Abstract
Mycobacterium tuberculosis protein pairs Rv1246c-Rv1247c, Rv2865-Rv2866, and Rv3357-Rv3358, here named RelBE, RelFG, and RelJK, respectively, were identified based on homology to the Escherichia coli RelBE toxin:antitoxin (TA) module. In this study, we have characterized each Rel protein pair and have established that they are functional TA modules. Overexpression of individual M. tuberculosis rel toxin genes relE, relG, and relK induced growth arrest in Mycobacterium smegmatis; a phenotype that was completely reversible by expression of their cognate antitoxin genes, relB, relF, and relJ, respectively. We also provide evidence that RelB and RelE interact directly, both in vitro and in vivo. Analysis of the genetic organization and regulation established that relBE, relFG, and relJK form bicistronic operons that are cotranscribed and autoregulated, in a manner unlike typical TA modules. RelB and RelF act as transcriptional activators, inducing expression of their respective promoters. However, RelBE, RelFG, and RelJK (together) repress expression to basal levels of activity, while RelJ represses promoter activity altogether. Finally, we have determined that all six rel genes are expressed in broth-grown M. tuberculosis, whereas relE, relF, and relK are expressed during infection of human macrophages. This is the first demonstration of M. tuberculosis expressing TA modules in broth culture and during infection of human macrophages.
Mycobacterium tuberculosis, the etiologic agent of tuberculosis, persists as one of the leading causes of death from a single infectious agent, accounting for an estimated 1.6 million deaths globally in 2006 (48). According to the World Health Organization, one person is newly infected by M. tuberculosis every second, resulting in one-third of the world's population being infected with the tuberculosis bacilli. With an increase in the incidence of drug-resistant M. tuberculosis and the desire to develop more effective antimycobacterial therapies, it is essential that this infectious agent be thoroughly studied.
As an intracellular pathogen that resides within human macrophages and dendritic cells, M. tuberculosis encounters challenging environmental conditions during host aerosolization, phagocytosis, active growth, latency, and reactivation. In general, bacteria exposed to a plethora of environments possess molecular responses that regulate the degradation of defective or unnecessary proteins and mRNA molecules. One well-described quality control mechanism of Escherichia coli involves toxin-antitoxin (TA) modules, protein pairs initially characterized as plasmid-addiction systems, which induce a postsegregational killing (PSK) program (8, 13, 23). In addition to PSK, TA modules have been associated with bacterial programmed cell death (PCD) and programmed cell survival (PCS) or persistence (2, 13, 24, 25, 27-29, 42). In bacteria, 11 different TA gene families located on the chromosome and plasmids have been identified: relBE, hipBA, ccdAB, mazEF, higBA, parDE, vapBC, phd/doc, dinJ/yafQ, yefM/yeoB, and ω-ɛ-ζ (36).
Generally, TA modules are defined as protein pairs where one protein, the toxin, is toxic to or inhibits the growth of the bacterial cell, and the second protein, the antitoxin, binds to and neutralizes the toxin's inhibitory effects. Typically, toxin-antitoxin pairs form stable complexes under normal growth conditions, while under stress or unfavorable growth conditions (starvation, antibiotic exposure, DNA damage, and/or translation/transcription inhibition), degradation of the antitoxin results in immediate toxin release, allowing the toxin to exert its effect on the cell (4, 6, 7). In addition, TA modules have other defining characteristics including: (i) organization—the antitoxin gene is upstream of the toxin gene, often overlapping or separated by a small intergenic region; (ii) TA genes are cotranscribed and, in most cases, cotranslated; (iii) regulation—the antitoxin autoregulates expression of the TA operon at the level of transcription, with the toxin acting as a corepressor of expression when bound to its cognate antitoxin; and (iv) the antitoxin is labile, while the toxin protein is much more stable.
relBE and mazEF are by far the best-studied TA modules in E. coli. The relB gene was identified in E. coli by mutations that conferred the “delayed-relaxed response,” characterized by the synthesis of stable RNA (tRNA and rRNA) molecules following amino acid starvation (10, 11). Since then, RelBE has been shown to promote a reversible cell cycle arrest program under conditions of starvation, by dramatically reducing the levels of translation through the cleavage of translating mRNAs in a sequence-specific manner, with a preference for stop codons (UAG > UAA > UGA), codons adjacent to the start codon, and codons with a G or C in the third position (38). Cell cycle arrest induced by relE overexpression increased the survival of E. coli 10- to 10,000-fold after treatment with cefotaxime, ofloxacin, tobramycin, and other antibiotics that target a variety of cell components (25). M. tuberculosis possesses three relBE-like gene pairs (36), and there is a high percentage of sequence similarity between M. tuberculosis and E. coli RelBE proteins. Since E. coli RelBE are involved in the survival of E. coli following stress conditions similar to what M. tuberculosis may encounter during infection (such as antibiotic treatment and nutrient deprivation), it is tempting to speculate that M. tuberculosis proteins homologous to E. coli RelBE function in a similar manner.
A whole-genome search of TA modules revealed an interesting phylogenetic pattern of TAs among 126 sequenced prokaryotic organisms (36). Of the 671 TAs found, 23% belonged to the relBE family and were present in bacteria and Archaea (36). Pandey and Gerdes observed that obligate host-associated organisms such as Mycobacterium leprae do not retain TA loci, whereas many TA modules were found in organisms that grow in nutrient-limiting environments. The most striking examples were Nitrosomonas europaea and M. tuberculosis, which encode 43 and 60 putative TA loci, respectively (Ken Gerdes, unpublished data). Pandey and Gerdes proposed that the number of TA loci is correlated with cell growth rate, such that slow-growing organisms characterized by low translation rates would benefit from having multiple TA loci (36). Since M. tuberculosis is a slow-growing obligate human pathogen, the involvement of TA modules in M. tuberculosis physiology is an intriguing possibility.
The goal of the studies presented here was to characterize three putative relBE gene pairs—Rv1246c-Rv1247c, Rv2865-Rv2866, and Rv3357-Rv3358—as bona fide TA modules. Since these genes are not alleles of a single gene but are separate genes occupying different sites on the chromosome, the gene pairs have been named relBE (Rv1247c-Rv1246c), relFG (Rv2865-Rv2866), and relJK (Rv3357-Rv3358) according to recommendations for nomenclature in bacterial genetics (9). The genetic organization and regulation were examined for relBE, relFG, and relJK, and it was concluded that each gene pair forms an autoregulatory operon, with regulation patterns different than typical TA modules. Importantly, it was determined that each of the three Rel toxins inhibits growth of Mycobacterium smegmatis when overexpressed in the absence of their cognate antitoxins, with growth inhibition completely reversed when both toxin and antitoxin were overexpressed, indicating toxin neutralization. We also show that RelB and RelE bind directly to each other, both in vitro and in vivo. Finally, in addition to demonstrating that all six rel genes are expressed in M. tuberculosis grown in liquid culture, the data presented here demonstrate for the first time that rel toxin-antitoxin genes are expressed in M. tuberculosis during infection of human macrophages.
MATERIALS AND METHODS
Media and chemicals.
M. tuberculosis and M. smegmatis were grown at 37°C in Middlebrook 7H9 liquid medium or on Middlebrook 7H10 agar (Difco) supplemented with ADS (0.5% bovine serum albumin-fraction V, 0.2% dextrose, 0.85% saline) enrichment, 0.05% Tween 80 (Tw) with or without 0.2% glycerol. For rel expression studies, M. smegmatis was grown at 37°C in Luria-Bertani (LB) broth supplemented with 0.05% Tw or on LB agar plates. For mycobacterial protein fragment complementation assays (44), M. smegmatis transformants were plated on Middlebrook 7H11 agar (Difco) supplemented with 0.5% glycerol, 0.5% glucose, and 0.2% Tw and grown at 37°C. E. coli was grown in LB broth or on LB agar plates at 37°C. The following concentrations of antibiotics or inducing supplement were added when appropriate: kanamycin (Kan), 50 μg/ml; hygromycin (Hyg), 50 μg/ml; ampicillin (Amp), 100 μg/ml; trimethoprim (TRIM), 15 μg/ml; IPTG (isopropyl-β-d-thiogalactopyranoside), 1 mM; and anhydrotetracycline (ATc), 200 ng/ml.
Strains and plasmids.
E. coli strain JM109 was used as a host for plasmid constructions (Stratagene, La Jolla, CA). T7 Express lysY-competent E. coli cells were used for protein purification (New England Biolabs, Ipswich, MA) and production of whole-cell lysates (WCL) for far-Western analysis. M. tuberculosis H37Rv (ATCC no. 25618) and M. smegmatis mc2155 (obtained from William R. Jacobs, Jr., Albert Einstein College of Medicine, New York, NY) were used in the present study. Table 1 summarizes the mycobacterial strains and plasmids used for the studies presented here. Detailed plasmid constructions are available upon request to S. B. Korch.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or featuresa | Source or referenceb |
|---|---|---|
| Strains | ||
| M. tuberculosis H37Rv | Virulent laboratory strain | ATCC |
| E. coli | ||
| JM109 | e14− (McrA−) recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) | Stratagene |
| T7 Express lysY | miniF lysY (Camr)/fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::mini-Tn10-Tets)2 [dcm] R(zgb-210::Tn10-Tets) endA1 Δ(mcrC-mrr)114::IS10 | NEB |
| SK140 | E. coli T7 Express lysY with pET28a(+) | This study |
| SK141 | E. coli T7 Express lysY with pET28::relB | This study |
| M. smegmatis mc2155 | ept-1 | 45 |
| LIX22 | mc2155 with pJEM15 | This study |
| LIX23 | mc2155 with pJEM15::PrelBE | This study |
| LIX24 | mc2155 with pJEM15::PrelBE-relB | This study |
| LIX25 | mc2155 with pJEM15::PrelBE-relBE | This study |
| LIX26 | mc2155 with pJEM15::PrelFG | This study |
| LIX27 | mc2155 with pJEM15::PrelFG-relF | This study |
| LIX28 | mc2155 with pJEM15::PrelFG-relFG | This study |
| LIX29 | mc2155 with pJEM15::PrelJK | This study |
| LIX30 | mc2155 with pJEM15::PrelJK-relJ | This study |
| LIX31 | mc2155 with pJEM15::PrelJK-relJK | This study |
| LIX32 | mc2155 with pYA1611 | This study |
| LIX33 | mc2155 with pYA1611::relE | This study |
| LIX34 | mc2155 with pYA1611::relBE | This study |
| LIX35 | mc2155 with pYA1611::relG | This study |
| LIX36 | mc2155 with pYA1611::relFG | This study |
| LIX37 | mc2155 with pYA1611::relK | This study |
| LIX38 | mc2155 with pYA1611::relJK | This study |
| Plasmids | ||
| pJEM15 | Kanr, pAL5000 replicon, cII-lacZ | 47 |
| pJEM15::PrelBE | PrelBE in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelBErelB | PrelBE + relB in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelBErelBE | PrelBE + relBE in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelFG | PrelFG in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelFGrelF | PrelFG + relF in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelFGrelFG | PrelFG + relFG in ApaI-SphI sites of pJEM15 | This study |
| pJEM15::PrelJK | PrelJK in ApaI-BamHI sites of pJEM15 | This study |
| pJEM15::PrelJKrelJ | PrelJK + relJ in ApaI-BamHI sites of pJEM15 | This study |
| pJEM15::PrelJKrelJK | PrelJK + relJK in ApaI-BamI sites of pJEM15 | This study |
| pSE100 | Hygr, Pmyc1tetO promoter, Myc ori | 19 |
| pYA1611 | tetR gene in NotI site of pSE100 | This study |
| pYA1611::relE | relE gene in PstI site of pYA1611 | This study |
| pYA1611::relBE | relBE gene in PstI site of pYA1611 | This study |
| pYA1611::relG | relG gene in PstI site of pYA1611 | This study |
| pYA1611::relFG | relFG gene in PstI site of pYA1611 | This study |
| pYA1611::relK | relK gene in PstI site of pYA1611 | This study |
| pYA1611::relJK | relJK gene in PstI site of pYA1611 | This study |
| pSH276 | rv0195 gene in pQE40ΔDHFR | 20 |
| pSH337 | clpC gene in pGEM-T | 20 |
| pSH317 | devR gene in pQE40ΔDHFR | 20 |
| pSH281 | rv2027c gene in pQE40ΔDHFR | 20 |
| pSH334 | regX3 gene in pZERO/Km | 20 |
| pGEM-T | Ampr, 3′-T overhang for PCR product ligation | Promega |
| pGEM::relE | relE gene in pGEM-T | This study |
| pGEM::relG | relG gene in pGEM-T | This study |
| pGEM::relK | relK gene in pGEM-T | This study |
| pGEM::relB | relB gene in pGEM-T | This study |
| pGEM::relF | relF gene in pGEM-T | This study |
| pGEM::relJ | relJ gene in pGEM-T | This study |
| pGEX-4T-2 | GST fusion expression vector | GE Healthcare |
| pGEX::relB | relB in EcoRI-XhoI sites of pGEX-4T-2 | This study |
| pGEX::relE | relE in EcoRI-XhoI sites of pGEX-4T-2 | This study |
| pET28a(+) | Kanr, T7 lac promoter-operator, N-terminal His6 | Novagen |
| pET28::relB | relB in NcoI-BamHI sites of pET28a(+) | This study |
| pUAB100 | Hygr, oriM oriE hsp60-GCN4-Gly10-mDHFR [F1,2] | 44 |
| pUAB200 | Kanr, attP int hsp60-GCN4-Gly10-mDHFR [F3] | 44 |
| pUAB300 | Hygr, oriM oriE hsp60-mDHFR [F1,2]-Gly10-MCS | 44 |
| pUAB400 | Kanr, attP int hsp60-mDHFR [F3]-Gly10-MCS | 44 |
| pUAB100::relB | relB in BamHI-ClaI sites of pUAB100, creating RelB[F1,2]-C | This study |
| pUAB200::relE | relE in MfeI-ClaI sites of pUAB200, creating RelE[F3]-C | This study |
| pUAB300::relB | relB in BamHI-ClaI sites of pUAB300, creating RelB[F1,2]-N | This study |
| pUAB400::relE | relE in MfeI-ClaI sites of pUAB400, creating RelE[F3]-N | This study |
GCN4 is the S. cerevisiae leucine-zipper sequence. MCS, multiple cloning site.
ATCC, American Type Culture Collection; NEB, New England Biolabs.
Protein purification of RelB and RelE.
To purify RelB-glutathione S-transferase (GST) and RelE-GST fusion proteins, relB and relE were separately cloned into the expression vector pGEX-4T2 (GE Healthcare, Pittsburgh, PA) to generate pGEX::relB and pGEX::relE, respectively. pGEX::relB or pGEX::relE were separately transformed into T7 Express lysY-competent E. coli cells. Briefly, cultures were grown in LB-Amp broth at 37°C with aeration to an optical density at 600 nm (OD600) of 0.2 to 0.4. Then, 1 mM IPTG was added to induce gene expression, cultures grown for an additional 3 h, and cells were harvested by centrifugation. Cell pellets were resuspended in lysis buffer (20 mM Tris [pH 7.5], 0.3 M KCl, 50 mM dithiothreitol), disrupted with a French pressure cell, and centrifuged to obtain clarified lysates. Proteins were isolated by fast-protein liquid chromatography (ÄTKA FPLC; Amersham Biosciences, Sweden) using GSTrap FF columns (GE Healthcare) according to the manufacturer's instructions. Proteins were eluted in elution buffer (50 mM Tris-HCl [pH 9.5], 10 mM reduced glutathione, 1.0 mM phenylmethylsulfonyl fluoride), followed by buffer exchange using either HiTrap desalting columns (for RelB; GE Healthcare) or 5,000- to 250,000-molecular-weight size exclusion columns (for RelE; GE Healthcare). Storage buffer was composed of 50 mM Tris-HCl (pH 8.0) and 20% glycerol. Both proteins were concentrated by using Amicon Ultra-15 centrifugal filter devices (10K MWCO; Millipore, Bedford, MA), and quantified by using the RC DC protein assay (Bio-Rad Laboratories, Hercules, CA). Purified recombinant RelB-GST and RelE-GST were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and stained with SimplyBlue SafeStain (Invitrogen, Carlsbad, CA).
RelB and RelE antibody generation and immunoblot analysis.
Purified RelB-GST or RelE-GST protein (140 μg) was emulsified in TiterMax Gold (Sigma-Aldrich, St. Louis, MO) and used to immunize two New Zealand White rabbits by subcutaneous injections. Rabbits were given booster injections of RelE or RelB antigens at 3-week intervals, and antisera were collected after one or two booster injections, respectively.
M. tuberculosis H37Rv was grown in 7H9-ADS-Tw medium at 37°C to an OD600 of ∼0.7. The culture was pelleted by centrifugation, and cells were washed with 1× phosphate-buffered saline (PBS), pelleted, and then resuspended in 1 ml of 2D-rehydration buffer (Bio-Rad) and mechanically lysed with 0.1-mm Zirconia silica beads by using a FastPrep FP120 bead beater. For Western analysis, 500 ng of purified RelE-GST or ∼50 μg of H37Rv WCL was resolved on an SDS-12% PAGE gel and then transferred onto a nitrocellulose membrane (Bio-Rad Laboratories). Membranes were blocked in 3% skim milk in TBST buffer (25 mM Tris, 140 mM NaCl, 3 mM KCl, 0.05% Tween 20), incubated with anti-RelE (1:15,000) rabbit polyclonal antiserum, and developed with a horseradish peroxidase-labeled anti-rabbit immunoglobulin G (IgG) antibody (Sigma-Aldrich) and LumiGLO chemiluminescent substrate system (KPL, Gaithersburg, MD).
RNA isolation.
M. tuberculosis H37Rv was grown in 7H9-ADS-Tw-glycerol to an OD600 of 0.5 to 0.6, and cells were harvested by centrifugation. Then, 1 ml of RNALater (Ambion, Inc., Texas) was added to prevent mRNA degradation. RNA isolation was performed using TRI-Reagent (Ambion). Briefly, in a 1:1 mixture of 0.1-mm zirconium-silica beads and 1 ml of TRI-Reagent, cells were mechanically lysed by using a FastPrep FP120 bead beater. RNA in the aqueous phase was extracted with chloroform and precipitated with isopropanol. To remove DNA contamination, RNA samples were treated with RNase-free Turbo DNase (Ambion), followed by PCR analysis of 16S DNA to verify the elimination of DNA. RNA was precipitated using sodium acetate (pH 5.2), glycogen, and 100% ethanol; resuspended in RNase-free water; and stored at −80°C. The quality of RNA was assessed by using an Experion system (Bio-Rad Laboratories), and the concentration was determined by using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Delaware).
Northern blot analysis.
To assess whether relBE genes were cotranscribed, total RNA was isolated from M. tuberculosis H37Rv grown in 7H9-ADS-Tw-glycerol to the logarithmic phase. A NorthernMax-Gly kit (Ambion) was used for electrophoresis of purified RNA samples, RNA transfer to a positively charged nylon membrane (BrightStar-Plus; Ambion), membrane-probe hybridization and membrane washing, in accordance with the manufacturer's instructions. Briefly, 30 μg of total RNA was resolved on 1% agarose gels and transferred to a BrightStar-Plus positively charged nylon membrane (Ambion). After transfer, RNA was cross-linked to the membrane by exposure to UV light, prehybridized, and then hybridized at 42°C with denatured end-labeled [γ-32P]ATP-relBE DNA probes to detect relBE mRNA. DNA probes consisted of the entire coding region of relBE and were generated by PCR amplification using the primers 5′-ATGGCTGTTGTCCCACTGGGC-3′ and 5′-TCACTATTAACGTGGCCGGCA-3′. Amplified PCR products were then end labeled with [γ-32P]ATP using T4 polynucleotide kinase according to the manufacturer's instructions (Epicentre). Membranes were exposed to film for 24 h at −80°C and hybridization signals visualized by autoradiography.
To assess whether relBE, relFG, and relJK genes were cotranscribed, mRNA isolated from M. tuberculosis H37Rv grown in 7H9-ADS-Tw-glycerol to the logarithmic phase was converted to cDNA by reverse transcriptase using an iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer's instructions. Using 1 μg of cDNA per reaction, cDNAs were PCR amplified using primer pairs that would anneal to the 3′ end of the antitoxin gene and the 5′ end of the toxin gene to amplify both genes entirely. The following primer pairs were used for amplification: relBE, 5′-ATGGCTGTTGTCCCACTGGGC-3′ and 5′-TCACTATTAACGTGGCCGGCA-3′; relFG, 5′-ATGCGGATACTGCCGATTTCG-3′ and 5′-TTATCACTATCGGCGGTAGAT-3′; and relJK, 5′-ATGAGCATCAGTGCGAGCGAG-3′ and 5′-TTACTATCAGTAGTGGTATCG-3′. For each amplification, M. tuberculosis H37Rv genomic DNA was used as a positive control, whereas RNA without reverse transcription served as a negative control to exclude DNA contamination. All reactions were resolved on a 1% agarose gel.
Analysis of β-galactosidase activity.
Autoregulatory experiments were performed in M. smegmatis using pJEM15, creating operon-cII-lacZ fusions (48). Strains LIX22, LIX23, LIX24, LIX25, LIX26, LIX27, LIX28, LIX29, LIX30, and LIX31 (Table 1) were grown in 7H9-ADS-Tw-glycerol-Kan medium at 37°C with aeration for ∼48 h. Each cell suspension was subcultured into 7H9-ADS-Tw-glycerol-Kan medium, grown at 37°C with aeration to an OD600 of 0.5 to 0.8. β-Galactosidase measurements were performed as previously described and expressed as Miller units (30).
Assay for toxin growth inhibition.
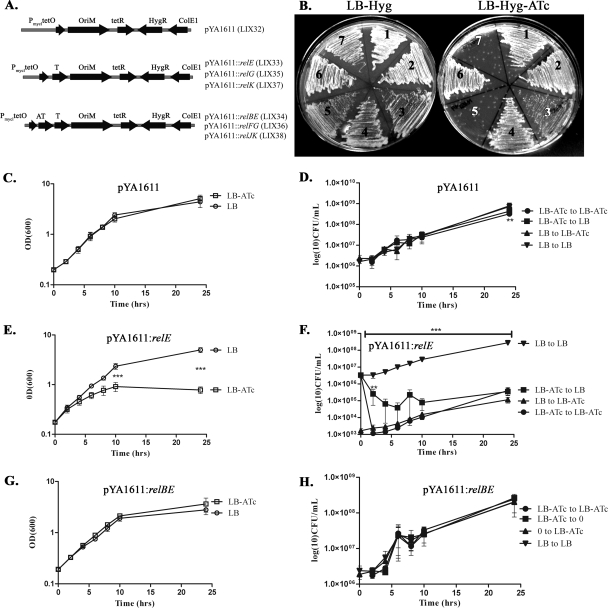
To analyze the ability of relE, relG, and relK to inhibit the growth of M. smegmatis, a TetR-controlled expression system was used (12). For controlled and inducible expression of toxin and antitoxin genes, pYA1611 (described in Table 1 and Fig. 3A) was constructed; this plasmid possesses a Hygr cassette, Myc ori, tetR gene, and Pmyc1tetO promoter to drive gene expression. pYA1611 was transformed into M. smegmatis mc2155 to generate LIX32. All three toxin genes—relE, relG, and relK—were cloned separately into pYA1611, generating pYA1611::relE, pYA1611::relG, and pYA1611::relK and transformed into M. smegmatis mc2155 generating strains LIX33, LIX35, and LIX37, respectively. Similarly, each rel gene pair was separately cloned into pYA1611 generating pYA1611::relBE, pYA1611::relFG, or pYA1611::relJK and transformed into M. smegmatis mc2155 generating strains LIX34, LIX36, and LIX38, respectively. Importantly, in all assays, ATc was used to induce gene expression since it has low toxicity and a high affinity to TetR (17).
FIG. 3.
Effect of toxin and antitoxin overexpression on growth of M. smegmatis. (A) Illustration of expression vectors showing important features used in overexpression experiments. AT, antitoxin; T, toxin. Strains generated by the transformation of each plasmid into M. smegmatis are shown in parentheses. (B) LIX32 to LIX38 strains were restreaked onto LB-Hyg plates (left) or LB-Hyg-ATc plates (right) and incubated at 37°C for 3 days. Sections: 1, LIX32; 2, LIX38; 3, LIX37; 4, LIX36; 5, LIX35; 6, LIX34; 7, LIX33. M. smegmatis strain LIX32(pYA1611) (C and D), LIX33(pYA1611::relE) (E and F), or LIX34(pYA1611::relBE) (G and H) was grown at 37°C with shaking to an OD600 of 0.2 and split into two cultures (time = 0), and 200 ng of ATc/ml was added to one culture to induce relE or relBE expression, while the second culture was used as the uninduced control. At selected time intervals, the OD600 of each culture was measured (C, E, and G), and sample dilutions were plated for CFU (D, F, and H) on LB agar plates to repress gene expression or on LB-ATc agar to continue gene overexpression. The values presented are the averages of three independent experiments; error bars represent the standard deviations. For statistical analysis, two-way analysis of variance with Bonferroni post tests was used to obtain P values for each time point, comparing the various growth conditions to the uninduced control. **, P < 0.01; ***, P < 0.001.
Strains LIX32(pYA1611), LIX33(pYA1611::relE), and LIX34(pYA1611::relBE) were analyzed for growth in the presence (inducing) or absence (no induction) of ATc. Cells were grown at 37°C with aeration in LB-Hyg-Tw to an OD of 1.5 to 2.0. Cells were diluted in LB-Hyg-Tw to an OD600 of 0.01 and grown at 37°C at 120 rpm to an OD600 of ∼0.2. At time zero, cultures were split, and 200 ng of ATc/ml was added to one culture to induce gene expression. Aliquots were taken at the indicated times, the OD600 was measured, and sample dilutions were plated on LB-Hyg or LB-Hyg-ATc. Plates were incubated at 37°C for 3 days. CFU were determined for each of the following conditions: (i) grown in LB-Hyg media and plated on LB-Hyg (LB to LB), (ii) grown in LB-Hyg media and plated on LB-Hyg-ATc (LB to LB-ATc), (iii) grown in LB-Hyg-ATc media and plated on LB-Hyg (LB-ATc to LB), and (iv) grown in LB-Hyg-ATc media and plated on LB-Hyg-ATc (LB-ATc to LB-ATc). Each assay was performed at least three times using triplicate plates at each time point.
To test the effects of relE, relBE, relG, relFG, relK, and relJK overexpression on mycobacterial growth, strains LIX33 to LIX38 were streaked onto LB-Hyg or LB-Hyg-ATc, and plates were incubated at 37°C for 3 days. Growth was compared between inducing and noninducing conditions.
Far-Western analysis.
To detect RelB-RelE protein-interactions in vitro, far-Western analysis was performed. Purified RelB-GST and RelB-His6 generated from WCL served as the “prey” protein, whereas purified RelE-GST was the “bait” protein for far-Western analysis. WCL of T7 Express lysY E. coli cells carrying the empty vector pET28a(+) (Novagen/EMD Chemicals, Inc., Darmstadt, Germany) (SK140) or pET28::relB (SK141) were used as a negative control or source of “prey” protein (RelB-His6), respectively. To generate the induced WCL, SK140 and SK141 cultures were grown at 37°C with aeration to an OD600 of ∼0.4, and then 1 mM IPTG was added to induce relB expression for 3 h. Cells were washed with PBS and centrifuged, and the pellet was resuspended in 1 ml of ProFound lysis buffer (Pierce, Rockford, IL) with 1 mM phenylmethylsulfonyl fluoride. Cell lysates were centrifuged, and the supernatant was filtered (0.2-μm-pore-size filter) to obtain clarified WCL. As negative controls, uninduced SK140 and SK141 WCL were prepared in the same manner, but omitting IPTG. WCL were quantified by using an RC DC protein assay (Bio-Rad Laboratories).
Western and far-Western analyses were performed by using SDS-15% PAGE gels and 1 μg each of purified RelB-GST, RelE-GST, induced SK140 WCL, uninduced SK140 WCL, induced SK141 WCL (source of RelB “prey”), and uninduced SK141 WCL. Samples were immobilized on polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories) for both Western and far-Western analyses. Western blots were developed as described above for immunoblot analysis using anti-RelB serum (1.5:100, to verify the position of RelB-His6; adsorbed against E. coli WCL) or anti-RelE serum (1:15,000, to demonstrate that anti-RelE does not recognize RelB-His6). Far-Western analysis was performed according to the method of Wu et al. (49), except that 10% Tween 80 was used instead of Tween 20. Briefly, 5 μg of RelE-GST protein (“bait”) was incubated with the immobilized WCL membrane and then incubated with anti-RelE (1:15,000) rabbit polyclonal serum to detect RelE bound to RelB-His6. Purified RelB-GST and purified RelE-GST served as a positive control for Western and far-Western analyses since both anti-RelB and anti-RelE rabbit polyclonal serum detect each purified protein due to the GST domain.
Mycobacterial protein fragment complementation.
To assess whether RelB-RelE interact in vivo, the mycobacterial protein fragment complementation (M-PFC) system was used (44). Briefly, when two mycobacterial interacting proteins are independently fused with domains of murine dihydrofolate reductase (mDHFR), functional reconstitution of the two mDHFR domains can occur in mycobacteria, allowing for the selection of mycobacterial resistance against TRIM. relB was cloned into pUAB100 and pUAB300, episomal mycobacterium-E. coli shuttle plasmids, giving rise to fusions of the [F1,2] mDHFR domains to relB (RelB[F1,2]). relE was cloned into the integrating mycobacterium-E. coli shuttle plasmids pUAB200 and pUAB400, resulting in the fusion of the [F3] mDHFR domain to relE (RelE[F3]). pUAB100 and pUAB200 generate C-terminal fusions (subscript “-C”), whereas pUAB300 and pUAB400 generate N-terminal fusions (subscript “-N”). For both pUAB100::relB and pUAB200::relE, the GCN4 domains from pUAB100 and pUAB200 were replaced with relB or relE DNA sequences, respectively. The following protein pairs were cotransformed into M. smegmatis mc2155: RelB[F1,2]-C/RelE[F3]-C, RelB[F1,2]-N/RelE[F3]-N, RelB[F1,2]-C/RelE[F3]-N, and RelB[F1,2]-N/RelE[F3]-C. As negative controls, each pUAB plasmid containing relB or relE was cotransformed with an empty vector (i.e., RelB[F1,2]-C/pUAB200), whereas a positive control was provided by the GCN4[F1,2]/GCN4[F3] (GCN4 leucine zipper, Saccharomyces cerevisiae [39]) interacting domains in pUAB100 and pUAB200. All transformants were plated on 7H11-Kan-Hyg plates and incubated at 37°C for 3 days. Transformants were restreaked onto 7H11-Kan-Hyg and 7H11-Kan-Hyg-TRIM and then incubated at 37°C for 3 to 5 days.
SCOTS analysis.
For analysis of relBE, relFG, and relJK expression during M. tuberculosis infection of human macrophages, selective capture of transcribed sequences (SCOTS) (18, 22) was performed. The M. tuberculosis SCOTS cDNA probes were developed from mRNAs expressed during intracellular growth within human macrophages for 18, 48, or 110 h as previously described (18). For Southern hybridizations with SCOTS probes, plasmids pGEM::relE, pGEM::relG, pGEM::relK, pGEM::relB, pGEM::relF, pGEM::relJ, pSH276, pSH337, pSH317, pSH281, and pSH334 were digested with the appropriate enzymes to isolate experimental genes relE, relG, relK, relB, relF, and relJ and the control genes Rv0195, clpC, devR, Rv2027c, and regX3, respectively (20). DNA was separated by agarose gel electrophoresis and transferred onto Magnacharge nylon transfer membranes (GE Osmonics Labstore, Minnetonka, MN). Transferred DNAs were then hybridized with SCOTS cDNA digoxigenin-labeled probes (18, 48, and 110 h) and detected by anti-digoxigenin-alkaline phosphatase and the CDP-Star chemiluminescent alkaline phosphatase substrate as described by the manufacturer (Roche, Indianapolis, IN). Membranes were exposed to X-ray film at room temperature for 15 to 60 min.
RESULTS
M. tuberculosis encodes three relBE-like gene pairs.
Three M. tuberculosis protein pairs, Rv1246c-1247c, Rv2865-2866, and Rv3357-3358, were identified as proteins homologous to the E. coli RelBE module (36). Compared to the amino acid sequence of E. coli RelB, M. tuberculosis RelB (Rv1247c), RelF (Rv2865), and RelJ (Rv3357) are 39, 37, and 46% similar, respectively. Correspondingly, their identified cognate M. tuberculosis toxins, RelE (Rv1246c), RelG (Rv2866), and RelK (Rv3358) are 41, 45, and 41% similar to E. coli RelE, respectively. Since these six proteins were identified based on amino acid similarity to E. coli RelBE, we sought to characterize each set of M. tuberculosis putative relBE genes as bona fide toxin-antitoxin modules.
Genetic organization and transcriptional analysis of relBE, relFG, and relJK.
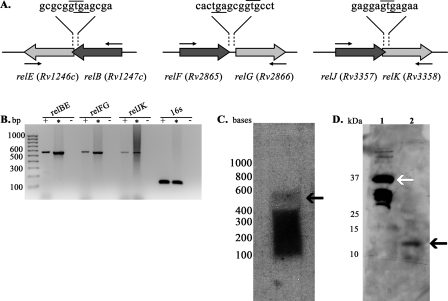
Toxin-antitoxin modules are generally organized into operons with the antitoxin gene located upstream of the toxin gene, often overlapping, or with a small intergenic region between the two genes. Typically, the small base pair overlap between the toxin and antitoxin genes includes the upstream antitoxin stop codon and the downstream toxin start codon, suggesting translational coupling (15). As seen in Fig. 1A, M. tuberculosis gene pairs relBE and relJK overlap (4 bp, GTGA), whereas relFG are separated by a small 3-bp region (GCG). For all three putative TA modules, the antitoxin genes relB, relF, and relJ are located upstream of the proposed toxin genes relE, relG, and relK, respectively. To assess whether each relBE, relFG, and relJK gene pair was cotranscribed, and thus formed individual operons, logarithmic-phase M. tuberculosis H37Rv mRNA was isolated and converted to cDNA by reverse transcriptase. The resulting cDNAs were PCR amplified using primer pairs that anneal to the 3′ end of the antitoxin gene and the 5′ end of the toxin gene (indicated by small arrows in Fig. 1A), or by primers specific for 16S RNA. Bands of the expected size (relBE, 564 bp; relFG, 546 bp; relJK, 534 bp) were observed after amplification of cDNA for relBE, relFG, and relJK (Fig. 1B, “+” lanes), indicating that all three gene pairs are cotranscribed and form operons. Likewise, as a positive control, bands of the expected sizes were observed for 16S RNA amplification using cDNA (Fig. 1B, 16S “+” lane) and for all amplifications using M. tuberculosis H37Rv chromosomal DNA as the template (Fig. 1B, “*” lanes). No amplification product was observed for the negative control reaction (without reverse transcription), eliminating the possibility of DNA contamination (Fig. 1B, “−” lanes). To confirm our cotranscription results, we performed Northern blot analysis using RNA isolated from M. tuberculosis grown to logarithmic phase and probes generated against relBE. As seen in Fig. 1C, a band of the expected size (564 bp, black arrow) was detected, confirming the cotranscription results that relBE does form an operon.
FIG. 1.
Genetic organization of the rel operons. (A) Schematic representation of relBE, relFG, and relJK. The GTG start codons are indicated with a line above the codons, and the TGA stop codons are underlined. Small arrows symbolize the location of the primers used for cotranscriptional analysis in panel B. (B) Transcription analysis by PCR amplification of M. tuberculosis rel genes using cDNA and gene-specific primers to amplify from the 3′ end of the antitoxin gene to the 5′ end of the toxin gene (amplifies both genes and the intergenic region). The positions of the standard DNA size markers are indicated on the left. Each set of three reactions consists of PCR-amplified products from cDNA prepared from logarithmic-phase M. tuberculosis (+), a positive control PCR assay with genomic DNA as the template (*), and a negative control assay without reverse transcriptase (−). The expected sizes for operon amplification are as follows: relBE, 564 bp; relFG, 546 bp; and relJK, 534 bp. (C) Northern blot analysis. Total RNA (30 μg) from M. tuberculosis was probed with an oligonucleotide specific for relBE. The positions of the standard RNA size markers are indicated on the left, and the position of the relBE transcript is indicated by a black arrow, at the expected size (564 bp). (D) Expression of RelE in M. tuberculosis. For Western analysis, purified RelE-GST (∼36 kDa, lane 1) and H37Rv WCL (lane 2) were analyzed. Membranes were incubated with anti-RelE (1:15,000) rabbit polyclonal serum and developed with a horseradish peroxidase-labeled anti-rabbit immunoglobulin G antibody. The positions of the standard protein size markers are indicated on the left, purified RelE-GST is indicated by a white arrow, and RelE from the WCL is indicated by a black arrow (11 kDa).
In addition, the size of each toxin protein is typical. The average size of RelE-like toxin proteins is 92 amino acids (36). In M. tuberculosis, RelE, RelG, and RelK are 97, 87, and 85 amino acids in length, respectively. Thus, the genetic organization and size of M. tuberculosis relBE, relFG, and relJK genes are similar to other TA modules.
Western blot analysis of RelB and RelE in M. tuberculosis cultures.
Cotranscription and Northern analysis established that all three rel gene pairs are expressed and cotranscribed in logarithmic-phase cultures of M. tuberculosis (Fig. 1B and C). To verify the transcriptional expression data and to determine RelB and RelE production, logarithmic-phase M. tuberculosis protein extracts were analyzed by Western blotting using either RelB or RelE polyclonal antiserum generated in rabbits. RelE sera reacted with protein bands from purified RelE-GST, as well as from M. tuberculosis protein samples that correlate with the expected size of RelE (Fig. 1D, lane 2, 11 kDa). The strong band observed in Fig. 1D, lane 1 (30 kDa), below purified RelE-GST (white arrow, 37 kDa), is likely due to degradation of RelE-GST. We also reproducibly detected very low levels of RelB protein at the appropriate size, using anti-RelB (1:15,000) rabbit polyclonal antiserum (data not shown). Since antitoxin proteins are inherently labile, subject to degradation during steady-state growth (1, 4, 6, 15), we anticipate that the low levels of RelB observed were due to protein instability. These results confirm that RelB and RelE are produced during logarithmic growth in M. tuberculosis.
rel operons are autoregulated.
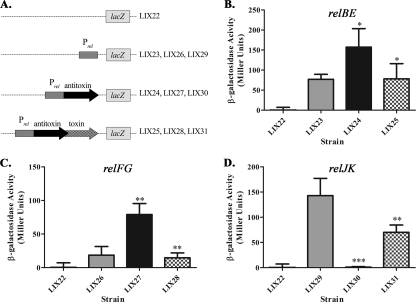
For TA regulation, antitoxins generally act as repressors and toxins act as corepressors of operon expression (15). To determine whether the six M. tuberculosis Rel proteins possess autoregulatory function, we constructed a series of transcriptional lacZ fusions (Table 1 and Fig. 2A) using pJEM15 (47). To study the promoter alone, 300 bp upstream of each antitoxin gene (designated PrelBE, PrelFG, or PrelJK) was cloned upstream of the promoterless lacZ gene. For analysis of antitoxin alone or antitoxin-toxin regulation, the promoter and respective antitoxin or antitoxin-toxin genes were cloned upstream of the promoterless lacZ gene, as shown in Fig. 2A. Each construct was individually transformed into M. smegmatis and analyzed for β-galactosidase activity as described in Materials and Methods. Since the toxin genes cannot be expressed in M. smegmatis in the absence of their cognate antitoxins (shown below), it was not possible to test toxin regulation independently. In contrast to the regulation of TA loci in other bacterial species, both RelB (Fig. 2B, LIX24) and RelF (Fig. 2C, LIX27) induce expression of their respective promoters above the basal level of the promoter. However, analogous with other TA gene pairs, the addition of RelE or RelG (for PrelBE-relBE-lacZ/PrelFG-relFG-lacZ fusions) reduced the β-galactosidase activity to basal levels (Fig. 2B, LIX25, and 2C, LIX28). Interestingly, the regulatory pattern of relJK was very unlike that of relBE or relFG. First, the activity of the relJK promoter (Fig. 2D, LIX29) was much higher than either PrelBE or PrelFG (146 Miller units for PrelJK versus 76 and 18.5 Miller units for PrelBE and PrelFG, respectively). Second, in the presence of RelJ, expression was repressed to levels observed in the promoterless lacZ construct (Fig. 2D, LIX30), an observation akin to antitoxin regulators in other bacteria. Surprisingly, RelJK increased expression 70-fold compared to the activity of PrelJK when only RelJ was present (Fig. 2D, LIX31) but at a level that was still less than that of the promoter alone. Overall, the Rel proteins are able to transcriptionally repress (RelBE, RelFG, RelJ, or RelJK) or activate (RelB or RelF) expression of their respective rel promoters in M. smegmatis, a unique regulatory pattern for TA modules.
FIG. 2.
Regulation of relBE, relFG, and relJK using transcriptional promoter-lacZ fusions in M. smegmatis. (A) Schematic representation of each clone used to generate strains LIX22-LIX31. To analyze the regulation of relBE (LIX23, LIX24, and LIX25) (B), relFG (LIX26, LIX27, and LIX28) (C), and relJK (LIX29, LIX30, and LIX31) (D), M. smegmatis was transformed with empty lacZ vector control (□), promoter-lacZ fusion plasmid (░⃞), promoter-antitoxin-lacZ fusion (▪), or promoter-antitoxin-toxin-lacZ fusion (▩). β-Galactosidase activity is expressed as Miller units (30). The values presented are the averages of three independent experiments; error bars represent the standard deviations. For statistical analysis, two-way analyses of variance with Bonferroni multiple comparison tests were performed using a P value of <0.05. Statistics for promoter-antitoxin-lacZ fusion activity (▪) were derived through comparisons to the promoter-lacZ fusion (░⃞). Statistics for the promoter-antitoxin-toxin-lacZ fusions (▩) were derived through comparison to the promoter-antitoxin-lacZ fusions (▪).
Mycobacterial growth is inhibited by Rel toxins.
The hallmark characteristic of TA modules is toxin-induced growth inhibition when in excess of their cognate antitoxins. To analyze whether M. tuberculosis Rel toxins were capable of inhibiting growth, the tetracycline (Tc)-regulated gene expression system was used to control endogenous expression of rel toxins and antitoxins (12). In this system, in the absence of Tc, the Tc repressor (TetR) binds to tetO and represses gene expression. Conversely, in the presence of Tc, Tc binds to TetR and induces a conformational change that results in dissociation of TetR from tetO, thus inducing the expression of TetR-controlled genes (12). Expression plasmids and strains used for toxin-antitoxin growth analysis are illustrated in Fig. 3A and described in Table 1. Importantly, all overexpression experiments were performed in M. smegmatis mc2155. Since M. smegmatis encodes only two putative TA modules (mazEF and phd/doc) (36), it provides a fairly clean genetic background and eliminates the possibility of cross-interaction between chromosomally encoded rel TAs and those expressed from a plasmid. To examine the effects of toxin overexpression on M. smegmatis growth, LIX33 (relE), LIX35 (relG), and LIX37 (relK) cells were streaked onto LB-Hyg (noninducing) and LB-Hyg-ATc (inducing) plates. As seen in Fig. 3B, overexpression of relE (section 7) and relG (section 5) resulted in the inability of cells to grow (LB-Hyg-ATc) compared to cells not overexpressing the toxin genes (LB-Hyg). relK overexpression (Fig. 3B, section 3, LB-Hyg-ATc) inhibited growth as well, but not to the same extent as relE and relG when overexpressed, indicating that RelK is not as potent a toxin as RelE or RelG in M. smegmatis. Overall, individual overexpression of any of the three M. tuberculosis toxin rel genes is sufficient to inhibit growth in M. smegmatis.
Mycobacterial antitoxins neutralize toxin-induced growth inhibition.
To determine whether RelB, RelF, and RelJ, identified as the cognate antitoxins for RelE, RelG, and RelK, respectively, were capable of neutralizing toxin-induced growth inhibition, the TetR system was used to analyze growth as described above. Compared to the overexpression of relE, relG, and relK in M. smegmatis, overexpression of relBE (LIX34, section 6), relFG (LIX36, section 4), or relJK (LIX38, section 2) did not generate growth defects in M. smegmatis (Fig. 3B, LB-Hyg-ATc). Thus, production of each antitoxin protein was sufficient to neutralize the growth effects induced by their cognate toxin protein.
RelE is a potent toxin protein in M. smegmatis and is effectively neutralized by RelB.
To determine the kinetics of M. smegmatis growth inhibition conferred by RelE and the rescue of growth inhibition by RelB, the TetR expression plasmids were used to examine the growth and CFU patterns of LIX32(pYA1611), LIX33(pYA1611::relE), and LIX34(pYA1611::relBE) under inducing and noninducing conditions. Importantly, no effect on the growth of LIX32 was observed in the presence of 200 ng of ATc/ml, indicating that 200 ng of ATc/ml does not adversely affect M. smegmatis growth (Fig. 3C and D). Conversely, LIX33 growth is dramatically inhibited when relE expression is induced (Fig. 3E, LB-ATc) compared to the uninduced control. At time zero, without the addition of ATC in the liquid media and thus no induction of relE expression, LIX33 exhibits a drop in CFU by 3 orders of magnitude when plated on LB-ATc plates compared to LB plates without inducer (Fig. 3F). This remarkable drop in CFU is attributed to the induced expression of relE, as the presence of 200 ng of ATc/ml was shown to have no effect on M. smegmatis growth (LIX32, Fig. 3C and D). Likewise, overexpressing relE for 2 h in liquid culture reduces the ability of LIX33 to grow on LB plates 10-fold (LB-ATc to LB), while 24 h of relE overexpression induces an 800-fold reduction in LIX33 CFU on LB plates compared to the uninduced control (Fig. 3F). Moreover, after 8 h of induced relE expression in liquid culture followed by relE overexpression on plates (Fig. 3F, LB-ATc to LB-ATc), M. smegmatis exhibits a dramatic 8,000-fold decrease in CFU compared to the uninduced control (Fig. 3F). The results of these experiments demonstrate that RelE is a potent inhibitor of mycobacterial growth.
In agreement with the results shown in Fig. 3B, no effect on M. smegmatis growth was observed 24 h after relBE overexpression in culture (LIX34, Fig. 3G) or on LB-ATc plates (Fig. 3H), with growth patterns analogous to LIX32 (Fig. 3C and D), but remarkably different than LIX33 (Fig. 3E and F). Thus, simultaneous overexpression of relB and relE abolishes mycobacterial growth inhibition conferred when relE is independently overexpressed, verifying the role of RelB as RelE's cognate antitoxin.
RelE induces a reversible dormant state.
Reversible toxin-induced bacterial dormancy has been demonstrated for RelE, MazF, and HipA toxins in E. coli (7, 25, 27, 37). Specifically, RelE or MazF toxin-induced growth arrest was reversible upon the expression of each toxin's cognate antitoxin, due to the ability of RelB or MazE to counteract RelE- or MazF-induced translation inhibition, respectively (37). Moreover, based on LIVE/DEAD analysis, it was determined that the decrease in E. coli's colony-forming ability after hipA induction was not correlated with cell death, but rather bacteriostasis, since the majority of cells (>95%) were viable (27). To address whether M. tuberculosis RelE was also capable of inducing a reversible bacteriostatic state, we compared the growth patterns of LIX33 (M. smegmatis with pYA1611::relE) grown in LB-ATc and then plated on either LB or LB-ATc. Of note, at 2 to 10 h after relE induction (LB-ATc), the same LIX33 population produced 10- to 100-fold more colonies when plated on LB than when plated on LB-ATc (Fig. 3D, compare “LB-ATc to LB” with “LB-ATc to LB-ATc”). Thus, the population of cells unable to form a colony when relE is continually expressed on plates are not dead, but rather, dormant, which is reversible upon inhibition of RelE production by the removal of ATc.
RelB and RelE interact via direct protein-protein interactions in vitro and in vivo.
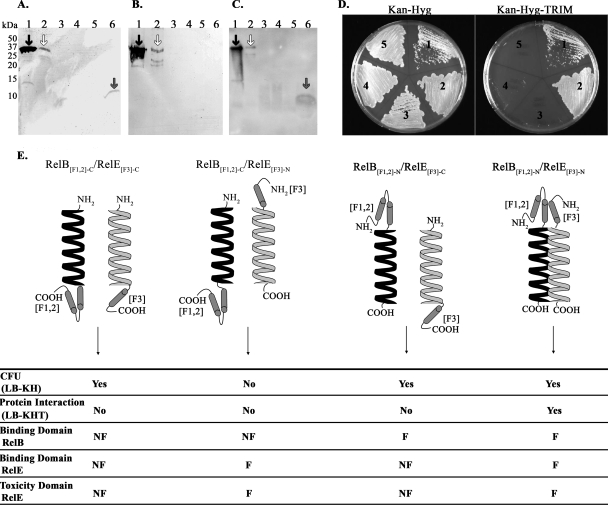
Antitoxins neutralize their cognate toxins through direct protein-protein associations. To ascertain whether RelB and RelE interact directly, far-Western analysis was performed as described in Materials and Methods. First, traditional Western analysis using anti-RelB antibody was performed to identify the position of the “prey” protein, RelB-His6 (Fig. 4A, lane 6, gray arrow, 10 kDa) from the induced SK141 WCL, which was not present in the uninduced SK141 WCL (Fig. 4A, lane 4). Second, as a control for anti-RelE antibody recognition, an additional Western blot was performed on the same set of WCL, using anti-RelE antibody. As expected, anti-RelE antibody recognized purified RelB-GST and RelE-GST (it recognizes both GST fusions; Fig. 4B, lanes 1 and 2), whereas bands were not visualized for any of the WCL at the expected size of RelB-His6, eliminating the possibility of nonspecific anti-RelE antibody interactions (Fig. 4B, lanes 3 to 6). Finally, to detect RelB-RelE binding using far-Western analysis, membranes containing immobilized WCL (SK140 and SK141, induced and uninduced) were incubated with purified RelE-GST “bait” protein and then immunoblotted with anti-RelE rabbit polyclonal serum. Using induced SK141 WCL to produce RelB-His6, anti-RelE antibody binds at the same position as RelB-His6 (∼10 kDa, Fig. 4C, lane 6, gray arrow), indicating an interaction between RelB and RelE in vitro. Importantly, no bands were visualized from the uninduced SK141 WCL (Fig. 4C, lane 4), or from SK140 WCL (Fig. 4C, lanes 3 and 5). Thus, RelB and RelE participate in protein-protein interactions in vitro.
FIG. 4.
RelB and RelE proteins interact directly in vitro and in vivo. For Western (A and B) and far-Western (C) analyses, the following samples were subjected to SDS-15% PAGE gel and transferred to PVDF membranes. Lane 1, purified RelB::GST; lane 2, purified RelE-GST; lane 3, uninduced SK140 WCL (E. coli T7 Express lysY with pET28a); lane 4, uninduced SK141 WCL (E. coli T7 Express lysY with pET28::relB); lane 5, induced SK140 WCL; lane 6, induced SK141 WCL. (A) Western analysis with anti-RelB-GST rabbit polyclonal serum to detect the size of RelB-His6 “prey” protein (∼10 kDa, gray arrow). (B) Western analysis performed with anti-RelE-GST rabbit polyclonal serum to demonstrate that anti-RelE-GST does not interact with RelB-His6 nonspecifically. (C) Far-Western analysis. PVDF membranes were prepared as described in Results, incubated with 5 μg of purified RelE-GST “bait” protein, and then probed with anti-RelE-GST rabbit polyclonal serum. The presence of RelE-GST bound to RelB-His6 is apparent in lane 6, with a band of expected size for RelB-His6 (∼10 kDa, gray arrow). For panels A, B and C, both purified proteins, RelB-GST (∼35 kDa, black arrow) and purified RelE-GST (∼36 kDa, white arrow), are recognized by anti-RelB-GST (A) and anti-RelE-GST (B and C) polyclonal serum, serving as the positive control. (D) Mycobacterial fragment complementation assay. To examine protein interaction, M. smegmatis was cotransformed with positive control pUAB100 + pUAB200 (section 1), RelB[F1,2]-N/RelE[F3]-N (section 2), RelB[F1,2]-C/RelE[F3]-C (section 3), RelB[F1,2]-N/RelE[F3]-C (section 4), and negative control RelB[F1,2]-C/pUAB200 (section 5). Transformants were streaked on 7H11-Kan-Hyg plates and 7H11-Kan-Hyg-TRIM plates to select for protein-protein interactions. (E) Diagram and table illustrating the MPF-C cotransformations, results, and interpretations. The entire RelB protein (black helix) and entire RelE protein (white helix) are fused to complementary mDHFR fragments [F1,2] and [F3], respectively, at the N or C termini. For each cotransformation, results are summarized for the ability to recover successful transformants (CFU), protein interaction as indicated by growth in the presence of TRIM, with inferences from the results regarding RelB and RelE binding and toxicity domains. KH, Kan-Hyg; KHT, Kan-Hyg-TRIM; F, functional; NF, nonfunctional.
The M-PFC assay (44) was used to verify the far-Western results, in addition to testing for RelB-RelE interaction in vivo. In the M-PFC assay, two mycobacterial interacting proteins are independently fused with domains of mDHFR. Interaction between the two proteins of interest results in the functional reconstitution of the two mDHFR domains, thereby allowing for the selection of mycobacterial resistance against TRIM. Figure 4E shows an illustration of four RelB and RelE cotransformations in M. smegmatis. As a control, M. smegmatis was cotransformed with the empty vector pUAB300/RelE[F3]-N or with the empty vector pUAB100/RelE[F3]-C. In agreement with the toxicity of RelE, we were unable to recover colonies after cotransformation of M. smegmatis with the empty vector pUAB300/RelE[F3]-N. However, after cotransformation with the empty vector pUAB100/RelE[F3]-C, M. smegmatis was able to grow but formed very small colonies, suggesting that the toxicity of RelE had been diminished by the C-terminal [F3] fusion. We then performed a series of cotransformations, using different combinations of C- and N-terminal fusion proteins. Unexpectedly, CFU were not obtained when RelB[F1,2]-C/RelE[F3]-N were cotransformed, suggesting that RelB[F1,2]-C was unable to bind to and neutralize RelE[F3]-N, possibly due to the mDHFR [F1,2] fusion at the C terminus of RelB. However, we were able to recover CFU after cotransformation of M. smegmatis with RelB[F1,2]-N/RelE[F3]-N, RelB[F1,2]-C/RelE[F3]-C, and RelB[F1,2]-N/RelE[F3]-C. To test for protein-protein interactions (growth in the presence of TRIM), the above successful cotransformants, along with the positive (pUAB100/pUAB200) and negative (RelB[F1,2]-C/pUAB200) controls, were streaked onto 7H11-Kan-Hyg and 7H11-Kan-Hyg-TRIM plates. As expected, no mycobacterial growth was observed on 7H11-Kan-Hyg-TRIM plates using the RelB[F1,2]-C/pUAB200 plasmid pair (Fig. 4D, section 5), whereas the positive control plasmid pair pUAB100/pUAB200 enabled M. smegmatis to grow in the presence of TRIM (Fig. 4D, section 1). M. smegmatis cotransformed with RelB[F1,2]-N/RelE[F3]-N was able to grow on 7H11-Kan-Hyg-TRIM plates, inferring direct protein interactions between RelB and RelE (Fig. 4D, section 2). This interaction suggests that the unsuccessful cotransformation of RelB[F1,2]-C/RelE[F3]-N (above) was not due to the inability of RelE[F3]-N to bind, since it can interact with RelB[F1,2]-N. Surprisingly, RelB[F1,2]-C/RelE[F3]-C and RelB[F1,2]-N/RelE[F3]-C (Fig. 4D, sections 3 and 4) protein fusions did not interact (no growth in the presence of TRIM) even though they were capable of producing M. smegmatis cotransformants. These results suggest that the C-terminal mDHFR [F3]-RelE fusion disturbs the RelB binding domain since no interaction was observed for RelB[F1,2]-N/RelE[F3]-C compared to RelB[F1,2]-N/RelE[F3]-N. Moreover, the data imply that the C-terminal mDHFR [F3] fusion interferes with RelE's toxicity domain, since RelB[F1,2]-C/RelE[F3]-C transformants were recovered (without RelB-RelE interaction), whereas RelB[F1,2]-C/RelE[F3]-N transformants were never recovered (also without RelB-RelE interaction). Cotransformation and protein interaction results and data interpretations are summarized in Fig. 4E. For schematic purposes only, RelB and RelE are represented by helices in Fig. 4E. This is not meant to suggest that only the helical regions of either protein were fused to mDHFR domains; in fact, the entire RelB or RelE protein was fused to the [F1,2] or [F3] mDHFR domains, respectively. Overall, in vivo, RelB and RelE directly bind, with interaction likely occurring between the free C termini of both proteins.
Analysis of rel gene expression in human macrophages infected with M. tuberculosis.
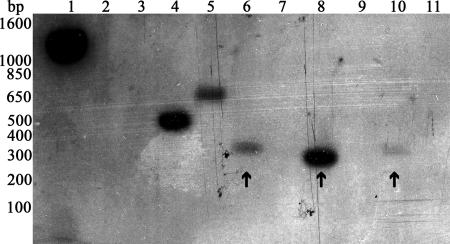
Amplification of each rel operon, as well as Northern and Western blot analyses, provided evidence that rel genes were expressed in broth-grown M. tuberculosis (Fig. 1B, C, and D). To determine whether rel genes are expressed in M. tuberculosis in response to phagocytosis by human macrophages, the selective capture of transcribed sequences (SCOTS) method was used (18, 22). Previously, SCOTS cDNA probes were generated from M. tuberculosis recovered 18, 48, and 110 h postinfection of peripheral blood mononuclear cell-derived human macrophages (18). Southern blot analysis of relB, relE, relF, relG, relJ, and relK using SCOTS cDNA probes from M. tuberculosis grown in human macrophages isolated 18 and 48 h postinfection did not show expression of any of the rel genes (data not shown). However, hybridizations using cDNA probes obtained after 110 h of infection revealed the expression of relE, relF, and relK at the expected sizes (Fig. 5, lanes 6, 8, and 10). Thus, the expression of three rel genes is evident in M. tuberculosis later in infection but is not observed during the early and middle stages of infection (18 and 48 h, respectively).
FIG. 5.
Southern blot hybridizations against rel genes using SCOTS prepared cDNA probes from M. tuberculosis grown in peripheral blood mononuclear cell-derived human macrophages for 110 h (18). Lanes 1 to 5 represent controls; lanes 6 to 11 are experimental. Lane 1, clpC (positive control); lane 2, Rv0195 (negative control); lane 3, Rv2027c (expression at 18 h); lane 4, regX3 (expression at 48 and 110 h); lane 5, devR (expression at 110 h); lane 6, relE; lane 7, relG; lane 8, relK; lane 9, relB; lane 10, relF; lane 11, relJ. The positions of the standard DNA size markers are indicated on the left. The expression of relE, relK, and relF is indicated by black arrows, and they are at the expected sizes of 364, 328, and 352 bp, respectively.
DISCUSSION
M. tuberculosis is an extraordinary pathogen that has evolved several stress response mechanisms enabling it to survive in a variety of environmental conditions. As stress response proteins, TA modules have attracted attention, due to their complex interactions and the debate regarding their role in bacterial physiology. M. tuberculosis encodes a remarkable number of putative TA modules, suggesting TA involvement in M. tuberculosis pathogenesis. Since TA modules are associated with translational maintenance, DNA replication, and bacterial persistence, it is reasonable that TAs may be involved in the persistent lifestyle of M. tuberculosis. On that basis, we sought to characterize three putative relBE modules as bona fide TA gene pairs and investigated their expression during growth in broth culture and in M. tuberculosis-infected human macrophages.
From the studies presented here, we have validated M. tuberculosis gene pairs relBE (Rv1247c-1246c), relFG (Rv2865-2866), and relJK (Rv3357-3358) as TA modules. We have found similarities between M. tuberculosis relBE, relFG, and relJK and other well-characterized E. coli TA modules, along with notable differences. Importantly, individual expression of RelE, RelG, or RelK toxins inhibits M. smegmatis growth, with growth arrest reversed upon expression of their cognate antitoxins, RelB, RelF, or RelJ, respectively. Toxin neutralization by antitoxin proteins suggests direct binding between the protein pairs. Indeed, we have demonstrated that RelB and RelE directly interact in vitro and in vivo using far-Western and M-PFC analysis, respectively. We also provide evidence that suggests that RelB-RelE binding requires the free C termini of both proteins and that RelE toxicity is likely conferred by its C terminus. Significantly, we have established that rel genes are expressed by M. tuberculosis in broth culture, as well as during infection of human macrophages. This is the first demonstration of TA gene expression in M. tuberculosis, both in broth culture and in human macrophages.
Toxin-antitoxin modules are generally organized in operons with the antitoxin gene upstream of the toxin gene, with a small overlap or intergenic region between them. We have shown that relBE, relFG, and relJK form operons with the antitoxin genes (relB, relF, and relJ) located upstream of the toxin genes (relE, relG, and relK), with little or no overlap (Fig. 1A to C). In terms of TA regulation, antitoxins typically function as transcriptional repressors, with the toxin acting as a corepressor of expression (15). We have, in part, observed this pattern of regulation for relJK, with RelJ repressing operon expression (Fig. 2D). However, RelB and RelF represent unique antitoxins as they act as transcriptional activators (Fig. 2B and C). Conversely, RelE and RelG toxins repress expression of their respective operons, in the presence of their cognate antitoxins, RelB and RelF, respectively, to basal levels of activity. From this, RelE and RelG may (i) act as transcriptional repressors when both toxin and antitoxin are in the cell, (ii) interfere with antitoxin binding to the promoter, or (iii) titrate the antitoxin away from the promoter. In E. coli, excess RelE destabilizes RelB2-RelE binding to the relBE operator, resulting in increased operon expression (35). Since M. tuberculosis inhabits a different niche within the human host than E. coli, the regulation differences observed may reflect different levels of toxin and antitoxin proteins in M. tuberculosis than in E. coli. In addition, RelB or RelF (repressor) binding to the promoter may be unstable without the toxin proteins (RelE or RelG), resulting in increased expression, or the absence of the toxin protein (RelE or RelG) is a cue to increase relBE or relFG expression. In either case, the increased RelE or RelG may enable M. tuberculosis to adapt to environmental changes. Overall, the alternative M. tuberculosis rel module regulation patterns suggest differential needs for Rel proteins during the bacillus’ unique pathogenic lifestyle.
All three Rel toxins inhibited mycobacterial growth. However, toxin-induced growth inhibition was differential, with RelK inhibiting mycobacterial growth moderately compared to RelE or RelG. This differential toxin inhibition has also been observed for M. tuberculosis MazF toxins expressed in E. coli. Of seven putative MazF M. tuberculosis toxins, four showed toxicity to E. coli (53). Importantly, growth arrest conferred by M. tuberculosis Rel toxins was completely abolished when both toxin and antitoxin were overexpressed, establishing RelB, RelF, and RelJ as cognate antitoxins to RelE, RelG, and RelK, respectively. Further, we have shown that RelB and RelE proteins directly interact, both in vitro and in vivo (Fig. 4C and D). Of interest, our in vivo protein interaction studies suggest that RelB-RelE binding requires the free C termini of both RelB and RelE and that RelE toxicity is conferred by the C terminus of the protein. In agreement with our interpretation, in E. coli, RelB neutralizes RelE via its C-terminal region (3). Moreover, the archaeon Pyrococcus horikoshii OT3 relE requires the Arg85 residue located in the C-terminal region for RelE-induced translation inhibition (46).
In E. coli, the TA modules hipBA and relBE mediate the development of persister cells (25, 27, 43). Mechanistically, HipA confers persistence by inducing bacteriostasis, a phenotype that is reversed when the hipA cognate antitoxin, hipB, is expressed (27). Similarly, we demonstrated that expression of M. tuberculosis relE in M. smegmatis induces a reversible dormant state to a fraction of the population. The same population of cells overexpressing relE in liquid culture formed CFU at a higher frequency (10- to 100-fold) on plates inhibiting relE expression compared to plates that induced relE expression (Fig. 4F). The mechanism by which relE-induced dormant cells were able to resume growth is unknown, since M. smegmatis does not possess any putative RelB homologues to neutralize RelE. However, the following scenarios would enable growth recovery in the absence of RelB: (i) the amount of RelE produced is insufficient to maintain dormancy; (ii) without de novo RelE synthesis, degradation of RelE would eliminate all RelE protein; or (iii) a fraction of cells is RelE or ATc insensitive. In a similar pattern, after 6 h of hipA overexpression in E. coli, outgrowth of a small fraction of cells was attributed to HipA- or inducer-insensitive cells or to the acquisition of a mutant expression plasmid (27). Overall, Rel toxins induce dormancy and may be involved in the transition into or maintenance of persistence in M. tuberculosis.
The mechanisms required for M. tuberculosis to persist within the human host are still largely undefined. Within pulmonary macrophages and human granulomas the bacilli are potentially exposed to diminished oxygen, reactive oxygen and nitrogen intermediates, and limiting nutrients. Since we hypothesize that relBE, relFG, and relJK are mediators of M. tuberculosis persistence, activated by alterations in their environment, we determined whether the six rel genes were expressed in M. tuberculosis-infected human macrophages. Using SCOTS analysis we identified relE, relK, and relF transcripts in M. tuberculosis-infected human macrophages at 110 h postinfection (Fig. 5). Interestingly, none of the rel genes were expressed at 18 or 48 h postinfection (data not shown), suggesting that Rel proteins are important at later times of infection. One gene from each operon was detected 110 h postinfection (Fig. 5), suggesting that the transcripts are unstable or that the expression of rel genes is driven from multiple promoters. Such complex regulation has been demonstrated for the Mycobacterium bovis BCG Rv3134c/devR/devS operon, with three promoters exhibiting differential activity under hypoxia and nutrient starvation conditions (40). Further, two toxins and one antitoxin are expressed 110 h after macrophage infection. The biological significance of this is unknown, but these results raise questions as to whether cross-neutralization between the Rel toxins and antitoxins occurs. Can RelF bind to RelE or RelK to neutralize their effects, or are RelE and RelK free to bind to their cellular targets, resulting in mycobacterial growth inhibition within the macrophage? Elucidating protein interactions between the three rel gene pairs is critical to understanding how Rel modules function as a family in M. tuberculosis.
Finally, of debate is how a bacterial cell or population of cells could benefit from carrying chromosomally encoded toxin proteins. However, TA modules could function in the following ways: (i) PCD, as initiators of altruistic suicide programs, and (ii) PCS, as macromolecular modulators, regulating DNA replication or protein synthesis during steady-state growth and/or in response to environmental stress. Depending on the experimental design, existing literature supports both hypotheses. First, the PCD model proposes that transcription and translation inhibition, DNA damage, nutritional and oxidative stress, thymine starvation, and high temperature initiate PCD in E. coli though the MazEF TA module (1, 21, 26, 41, 42). In addition, in Myxococcus xanthus, MazF is required for the cell death stage during fruiting body formation and sporulation (34). Alternatively, the PCS model suggests that toxins modulate macromolecular synthesis following nutritional stress (6, 7, 14, 37). Through specific cleavage of mRNA, RelE and MazF inhibit translation, resulting in cell cycle arrest (5, 7, 33, 50), which was reversed upon expression of their cognate antitoxins (37), or through increased production of tmRNA (for tRNA- and mRNA-like) (5, 7). Moreover, the HipA and RelE toxins mediate the development of persister E. coli cells after exposure to lethal concentrations of β-lactam and fluoroquinolone antibiotics (24, 25, 27, 28, 31, 32, 43). Resumption of persister cell growth was dependent upon production of HipB (HipA) and RelB (RelE) antitoxins (25, 27).
In support of PCS, independent overexpression of four putative M. tuberculosis MazF homologues resulted in growth inhibition of E. coli (53). In addition, overexpression of MazF homologue Rv1991c inhibited the growth of M. smegmatis, which was moderately reversed after expression of its cognate antitoxin, Rv1991a (51). Mechanistically, M. tuberculosis MazF-mt3 and MazF-mt7 act as endoribonucleases, targeting the unique sequences CUĈCU/UUĈCU (MazF-mt3) and UĈGCU (MazF-mt7) in single-stranded RNA (52). Conceivably, both MazF proteins could regulate protein expression, and thus growth rate, through differential mRNA degradation.
Although the exact mechanism of RelE-, RelG-, and RelK-induced growth inhibition has yet to be elucidated, the studies presented here validate a role for Rel proteins in M. tuberculosis growth regulation and suggest that Rel proteins are required for survival within human macrophages. Thus, it is plausible that Rel modules function as modulators of PCS and persistence of M. tuberculosis.
Acknowledgments
We are grateful to Sabine Ehrt for use of the pSE100 vector, to Adrie Steyn for use of the M-PFC vectors, and to Shelley Haydel for providing the SCOTS control plasmids. We thank Shelley Haydel and Kenneth Roland for critical review of the manuscript and for helpful suggestions and discussions.
This study was supported by Public Health Service grant AI46428 from the U.S. National Institutes of Health awarded to J.E.C.-C. H.C. was supported by Arizona State University PREP for Biomedical Research and Public Health Service grant GM071798 from the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 936059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 1868295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherny, I., M. Overgaard, J. Borch, Y. Bram, K. Gerdes, and E. Gazit. 2007. Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry 4612152-12163. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, S. K., and K. Gerdes. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 53587-597. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, S. K., and K. Gerdes. 2003. RelE toxins from bacteria and archea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 481389-1400. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 9814328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332809. [DOI] [PubMed] [Google Scholar]
- 8.Couturier, M., E. M. Bahassi, and L. Van Melderen. 1998. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 6269. [DOI] [PubMed] [Google Scholar]
- 9.Demerec, M., E. A. Adelberg, A. J. Clark, and P. E. Hartman. 1965. A proposal for a uniform nomenclature in bacterial genetics. Genetics 5461-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diderichsen, B., and L. Desmarez. 1980. Variations in phenotype of relB mutants of Escherichia coli and the effect of pus and sup mutations. Mol. Gen. Genet. 180429-437. [DOI] [PubMed] [Google Scholar]
- 11.Diderichsen, B., N. P. Fiil, and R. Lavalle. 1977. Genetics of the relB locus in Escherichia coli. J. Bacteriol. 13130-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 5343-70. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3371-382. [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Gossen, M., and H. Bujard. 1993. Anhydrotetracycline, a novel effector for tetracycline controlled gene expression systems in eukaryotic cells. Nucleic Acids Res. 214411-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 9611554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, X. V., M. Monteleone, M. Klotzsche, A. Kamionka, W. Hillen, M. Braunstein, S. Ehrt, and D. Schnappinger. 2007. Silencing essential protein secretion in Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 1894614-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haydel, S. E., and J. E. Clark-Curtiss. 2004. Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages. FEMS Microbiol. Lett. 236341. [DOI] [PubMed] [Google Scholar]
- 21.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 1863663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou, J. Y., J. E. Graham, and J. E. Clark-Curtiss. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect. Immun. 703714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17205-210. [DOI] [PubMed] [Google Scholar]
- 24.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 23013. [DOI] [PubMed] [Google Scholar]
- 25.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 1868172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 1883420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 1883826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korch, S. B., T. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 501199-1213. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muñoz-Gómez, A. J., S. Santos-Sierra, A. Berzal-Herranz, M. Lemonnier, and R. Díaz-Orejas. 2004. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567316. [DOI] [PubMed] [Google Scholar]
- 34.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular myxococcus development. Cell 13255. [DOI] [PubMed] [Google Scholar]
- 35.Overgaard, M., J. Borch, M. Jorgensen, and K. Gerdes. 2008. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69841-857. [DOI] [PubMed] [Google Scholar]
- 36.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45501-510. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112131. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier, J. N., F. X. Campbell-Valois, and S. W. Michnick. 1998. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. USA 9512141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, J. G., C. S. Burbano, C. Nuñez, C. E. González, M. M. Zambrano, M. J. García, and P. Del Portillo. 2008. Rv3134c/devR/devS operon of Mycobacterium bovis BCG is differentially transcribed under “in vitro” stress conditions. Tuberculosis 88273. [DOI] [PubMed] [Google Scholar]
- 41.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 1832041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 1851803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 1703321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, A., D. Mai, A. Kumar, and A. J. C. Steyn. 2006. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. USA 10311346-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snapper, S. B., R. E. Melton, S. Mustafa, T. Keiser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1999. [DOI] [PubMed] [Google Scholar]
- 46.Takagi, H., Y. Kakuta, T. Okada, M. Yao, I. Tanaka, and M. Kimura. 2005. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 12327. [DOI] [PubMed] [Google Scholar]
- 47.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 1766749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO Programme. 2006. Fact sheet no. 104: tuberculosis. World Health Organization, Geneva, Switzerland.
- 49.Wu, Y., Q. Li, and X.-Z. Chen. 2007. Detecting protein-protein interactions by far Western blotting. Nat. Protoc. 23278. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y., J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing, and M. Inouye. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12913-923. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, L., and J. Zhang. 2008. Biochemical characterization of a chromosomal toxin-antitoxin system in Mycobacterium tuberculosis. FEBS Lett. 582710. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, L., S. Phadtare, H. Nariya, M. Ouyang, R. N. Husson, and M. Inouye. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, L., Y. Zhang, J.-S. Teh, J. Zhang, N. Connell, H. Rubin, and M. Inouye. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 28118638-18643. [DOI] [PubMed] [Google Scholar]