagocytosis by human monocytes/macrophages. Infect. Immun. 711604-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1816223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaballa, A., and J. D. Helmann. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45997-1005. [DOI] [PubMed] [Google Scholar]
- 14.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 1805815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 1846508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hantke, K. 2001. Bacterial zinc transporters and regulators. Biometals 14239-249. [DOI] [PubMed] [Google Scholar]
- 17.Hardham, J. M., L. V. Stamm, S. F. Porcella, J. G. Frye, N. Y. Barnes, J. K. Howell, S. L. Mueller, J. D. Radolf, G. M. Weinstock, and S. J. Norris. 1997. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene 19747-64. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, R. K. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181(Suppl. 1)S156-S167. [DOI] [PubMed] [Google Scholar]
- 19.Huang, D. L., D. J. Tang, Q. Liao, H. C. Li, Q. Chen, Y. Q. He, J. X. Feng, B. L. Jiang, G. T. Lu, B. Chen, and J. L. Tang. 2008. The Zur of Xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res. 364295-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jobling, M. G., and R. K. Holmes. 2000. Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system. Proc. Natl. Acad. Sci. USA 9714662-14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloosterman, T. G., M. M. van der Kooi-Pol, J. J. Bijlsma, and O. P. Kuipers. 2007. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 651049-1063. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. W., and J. D. Helmann. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20485-499. [DOI] [PubMed] [Google Scholar]
- 23.Maciag, A., E. Dainese, G. M. Rodriguez, A. Milano, R. Provvedi, M. R. Pasca, I. Smith, G. Palu, G. Riccardi, and R. Manganelli. 2007. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milano, A., M. Branzoni, F. Canneva, A. Profumo, and G. Riccardi. 2004. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155192-200. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Mills, D. A., B. Schmidt, C. Hiser, E. Westley, and S. Ferguson-Miller. 2002. Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J. Biol. Chem. 27714894-14901. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, J. R., J. L. Michel, and M. Teng. 1978. Evidence that the regulation of diphtheria toxin production is directed at the level of transcription. J. Bacteriol. 135511-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2002. Construction and characterization of transposon insertion mutations in Corynebacterium diphtheriae that affect expression of the diphtheria toxin repressor (DtxR). J. Bacteriol. 1845723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oram, D. M., A. D. Jacobson, and R. K. Holmes. 2006. Transcription of the contiguous sigB, dtxR, and galE genes in Corynebacterium diphtheriae: evidence for multiple transcripts and regulation by environmental factors. J. Bacteriol. 1882959-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oram, M., J. E. Woolston, A. D. Jacobson, R. K. Holmes, and D. M. Oram. 2007. Bacteriophage-based vectors for site-specific insertion of DNA in the chromosome of corynebacteria. Gene 39153-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 2922488-2492. [DOI] [PubMed] [Google Scholar]
- 32.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 1009912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patzer, S. I., and K. Hantke. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 1834806-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 281199-1210. [DOI] [PubMed] [Google Scholar]
- 35.Pennella, M. A., and D. P. Giedroc. 2005. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals 18413-428. [DOI] [PubMed] [Google Scholar]
- 36.Schafer, A., J. Kalinowski, and A. Puhler. 1994. Increased fertility of Corynebacterium glutamicum recipients in intergeneric matings with Escherichia coli after stress exposure. Appl. Environ. Microbiol. 60756-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, M. P. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J. Bacteriol. 1846882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 591899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon, R., U. Reifer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1784-791. [Google Scholar]
- 40.Smith, K. F., and D. M. Oram. Corynebacteria (including diphtheria), in press. In M. Schaechter (ed.), The encyclopedia of microbiology, 3rd ed. Elsevier, New York, NY.
- 41.Sun, H. W., and B. V. Plapp. 1992. Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J. Mol. Evol. 34522-535. [DOI] [PubMed] [Google Scholar]
- 42.Tai, S. P., A. E. Krafft, P. Nootheti, and R. K. Holmes. 1990. Coordinate regulation of siderophore and diphtheria toxin production by iron in Corynebacterium diphtheriae. Microb. Pathog. 9267-273. [DOI] [PubMed] [Google Scholar]
- 43.Tang, D. J., X. J. Li, Y. Q. He, J. X. Feng, B. Chen, and J. L. Tang. 2005. The zinc uptake regulator Zur is essential for the full virulence of Xanthomonas campestris pv. campestris. Mol. Plant-Microbe Interact. 18652-658. [DOI] [PubMed] [Google Scholar]
- 44.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 501429-1438. [DOI] [PubMed] [Google Scholar]