Abstract
It is well established that in nature, bacteria are found primarily as residents of surface-associated communities called biofilms. These structures form in a sequential process initiated by attachment of cells to a surface, followed by the formation of matrix-enmeshed microcolonies, and culminating in dispersion of the bacteria from the mature biofilm. In the present study, we have demonstrated that, during growth, Pseudomonas aeruginosa produces an organic compound we have identified as cis-2-decenoic acid, which is capable of inducing the dispersion of established biofilms and of inhibiting biofilm development. When added exogenously to P. aeruginosa PAO1 biofilms at a native concentration of 2.5 nM, cis-2-decenoic acid was shown to induce the dispersion of biofilm microcolonies. This molecule was also shown to induce dispersion of biofilms, formed by Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Streptococcus pyogenes, Bacillus subtilis, Staphylococcus aureus, and the yeast Candida albicans. Active at nanomolar concentrations, cis-2-decenoic acid appears to be functionally and structurally related to the class of short-chain fatty acid signaling molecules such as diffusible signal factor, which act as cell-to-cell communication molecules in bacteria and fungi.
Biofilms are comprised of microorganisms enmeshed in a hydrated polymer matrix attached to a solid surface. Biofilm growth is a leading cause of materials damage, product quality degradation, and risk to public health. Bacterial biofilms are an important cause of chronic inflammatory and infectious diseases in plants and animals. In humans, biofilms have been implicated in chronic otitis media, native valve endocarditis, gastrointestinal ulcers, urinary tract and middle ear infections, and chronic lung infections in patients with cystic fibrosis (8, 11, 13, 15, 16, 29). Unfortunately, the control of biofilm growth and persistence has been problematic due to the enhanced resistance of biofilms to treatment with microbicides and antibiotics when compared to planktonic cells (30).
Biofilm formation has been most intensively studied in the bacterium Pseudomonas aeruginosa, which has been shown to progress through multiple developmental stages, beginning with reversible attachment to a surface, followed by irreversible attachment and the development of microcolonies, which continue to grow to the final stage of development when dispersion occurs, releasing cells into the bulk liquid (27, 32). Bacteria have been shown to display unique phenotypes at each stage of biofilm development and possess properties that are markedly different from planktonic cells of the same species (27, 28, 32, 33, 36, 38). As a behavioral characteristic of bacteria, biofilm dispersion is of major significance because of its promise to provide a mechanism for the control of the growth and persistence of biofilms, particularly in household, medical, and industrial settings.
The search for an extracellular signal responsible for biofilm dispersion has uncovered a range of factors that have been shown to stimulate biofilm disruption. In 2000, Chen and Stewart (6) reported that reactive chemicals (e.g., NaCl, CaCl2, hypochlorite, monochloramine, and concentrated urea), chelating agents, surfactants (e.g., sodium dodecyl sulfate, Tween 20, and Triton X-100), and lysozyme, as well as a number of antimicrobial agents, when added to mixed biofilms of P. aeruginosa and Klebsiella pneumoniae, resulted in the removal of more than 25% of protein from the surface, indicating cell release from the biofilms. Sauer et al. (27) have shown that a sudden increase in the concentration of organic carbon causes bacteria to disaggregate from a biofilm. Thormann et al. (33) reported that a rapid reduction in oxygen could induce biofilm dispersion after cessation of flow in an oxygen-limited growth medium. Other studies have shown that starvation may be a trigger for dispersion (14), that a prophage in P. aeruginosa may mediate cell death and provide a vehicle for cell cluster disaggregation (37), and that nitric oxide may play a role in the biofilm dispersion process (3). Finally, the chelator EDTA has been shown to induce killing and dispersion in P. aeruginosa biofilms (1). Although the mechanism of dispersion induction is unknown in these cases, a common thread throughout these studies is that they induce major perturbations of cellular metabolism and likely also activate stress regulons, which may be involved in biofilm dispersion.
The identification of a cell-to-cell communication molecule responsible for biofilm dispersion has been the focus of a number of researchers over the past decade. Recently, indole has been shown to act as an intercellular messenger, inhibiting biofilm formation in Escherichia coli but enhancing biofilm formation in P. aeruginosa (19, 20). To date, however, indole has not been shown to activate a dispersion response in existing biofilms. Rice et al. (23) described a limited role for N-butanoyl-l-homoserine lactone in modulating detachment, or sloughing, of Serratia marcescens; however, the role of quorum-sensing molecules in biofilm dispersion remains controversial. Dow et al. (10) have characterized a substituted fatty acid messenger, cis-11-methyl-2-dodecenoic acid, called diffusible signal factor (DSF), recovered from Xanthomonas campestris and shown it to be responsible for virulence, as well as induction of the release of endo-β-1,4-mannanase. Intriguingly, DSF was shown to be able to disaggregate cell flocs formed in broth culture by X. campestris, although no activity against extracellular xanthan was detected (10).
In the present study we demonstrate that an unsaturated fatty acid, cis-2-decenoic acid, produced by P. aeruginosa both in batch and biofilm cultures is responsible for inducing a dispersion response in biofilms formed by a range of gram-negative bacteria, including P. aeruginosa, and by gram-positive bacteria. Furthermore, cis-2-decenoic acid was also capable of inducing dispersion in biofilms of Candida albicans, indicating that this molecule has cross-kingdom functional activity.
MATERIALS AND METHODS
Bacterial strains and media.
The microorganisms used in the present study included P. aeruginosa PAO1 from B. H. Holloway, E. coli (ATCC 10798), Proteus mirabilis (ATCC 25933), K. pneumoniae (ATCC 10273), Staphylococcus aureus (ATCC 12602), Streptococcus pyogenes (ATCC 19615), Bacillus subtilis (ATCC 6633), C. albicans (ATCC 20260), and a mixed undefined culture collected on agar plates via airborne contamination. Except where indicated, all experiments were performed in modified EPRI medium containing 0.005% ammonium nitrate, 0.00019% KH2PO4, 0.00063% K2HPO4 (pH 7.0), and 0.001% Hutner salts (7), supplemented with 0.2% glucose. C. albicans was grown in modified EPRI medium supplemented with 0.2% glucose and 0.1% peptone. K. pneumoniae, P. mirabilis, S. aureus, and B. subtilis were grown in modified EPRI medium supplemented with 0.1% peptone. S. pyogenes was grown in 10% brain heart infusion broth.
Preparation of P. aeruginosa spent medium.
To prepare cell-free spent culture medium, 6 ml of an overnight culture of P. aeruginosa PAO1, grown in modified EPRI medium at 30°C, was inoculated into 4 liters of modified EPRI medium, followed by incubation for 10 days at room temperature with continuous stirring. Bacterial cells were sedimented by centrifugation (Sorvall RC 5B Plus centrifuge, GSA Rotor; Thermo Electron Co., Ashville, NC) at 13,000 × g for 15 min at 4°C. The supernatant was removed and filtered under vacuum through a 0.45-μm-pore-size type HA filter (Millipore Co., Billerica, MA) and, subsequently, through a 0.2 μm-pore-size, Acrodisc 32-mm syringe filter (Pall Co., East Hills, NY). Spent medium was stored at 4°C. Prior to use, spent medium was aerated and supplemented with 2 g of glucose liter−1, and its pH was adjusted to neutrality to ensure that starvation, oxygen depletion, or a change in pH was not responsible for the induction of biofilm dispersion.
Preparation of CSM.
Spent culture medium was prepared from 10-day batch cultures of P. aeruginosa PAO1, grown in modified EPRI medium at room temperature, filtered to remove cells, and stored at 4°C. The organic components of spent medium were extracted by adding 80 ml of chloroform to 250 ml of filtered spent medium in a separatory funnel. The chloroform fraction was removed after a separation time of 1 h. Chloroform was evaporated at 40°C using a Rotavapor R-3000 rotary evaporator (Büchi Laboratories, Flawil, Switzerland), and the remaining organic material was resuspended in 6 ml of filtered Nanopure water and evaporated to dryness by using a Speed-Vac evaporator system (Savant Instruments, Inc., Hicksville, NY) or lyophilized. These samples were then resuspended in 6 ml of culture medium or purified water. The final product is referred to as chloroform-extracted spent medium (CSM). Except where indicated, CSM was used in experiments at a final chloroform-extracted organic carbon concentration 125-fold greater than that found in spent medium.
HPLC fractionation of CSM.
CSM was fractionated by high-performance liquid chromatography (HPLC; Varian Prostar model 320; Varian, Inc., Palo Alto, CA) using a C18 Microsorb-mv reversed-phase column (250 by 4.6 mm; Varian, Inc.). The column was loaded with 100 μl of CSM and eluted in an acetonitrile-water gradient (2 to 75%) with a flow rate of 1 ml min−1 for 29 min. Samples were collected every minute, starting at 2 min. HPLC fractions were pooled, concentrated in a Speed-Vac concentrator (Savant Instruments), and resuspended in 0.5 ml of modified EPRI medium or purified water. The active HPLC fraction was found to elute from the column in 75% acetonitrile-25% water. The active fraction of each HPLC separation was determined by microtiter plate dispersion bioassay.
Microtiter plate dispersion bioassay.
Microtiter plate dispersion bioassays were used to test various preparations for their ability to exogenously induce biofilm dispersion. Biofilms were grown on the inside surface of microtiter plate wells by using a semi-batch culture method in which the medium within each well was replaced periodically to reduce the accumulation of native dispersion inducing factors. Biofilms grown in this manner were treated with dispersion inducer or sterile medium to release cells into the bulk liquid and evaluate dispersed cell number by measuring the optical density (OD). Briefly, sterile polystyrene 96-well plates were etched with acetone for 10 s to create a rough surface for the attachment of microbial cells. After drying for 24 h, plates were inoculated with 150 μl/well of overnight culture containing the test organism, previously diluted 1:20 in growth medium, and incubated at 30°C with shaking at 200 rpm. Medium in the wells was replaced every 24 h for 5 days and every 12 h on days 6 and 7. Medium was then replaced after 7 h. Dispersion induction was tested by adding 150 μl of growth medium containing dispersion inducer for 1 h at 30°C or sterile medium as a control. Medium containing dispersed cells was then transferred by pipette to a nonetched microtiter plate, and the OD at 570 nm (OD570) was determined (ELx808 Absorbance Microplate Reader; BioTek Instruments, Inc., Winooski, VT). Treatments consisted of spent medium, CSM, cis-2-decenoic acid, trans-2-decenoic acid, decanoic acid, and DSF at various concentrations. Ethanol (10%) was used as a carrier for fatty acid inducer samples and was determined to have no influence on dispersion. The results from use of this method are meaningful in making comparisons of different treatments and to determine whether dispersion activity is statistically significant.
Dispersion bioassays in biofilm tube reactors.
P. aeruginosa PAO1 biofilm cultures were grown in tube reactors as described previously by Davies et al (9) and Sauer et al. (27). A continuous once-through tube reactor system was configured by using eight silicone reactor tubes (81.5-cm length by 14-mm inner diameter), connected to an eight-roller head peristaltic pump and medium reservoir, via additional silicone tubing. Medium was pumped through the tubing to a closed effluent medium reservoir. The assembled system was sterilized by autoclaving prior to inoculation. The silicone tubes were inoculated by syringe injection through a septum 1 cm upstream from each reactor tube, with 2 ml of overnight cultures of P. aeruginosa (containing ∼108 CFU ml−1). Bacterial cells were allowed to attach (static incubation) to the tubing for 1 h, after which the flow was started at an elution rate of 10.8 ml h−1. Treatments were carried out after 96 h of P. aeruginosa PAO1 biofilm culture. The treatments were performed under continuous and static conditions in triplicate.
Under continuous treatment the influent medium was switched from fresh medium in the test lines to spent medium. Control lines were switched to new lines containing fresh modified EPRI medium. Samples were collected in test tubes on ice and were subsequently homogenized for 30 s at 5,000 rpm with a Tissue-Tearor model 985370 (Biospec Products, Inc.) to ensure the separation of cells. The cell density was determined based on the OD600 with an Ultrospec 3000 spectrophotometer (Amersham Pharmacia Biotech, Inc.).
Under conditions of static treatment, CSM, spent medium, or cis-2-decenoic acid was added in modified EPRI medium as a bolus by syringe injection through the inoculation port, 2 cm upstream from the beginning of the tube reactor, displacing the reactor volume with medium containing inducer. CSM or synthetic dispersion inducer (e.g., cis-2-decenoic acid) was prepared in modified EPRI and added. After 1 h of exposure under nonflowing conditions, an 81.5-cm length of each silicone tube reactor was cut out, the liquid fraction containing released biofilm cells, and the biofilm fractions were collected in test tubes on ice as described previously (27). Cell numbers were determined by spread plate method on standard plate count agar medium (Difco, Detroit, MI) or by evaluating the OD600. The dispersion efficacy was calculated by using either the OD adjusted to the cell number or by viability measurements as follows: dispersion efficacy = (the number cells from bulk liquid × 100)/(the number of cells from bulk liquid + the number of cells from biofilm).
Microscopic analysis.
A continuous-culture once-through flow cell was configured to observe the growth and development of biofilms attached to a glass substratum. The flow cell was constructed of anodized aluminum containing a chamber 1.0 mm by 1.4 cm by 4.0 cm, with a glass top and bottom. Sterile modified EPRI medium was pumped from a 10-liter vessel through silicone tubing to the flow cell by using a Masterflex 8-roller-head peristaltic pump at a flow rate of 0.13 ml min−1. Flow through the chamber was laminar, with a Reynolds number of 0.17, having a fluid residence time of 4.3 min. Medium leaving the flow cell was discharged to an effluent reservoir via silicone tubing. The entire system was closed to the outside environment but maintained in equilibrium with atmospheric pressure by a 0.2-μm-pore-size gas-permeable filter fitted to each vessel. Log-phase P. aeruginosa (∼108 CFU ml−1) were inoculated as a 3.0-ml slug dose through a septum 4 cm upstream from the flow cell, under flowing conditions. Cells attached to the inner surface of the glass substratum were viewed by transmitted light or epi-UV illumination using an Olympus BX60 microscope and a ×100 magnification A100PL objective lens or a ×50 magnification ULWD MSPlan long-working-distance Olympus objective lens. All images were captured by using a Magnafire cooled three-chip charge-coupled device camera (Optronics, Inc., Galena, CA) and stored as separate digital files for subsequent retrieval and analysis. P. aeruginosa was grown in the flow cell for up to 12 days. Previous work in our laboratories has shown P. aeruginosa to develop steady-state biofilms following a continuous culture period of 7 to 9 days. Steady state is defined by no change in effluent cell counts (in CFU) resulting from detached biofilm cells; in steady state, growth of the biofilm is balanced by the loss of cells through dispersion or detachment. Individual cell clusters were examined during the course of each experiment and assigned grid coordinates, which were reexamined periodically during the course of the experiments. Size measurements were taken of random microcolonies by locating the microcolony nearest to a randomly selected microscope stage coordinate. Each microcolony was measured to determine its height by focusing from the substratum through to its apex and its width by measurement at the base of the microcolony using a stage micrometer. Microcolonies were defined as cells embedded within an exopolysaccharide matrix attached to the substratum and lacking motility; void areas were determined by the observation of free-swimming bacteria within a space inside a microcolony.
Inhibition of biofilm development.
A flow cell was used to culture bacteria on the surface of a glass substratum (as described above). Biofilms of P. aeruginosa were grown in modified EPRI medium at room temperature in the presence or absence of CSM (diluted 1:125 to match the concentration of the CSM found in spent medium). During the course of the experiment, the total cell coverage of the bacteria on the surface and average biofilm thickness were determined by counting 20 microscope fields using a ×50 ULWD MSPlan objective lens for each time point. Using the image analysis software, ImagePro Plus (Media Cybernetics, Bethesda, MD), the total area of cells per cm2 and the average maximum height of 20 random microcolonies was determined at 72 and 99 h of growth. Control samples were grown and tested in the presence of modified EPRI medium with no added CSM.
Spectral analysis of P. aeruginosa CSM and cis-2-decenoic acid.
All CSM samples prepared in purified water were lyophilized and resuspended in appropriate carriers for each spectroscopic analysis. CSM controls in all experiments consisted of CSM HPLC products that did not induce dispersion, as determined by microtiter plate dispersion bioassay, and carrier solution not containing CSM.
Mass spectroscopy.
Samples were resuspended in carrier solution (50% water, 50% methanol, and 0.01% formic acid). Mass spectroscopy was performed by using a high-performance, hybrid quadrupole time-of-flight mass spectrometer (QSTAR XL Hybrid LC/MS/MS system; Applied Biosystems, Foster City, CA) in positive ion mode, at room temperature, with an IonSpray source for the API 150EX, API 3000, and QSTAR systems (Applied Biosystems). The data were analyzed by using Analyst QS (version 1.1).
NMR.
Samples of CSM and cis-2-decenoic acid were resuspended in 1 ml of deuterated acetonitrile and inserted into a thin-walled nuclear magnetic resonance (NMR) sample tube (VWR). Analyzes was performed in a 300 MHz Proton NMR-Bruker AC 300 (Bruker Daltonics, Inc., Vilarica, MA). Spectra were accumulated for 24 h.
GC-MS.
Samples of CSM and concentrations of cis-2-decenoic acid from 0.01 to 10 mg ml−1 were resuspended in 2 ml of acetonitrile. A three-step sequential hexane extraction was performed to remove soluble organic sample material. Hexane was evaporated to dryness in a water bath (55 to 70°C). Puridine (250 μl) was subsequently added to solubilize samples for injection into GC. Spectra were obtained with a Shimadzu QP5050A gas chromatography-mass spectrometry (GC-MS) system, using helium as a carrier and a Restek (Columbia, MD) XTI-5 GC column (30 m, 0.25 mm [inner diameter], 0.25-μm film thickness) with a flow rate of 1 ml min−1. All analyses incorporated splitless injection and electron impact ionization. The interface temperature between the GC and the MS was maintained at 310°C. The data were analyzed by using the Lab Solutions program GC-MS solution (version 1.2).
IR.
Samples of CSM and cis-2-decenoic acid were weighed before and after lyophilization to determine the amount of KBr to add to each sample. KBr was added at 10 times the sample mass and mixed by using a mortar and pestle. The resulting powder was formed into a pellet by using a Carver 4350 manual pellet press (Carver, Inc., Wabash, IN). Pressure was applied at 10 tons for 10 min. Infrared (IR) spectroscopy spectra were obtained by using a Bruker Equinox 55 FT-IR spectrometer at room temperature in the range of 3,500 to 400 cm−1 at a resolution of 1 cm−1. The final spectra represent the means of 128 scans. Each sample was measured in triplicate.
RESULTS
Formation of voids in biofilm microcolonies.
In our laboratory, we have observed that P. aeruginosa PAO1 will disperse from a continuous culture biofilm grown on a glass substratum in a flow-cell reactor after medium flow had been stopped for several hours. This observation has led to the hypothesis that biofilm dispersion may result from the accumulation of an extracellular messenger, which acts as an inducer of the release of cells from the biofilm. This hypothesis is supported by observations in our laboratory that biofilms of P. aeruginosa do not form in batch culture flasks but will form on the walls of a chemostat, indicating that accumulation of a signal for dispersion may prevent biofilm development.
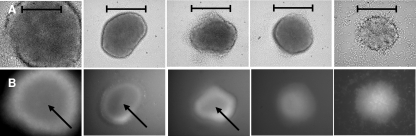
To test this hypothesis, biofilms of P. aeruginosa PAO1 were cultured in modified EPRI defined medium in continuous culture in flow cells and monitored over the course of 12 days. Biofilm dispersion was observed to occur as the periodic release of bacteria from the center of maturing microcolonies; the bacteria entering the bulk liquid through breaches in the microcolony walls. These dispersion events were observed to originate at the center of microcolonies near the substratum, but only within microcolonies that had attained a minimum diameter of 40 μm and a minimum thickness of 10 μm (Fig. 1). The incidence of dispersion was found to be independent of microcolony age. Interestingly, the microcolony size within which dispersion occurred was found to be dependent on the fluid flow rate. When flow in our biofilm reactor was increased, the diameter and thickness of microcolonies in which dispersion occurred also increased, indicating a relationship between dispersion induction and transport. These observations hinted that an extracellular substance produced by P. aeruginosa may be responsible for inducing biofilm dispersion. Similar observations have been made in the laboratory of Paul Stoodley (22), who has reported that under different experimental conditions, microcolonies of P. aeruginosa formed central voids, indicating dispersion, when the microcolonies attained a diameter of 80 microns. The fluid flow rate for experiments in our laboratory was 0.13 ml min−1, while the flow rate used in the study by Stoodley was 1.0 ml min−1, supporting the observation that increased fluid flow results in increased microcolony size prior to dispersion and void formation.
FIG. 1.
Microcolonies of P. aeruginosa biofilms grown in continuous culture demonstrating native dispersion response. Transmitted light image (A) and fluorescent image (B) showing the size dependence of the dispersion response. Biofilm microcolonies growing in continuous culture with dimensions of greater than 40 μm in diameter by 10-μm thickness show dispersion (left 3). Microcolonies below this minimum dimension remain “solid” (right two photomicrographs). Fluorescence indicates presence of cells (lacZ reporter on chromosome). All images are the same relative size at ×500 magnification. Bars, 40 μm. Arrows indicate void areas within a microcolony.
Spent medium induces biofilm dispersion.
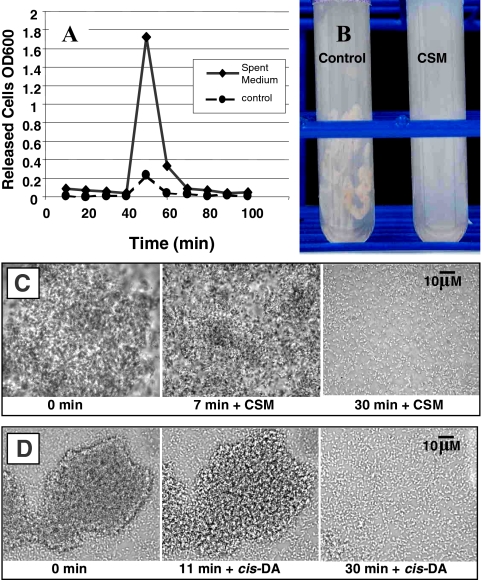
We hypothesized that if P. aeruginosa produces an extracellular dispersion-inducing compound, the cell-free spent medium recovered from P. aeruginosa cultures should be able to induce the release of cells from a P. aeruginosa biofilm. After challenge with spent medium, dispersion should be detectable as an increase in the number of cells recovered in the reactor effluent of biofilms grown on the inner surface of silicone tubing. To test this, biofilms of P. aeruginosa PAO1 were grown in modified EPRI medium in continuous culture at room temperature in a biofilm tube reactor (27). These biofilms were treated after 4 days of growth, by switching from fresh medium to spent culture medium from a 24-h batch culture of P. aeruginosa grown in EPRI medium. The results from a representative experiment are illustrated in Fig. 2A, in which cell-free spent medium or fresh medium were added at t = 30 min. A large spike in the effluent cell number was detectable compared to control lines within 20 min of spent medium addition, indicating the release of biofilm bacteria into the effluent of cultures treated with spent medium. A small spike of released cells was also detectable in control samples, likely representing a response to the physical or mechanical effects associated with switching lines to a fresh medium reservoir.
FIG. 2.
Treatment of P. aeruginosa mature biofilms with spent medium, CSM, and cis-2-decenoic acid. (A) At 30 min, biofilms grown in silicone tubing were exposed to spent medium or fresh medium. Bacteria in effluent were collected continuously for 100 min, and the cell density determined by OD600. (B) Biofilm grown in continuous culture in silicone tubing for 4 days and switched either to fresh medium for 1 h or CSM for 1 h. Extruded contents of control tube shows intact biofilm. Extruded contents of CSM-treated biofilm show dispersion. (C) Photomicrographs show the addition of CSM to mature biofilm grown in continuous culture in a microscope-mounted flow cell. Microcolony disaggregation is shown to begin at 7 min. After 30 min of exposure, the microcolony had completely disaggregated. Dispersed cells were actively motile (not visible in static image), indicating a change in phenotype compared to cells in intact microcolony (prior to CSM addition). (D) Addition of 10 μM cis-2-decenoic acid (cis-DA) to mature biofilm grown in continuous culture in a microscope-mounted flow cell. Microcolony disaggregation is shown to begin at 11 min. Complete microcolony disaggregation is shown within 30 min of exposure. Control biofilms treated with carrier fluid were not affected by treatment up to 1 h (data not shown).
It was not clear whether enzymes or potential virus particles in spent medium were responsible for disaggregating the biofilm cells or whether a signaling molecule was stimulating a dispersion response in the biofilm bacteria. To test this, we ran dispersion experiments using spent medium that had been filtered through Centricon molecular filters with a 5,000-molecular-weight cutoff. In these experiments, filtered spent culture medium did not result in a significant reduction in the number of cells released from treated biofilms. Alterations in the concentration of medium salts and pH were also tested, and negative results confirmed that these did not contribute to an increase in the release of bacteria from the biofilm (data not shown). These experiments suggested that spent medium-induced dispersion resulted from the organic fraction of metabolites produced by P. aeruginosa during growth in EPRI medium, with a molecular mass of <5,000 Da.
Organic fraction of spent medium causes biofilm dispersion.
To purify the organic fraction of spent medium, we extracted cell-free stationary-phase batch cultures of P. aeruginosa grown in modified EPRI medium using ethanol, ethyl acetate, butanol, and chloroform and challenged continuous culture biofilms with EPRI medium amended with these extracts. Butanol and chloroform extracts performed the best in these studies, indicating that dispersion induction resided within the hydrophobic organic fraction of spent medium. Chloroform was used as the preferred solvent to continue an investigation of the dispersion response in P. aeruginosa. After extraction of spent medium, the chloroform-soluble fraction was evaporated to dryness, and the residual organic material was resuspended in fresh medium or buffer solution, resulting in a 125-fold increase in the chloroform-soluble organic fraction compared to levels found in spent medium. We refer to this preparation as CSM. To test the dispersion-inducing activity of CSM, P. aeruginosa biofilms were grown in continuous culture in silicone tubing and exposed for 1 h to medium amended with CSM. The extruded contents of the tube reactors showed a largely intact biofilm in the control line treated with fresh medium, while the contents of the tubes treated with fresh medium containing CSM showed the biofilm to have completely disaggregated (Fig. 2B). Studies of 4-day-old biofilms grown in continuous culture in silicone tubing revealed that treatment with CSM-containing medium for 1 h was effective in releasing an average 87.4% (±1.4%) of biofilm cells, as determined by CFU released into the effluent. In contrast, spent medium had an average dispersion efficacy of 32.4% (±5.5%). The lower dispersion efficacy of spent medium reflects a correspondingly lower concentration of dispersion inducer.
Microscopic analysis of extracted spent medium activity.
Microscopy was used to evaluate the effect of CSM on biofilm microcolonies grown for 6 days in continuous culture on the glass substratum of a flow-cell mounted to a microscope (27). Prior to the addition of CSM, a well-developed microcolony was observed to contain cells that were stationary and showed no sign of motility. After 7 min of contact with CSM-containing medium, cells within the microcolony began to display active motility. After 30 min, the microcolony had become completely disaggregated, and cells were observed to swim freely through the medium (Fig. 2C). Compared to natural dispersion, which is initiated in the interior of biofilm microcolonies, exogenous dispersion induction was observed to progress from the outside of the microcolony toward the interior and, instead of creating a central void, resulted in complete disaggregation of the microcolony.
Extracted spent medium inhibition of biofilm development.
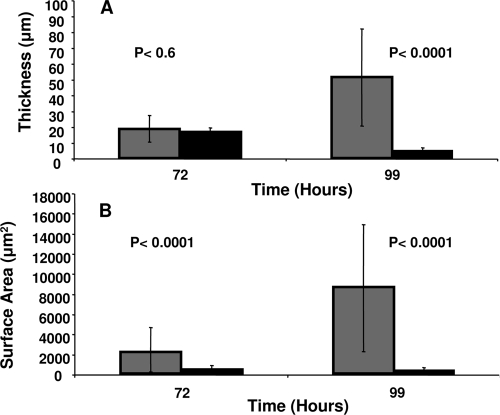
When added continuously to flow cells, CSM adjusted to the concentration of spent medium (125-fold diluted) showed a significant inhibition of biofilm development over a period of 99 h. The results from these experiments showed that the addition of CSM caused a significant reduction in the average biofilm cell cluster thickness after 99 h growth compared to samples not treated with CSM (Fig. 3A). Surface area coverage of the growing biofilm was also significantly reduced when biofilms were grown in the presence of CSM compared to biofilms grown in modified EPRI medium alone (Fig. 3B). Exogenous dispersion induction of preformed biofilms by CSM was measurable at all time points from day 1 (beginning of biofilm microcolony formation) through day 6, after which natural dispersion became detectable. Activity of CSM was shown to persist up to 6 months with no significant reduction when stored under refrigeration (data not shown). Extraction of spent medium by ethyl acetate (to recover acyl-homoserine lactones) did not result in a preparation with dispersion activity (data not shown).
FIG. 3.
Biofilm development in the continuous presence of CSM diluted in modified EPRI to concentration of spent medium. The average thickness (A) and surface area (B) of biofilms grown in the presence of CSM is significantly less than for untreated biofilms. Gray bars indicate biofilms grown in the absence of dispersion inducer. Black bars indicate biofilms grown in the presence of CSM. Error bars represent the standard deviation of 20 randomly selected microcolonies.
Effect of extracted spent medium on species other then P. aeruginosa.
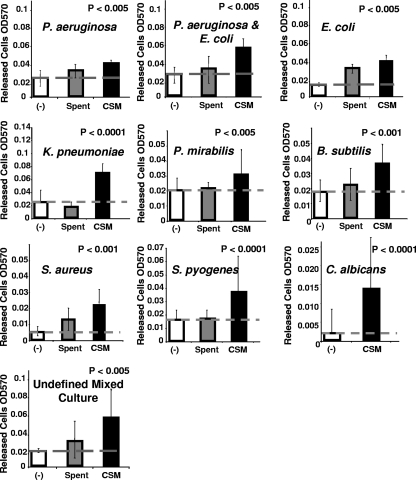
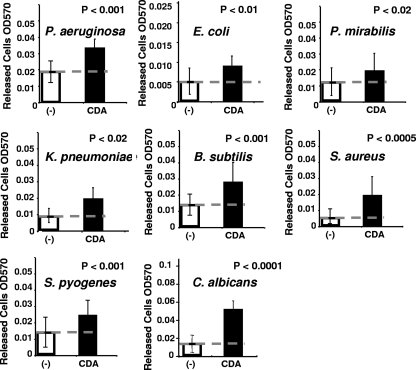
In nature, most biofilms contain multiple species, and it is unlikely that one species can disperse from its nonspecific neighbors if these neighbors do not also disperse. This led to the question of whether P. aeruginosa CSM may be able to stimulate dispersion in other species of bacteria and perhaps also fungi, which are commonly found in mixed-species biofilms. To examine this possibility, dispersion bioassays were performed using 96-well polystyrene microtiter plates etched with acetone to allow an increased surface area for the attachment of bacteria. Cultures were grown in semibatch, by exchanging medium every 12 h and again 7 h prior to testing. This was done to allow the accumulation of sufficient biofilm biomass while reducing the native dispersion due to the build-up of endogenous inducer. Using this bioassay system, P. aeruginosa spent medium and CSM were tested to determine whether they induced dispersion in biofilms formed by E. coli, E. coli mixed with P. aeruginosa, and an undefined mixed bacterial biofilm derived from airborne contaminants and against biofilms formed by K. pneumoniae, P. mirabilis, S. pyogenes, B. subtilis, S. aureus, and the yeast, C. albicans. The results from these experiments are summarized in Fig. 4. The data generated by using the microtiter plate dispersion bioassay tended to be variable from one plate well to the next, due to differences in the amount of biofilm developed in each well, and to the level of endogenously induced dispersion in both the control and test samples against which the exogenous dispersion activity was measured. This variability is indicated by error bars, which represent the standard deviation of the number of cells released into the bulk liquid of 48 wells for each type of biofilm culture tested. In order to indicate whether the released cell number from biofilm populations treated with CSM was statistically different from the biofilm populations treated with fresh medium, a Student t test was performed comparing these two populations. In all cultures tested, these populations were found to be significantly different, as indicated by the P values provided with each graph (CSM-treated versus control). The results from these assays demonstrated the ability of P. aeruginosa CSM to stimulate the release of cells from biofilms formed by different species of bacteria and by C. albicans and indicated that CSM possesses cross-phylum and cross-kingdom dispersion activity.
FIG. 4.
Dispersion of different bacterial biofilms by P. aeruginosa CSM using microtiter plate dispersion bioassay. The y axis indicates the number of cells released into the bulk liquid of 16 replicate wells in three replicate experiments, after treatment for 1 h with CSM or carrier control (−), containing sterile medium. Hatched line indicates level of dispersion in carrier control samples. All differences between CSM samples and controls are statistically significant at the indicated P value as determined by using the Student t test.
Isolation of active fraction of extracted spent medium.
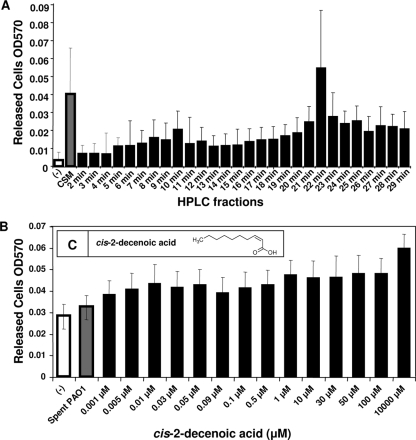
Having established the role of CSM as an inducer of biofilm dispersion, we proceeded to identify the active molecule or molecules present in CSM. We began by assaying the dispersion activity of multiple fractions of CSM separated by isocratic gradient in acetonitrile and water by using C18 reversed-phase HPLC. Eluted HPLC fractions (collected at 1-min intervals) were desiccated in a Speed-Vac to remove residual acetonitrile, resuspended in purified water, and tested by microtiter plate dispersion bioassay to determine the dispersion activity. Figure 5A shows the results of CSM fractionation biofilm dispersion assays. The results indicated that the HPLC fraction of CSM showing the highest activity eluted at 22 min, an acetonitrile/water ratio of 75 to 25%.
FIG. 5.
Microtiter plate dispersion bioassay. (A) ODs of cells released from biofilm-containing microtiter plate wells. White bar, control sample treated with EPRI alone; gray bar, sample treated with CSM. Black bars represent biofilms treated with C18 reversed-phase HPLC fractions of CSM eluted in an acetonitrile gradient from 2 to 75%. The results are the average of 16 replicate wells; error bars represent one standard deviation. The results from Student t test show P < 0.001 for CSM and 22-min HPLC samples. (B) Microtiter plate biofilm dispersion bioassay comparing various concentrations of cis-2-decenoic acid to spent medium. ODs of cells released from biofilm-containing microtiter plate wells. Negative control wells contained P. aeruginosa treated with 10% ethanol in EPRI. The gray bar represents biofilms treated with spent medium. The black bars represent biofilms treated with increasing concentrations of cis-2-decenoic acid in 10% ethanol. The results are the average of 16 replicate wells; error bars represent one standard deviation. Student t test indicated P < 0.001 for all cis-2-decenoic acid samples compared to control. (C [inset]) Structure of cis-2-decenoic acid.
Chemical identification of CSM active fraction.
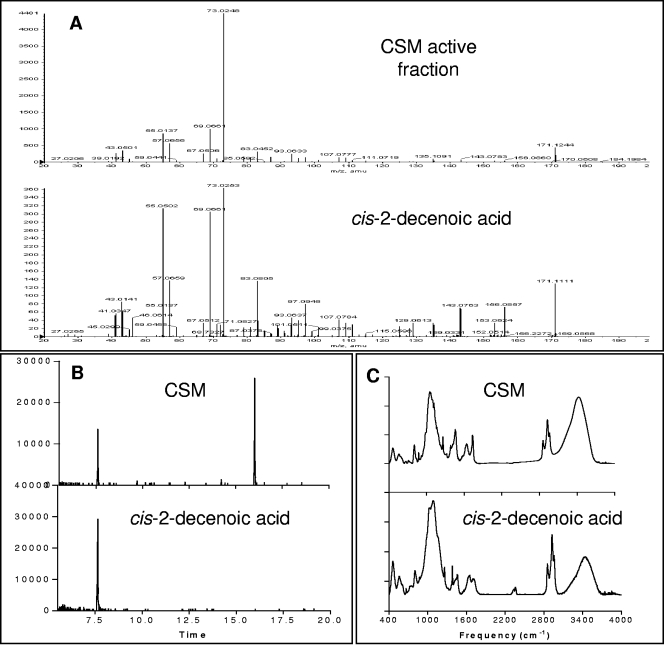
MS of the active HPLC CSM fraction showed a consistent molecular peak with low ionization activity at 171 m/z (Mw = 170). This peak was present in samples showing dispersion activity and missing from samples lacking dispersion activity (data not shown). This peak was also shown to be missing from all carrier liquids and solvents used in preparing CSM (including fresh culture medium). MS product ion analysis of the 170-Mw peak, solubility analysis, H1- and C13 NMR spectroscopy, and IR spectroscopy have demonstrated that the 170-Mw molecule was a mono-unsaturated C10 fatty acid, with a double bond located at the number 2 carbon (2-decenoic acid).
In order to confirm that the 170-Mw molecule (m/z = 171) from the 22-min CSM HPLC fraction was identical to 2-decenoic acid, we fragmented the original molecule in the mass spectrometer to generate product ion peaks. The product ions from the active CSM fraction and 2-decenoic acid were analyzed by tandem MS to evaluate fragmentation differences between these two molecules. Figure 6A shows that the 171 m/z CSM sample had identity with 2-decenoic acid. When analyzed by GC-MS, unfractionated CSM displayed a single major peak with a retention time of 7.6 min, identical to that of 2-decenoic acid (Fig. 6B). IR spectroscopy confirmed that the cis isomer of 2-decenoic acid was the organic compound isolated from CSM (Fig. 6C).
FIG. 6.
Spectral analysis of P. aeruginosa CSM and cis-2-decenoic acid. (A) Product ion mass peaks for the 171 m/z molecule detected in active HPLC CSM fraction and for synthetic cis-2-decenoic acid. The y axis indicates the intensity; The x axis indicates m/z in positive ion mode. CSM sample matches peaks from synthetic cis-2-decenoic acid. Note that in MS, the peak intensity is not a direct indication of concentration. (B) GC-MS spectrum of P. aeruginosa CSM and cis-2-decenoic acid. CSM sample peak at 15.9 min, indicates solvent carrier. The y axis indicates intensity; the x axis indicates time in minutes. (C) FT-IR spectrum of P. aeruginosa CSM and cis-2-decenoic acid. The y axis indicates absorbance; the x axis indicates reciprocal centimeters.
Activity of cis-2-decenoic acid as an inducer of biofilm dispersion.
Following this identification, commercially synthesized mono-unsaturated fatty acids of various molecular weights were tested for dispersion activity against P. aeruginosa. DSF, which has been shown to disrupt cell flocs of X. campestris (10), was shown not to promote statistically significant dispersion of P. aeruginosa biofilms. The compounds with the highest activity were two isomers of 2-decenoic acid. The trans isomer (trans-2-decenoic acid) was shown by microtiter plate dispersion bioassay to have activity only at millimolar concentrations, typically not low enough to qualify as a cell-cell signaling molecule. Figure 5B shows the dispersion activity of increasing concentrations of cis-2-decenoic acid against biofilm cultures of P. aeruginosa grown in microtiter plates. These results demonstrated that the cis isomer (cis-2-decenoic acid) was active over a concentration range from 1.0 nM to 10 mM. Microscopy revealed that the activity of cis-2-decenoic acid as a dispersion inducer was similar to CSM activity, completely disrupting a biofilm microcolony, as shown in Fig. 2D. The activity of cis-2-decenoic acid was also tested against E. coli, K. pneumoniae, P. mirabilis, S. pyogenes, B. subtilis, S. aureus, and C. albicans biofilm cultures, yielding results similar to those obtained for CSM (Fig. 7). The concentration of cis-2-decenoic acid as found in spent medium has been determined by GC-MS to be 2.5 nM. This concentration is consistent with other known cell-to-cell signaling molecules, which typically act at nanomolar and low-micromolar concentrations. When added to 4-day biofilm cultures of P. aeruginosa, grown in tube reactors, commercially synthesized cis-2-decenoic acid was shown to induce dispersion with an efficacy of 24.6% (±4.2%).
FIG. 7.
Dispersion of different bacterial biofilms by cis-2-decenoic acid using microtiter plate dispersion bioassay. The y axis indicates number of cells released into the bulk liquid of 16 replicate wells in three replicate experiments, following treatment for 1 h with 0.01 μM cis-2-decenoic acid (CDA), or carrier control (−), containing medium plus 10% ethanol. The hatched line indicates level of dispersion in carrier control samples. All differences between cis-2-decenoic acid-treated samples and controls are statistically significant at the indicated P value as determined by using a Student t test.
DISCUSSION
The regulation of biofilm dispersion has been investigated by several laboratories over the past decade resulting in findings on the intracellular mechanisms involved in some species of bacteria. In 2002, Jackson et al., described an RNA-binding protein CsrA (carbon storage regulator) that acted as an activator of biofilm dispersal in E. coli (18). The effects of CsrA were believed to be mediated by regulation of intracellular glycogen biosynthesis and catabolism. In more recent work, Thormann et al. (34) reported that by modulating the intracellular cyclic di-GMP (c-di-GMP) pool via overexpression of a diguanylate cyclase or phosphodiesterase it was possible to induce the dispersion of Shewanella oneidensis biofilms. An intracellular transducer protein, BdlA, that apparently is located in the cytoplasm has been shown to act as a regulator of biofilm development in P. aeruginosa (21). Biofilms defective in bdlA were found to have increased adherent properties and increased intracellular levels of c-di-GMP, suggesting that BdlA may be a link between sensing environmental cues, c-di-GMP levels, and biofilm dispersion.
The present study has shown that a small messenger fatty acid molecule, cis-2-decenoic acid, produced by P. aeruginosa in batch and biofilm cultures induces a dispersion response in biofilms formed by a range of gram-negative and gram-positive bacteria and yeast, as well as in P. aeruginosa. Cell-to-cell signaling mediated by fatty acid derivatives has previously been described for a number of bacterial species, including Xanthomonas sp., P. aeruginosa, Mycobacterium sp., Stenotrophomonas maltophilia, Xylella fastidiosa, and Burkholderia cenocepacia (4, 5, 17, 25, 35). The best characterized of these low-molecular-weight fatty acids is DSF, which is responsible for the regulation of virulence in X. campestris (2). DSF signaling has also been shown to regulate the production of extracellular proteases and exopolysaccharide production, aggregative behavior, biofilm formation, flagellum synthesis, resistance to toxins, activation of oxidative stress, and activation of aerobic respiration (5, 12, 17). DSF is structurally related to cis-2-decenoic acid, having a double bond at position 2, but it contains a 12-carbon chain with a branched methyl group at the number 11 position. The synthesis and detection of DSF have been shown to require products of the rpf (for regulation of pathogenicity factors) gene cluster in X. campestris (2). Synthesis of DSF requires RpfF, a putative enoyl coenzyme A (CoA) hydratase, and RpfB, a putative long-chain fatty acyl CoA ligase. DSF perception is dependent on the two-component system comprising the sensor kinase, RpfC, and the response regulator, RpfG (2, 10, 26, 31).
While a BLAST search of the P. aeruginosa genome does not reveal the presence of an rpf gene cluster or protein closely related to RpfF, it does show that P. aeruginosa possess 12 enoyl CoA hydratases with some sequence homology to RpfF, indicating the potential for synthesis of a DSF-related fatty acid signal. The P. aeruginosa protein PA0745 is shown by BLASTp search to have greatest amino acid sequence homology to RpfF, with ca. 30% identity with the X. campestris protein, with an expect value of 6.0 E −14 over 210 amino acids. More closely related to cis-2-decenoic acid is cis-2-dodecenoic acid, a functional analog of DSF produced by B. cenocepacia and named BDSF (4). The protein responsible for BDSF synthesis is Bcam0581, which has 30% sequence identity with PA0745 over a length of 137 amino acids. This is similar to the 37% sequence identity between RpfF and Bcam0158, and both PA0745 and Bcam0158 are 30-kDa proteins with a pI of 6.41, indicating a potential for functional relatedness between these two enoyl CoA hydratases.
Sensing of DSF has been shown by Slater et al. (31) to be mediated in X. campestris by the sensor kinase RpfC. A structural analog to this sensor in P. aeruginosa is PA1396, which was shown by Ryan et al. to respond to DSF produced by S. maltophilia (24). The addition of DSF to P. aeruginosa cultures was found to result in an increase in filamentation and to activate genes involved in resistance to cationic antimicrobial peptides. These phenotypes were abolished in PA1396 knockouts and recovered when PA1396 was restored in the mutants. It is likely that cis-2-decenoic acid is perceived by P. aeruginosa in a similar manner, perhaps by PA1396 or another closely related sensor kinase, of which 155 can be identified by BLAST search. Of these, PA4982 has the highest sequence identity with RpfC with 32% identity over 538 amino acids and an expect value of 2.0 E −72, indicating that there may be more than one sensor for fatty acid signal reception in P. aeruginosa.
The similarity between cis-2-decenoic acid, DSF and BDSF suggests that as a class of molecules, small messenger cis-monounsaturated fatty acids have activity across a wide range of bacteria as extracellular signals. As an inducer of biofilm dispersion, there are clear advantages to having an extracellular signal that is recognized by diverse species. In order to release cells from the biofilm matrix during a dispersion response, microorganisms must rely on the degradation of extracellular polymers produced by neighboring microorganisms of other species, as well as their own species. It is unlikely that a bacterium will produce enzymes to degrade all of the matrix polymers in which it may be enmeshed within a multispecies biofilm. Consequently, in order to disperse from a multispecies biofilm, it must be necessary for the resident organisms to release enzymes in a coordinated fashion. This can be effectively achieved when a cell-to-cell communication molecule is used to orchestrate a coordinated response by unrelated organisms. In addition to prokaryotes, many biofilms also contain eukaryotes, indicating an advantage to cross-kingdom activity for an extracellular inducer of biofilm dispersion. Cross-kingdom activity has been proposed previously for fatty acid messengers from evidence that DSF is recognized by C. albicans binding to the receptor of farnesoic acid, leading to an arrest in filamentation (35). This is further supported by the observation that cis-2-decenoic acid is able to induce the dispersion of C. albicans biofilms, as shown here.
We believe that the dispersion response is a mechanism to escape starvation conditions or overcrowding within a population, allowing fixed cells the opportunity to migrate to a more favorable environment and thin out the population that remains. When biofilm microcolonies are small, the inducer, which accumulates in the extracellular matrix, is removed by diffusive and advective transport. This removal is not possible in small-scale batch systems and may explain why biofilms typically fail to develop at the solid-liquid interface in these systems. When cell clusters attain a dimension where the inducer is not adequately washed out from the interior (the rate of diffusion being exceeded by the rate of production), the inducer is able to attain a concentration necessary for activation of the dispersion response, releasing cells from the biofilm.
The discovery of a signaling molecule responsible for biofilm dispersion has important implications for the exogenous induction of the transition of biofilm bacteria to a planktonic state. The unusual resistance of biofilm bacteria to treatment with antimicrobial agents and the persistence and chronic nature of biofilm infections could potentially be reversed if, in treatment, biofilm bacteria could be forced to transition to a planktonic phenotype. The application of a dispersion inducer prior to, or in combination with, treatment by antimicrobial agents provides a novel mechanism for enhancing the activity of these treatments through the disruption of existing biofilms. In situations where microbicides are unwanted or unnecessary, dispersion induction could be used as an alternative to toxic compounds or reactive chemicals.
The broad-spectrum activity of cis-2-decenoic acid suggests that this and other short-chain cis-2-monounsaturated fatty acids likely have deep evolutionary roots. Therefore, the discovery of additional small fatty acid messengers is anticipated in other organisms. It is interesting that fatty acid communication has been found to be present in many plant and animal species, and the connection to cell dispersion in these systems may be an interesting area for future investigation.
Acknowledgments
We thank Karin Sauer and Sid Mitra for their valuable assistance and for the use of their laboratory facilities. We also thank Szu-Wei Yang, Susan Bane, Barry Jones, Damian Tagliente, Julie Silverman, Sarah Guttenplan, Yuta Okkotsu, Danielle Saldin, and Diana Amari for their contributions to this work.
This study was supported in part by NSF grant 0321672 (MCB) and NIH grant AI055521-01 (NIAID).
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Banin, E., K. M. Brady, and E. P. Greenberg. 2006. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 722064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, C. E., J. L. Tang, J. X. Feng, M. O. Pan, T. J. G. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24555-566. [DOI] [PubMed] [Google Scholar]
- 3.Barraud, N., D. J. Hassett, S.-H. Hwang, S. A. Rice, S. Kjelleberg, and J. S. Webb. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 1887344-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon, C., Y. Deng, L.-H. Wang, Y. He, J.-L. Xu, Y. Fan, S. Q. Pan, and L.-H. Zhang. 2007. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 227-36. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, S., K. L. Newman, and S. E. Lindow. 2008. Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant-Microbe Interact. 211309-1315. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X., and P. S. Stewart. 2000. Biofilm removal caused by chemical treatments. Water Res. 344229-4233. [Google Scholar]
- 7.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell Comp. Physiol. 4925-68. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infection. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 10.Dow, J. M., L. Crossman, K. Findlay, Y.-Q. He, J.-X. Feng, and J.-L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 10010995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2871710-1715. [DOI] [PubMed] [Google Scholar]
- 12.Fouhy, Y., K. Scanlon, K. Schouest, C. Spillane, L. Crossman, M. B. Avison, R. P. Ryan, and J. M. Dow. 2007. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 1894964-4968. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Gilbert, P., T. Maira-Litran, A. J. McBain, A. H. Rickard, and F. W. Whyte. 2002. The physiology and collective recalcitrance of microbial biofilm communities. Adv. Microb. Physiol. 46202-256. [PubMed] [Google Scholar]
- 14.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7894-993. [DOI] [PubMed] [Google Scholar]
- 15.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, T.-P., and A. C. L. Wong. 2007. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 735034-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersion under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J., A. Jayaraman, and T. K. Wood. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 71-15, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J., T. Bansal, A. Jayaraman, W. E. Bentley, and T. K. Wood. 2007. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydrozyindole and stimulated by isatin. Appl. Environ. Microbiol. 734100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan, R., S. Kohn, S.-H. Hwang, D. J. Hasset, and K. Sauer. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 1887335-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purevdorj-Gage, B., W. J. Costerton, and P. Stoodley. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 1511596-1597. [DOI] [PubMed] [Google Scholar]
- 23.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 1873477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan, R. P., Y. Fouhy, B. F. Garcia, S. A. Watt, K. Niehaus, L. Yang, T. Tolker-Nielsen, and J. M. Dow. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 6875-86. [DOI] [PubMed] [Google Scholar]
- 25.Ryan, R. P., and J. M. Dow. 2008. Diffusible signals and interspecies communication in bacteria. Microbiology 1541845-1858. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y.-W. He, L.-H. Zhang, S. Heeb, M. Cámara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 1036712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 1841140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48253-267. [DOI] [PubMed] [Google Scholar]
- 29.Schmidtchen, A., E. Holst, H. Tapper, and L. Bjorck. 2003. Elastase-producing Pseudomonas aeruginosa degrade plasma proteins and extracellular products of human skin and fibroblasts, and inhibit fibroblast growth. Microb. Pathog. 3447-55. [DOI] [PubMed] [Google Scholar]
- 30.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 31.Slater, H., A. Alvarez-Morales, C. E. Barber, M. J. Daniels, and J. M. Dow. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38986-1003. [DOI] [PubMed] [Google Scholar]
- 32.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56187-209. [DOI] [PubMed] [Google Scholar]
- 33.Thormann, K. M., R. M. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 1871014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thormann, K. M., S. Duttler, R. M. Saville, M. Hyodo, S. Shukla, Y. Hayakawa, and A. M. Spormann. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 1882681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51903-912. [DOI] [PubMed] [Google Scholar]
- 36.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21319-346. [DOI] [PubMed] [Google Scholar]
- 37.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 1854585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]