Abstract
The gram-negative, strictly anaerobic epsilonproteobacterium Sulfurospirillum multivorans is able to gain energy from dehalorespiration with tetrachloroethene (perchloroethylene [PCE]) as a terminal electron acceptor. The organism can also utilize fumarate as an electron acceptor. Prolonged subcultivation of S. multivorans in the absence of PCE with pyruvate as an electron donor and fumarate as an electron acceptor resulted in a decrease of PCE dehalogenase (PceA) activity. Concomitantly, the pceA transcript level equally decreased as shown by reverse transcriptase PCR. After 35 subcultivations (approximately 105 generations), a pceA transcript was not detectable and the PceA protein and activity were completely absent. In such long-term subcultivated S. multivorans cells, the biosynthesis of catalytically active PceA was restored to the initial level within about 50 h (approximately three generations) by the addition of PCE or trichloroethene. Single colonies obtained from PceA-depleted cultures were able to induce PCE dechlorination, indicating that long-term subcultured cells still contained the functional pceA gene. The results point to a novel type of long-term regulation of PCE dehalogenase gene expression in S. multivorans.
Sulfurospirillum multivorans is a strictly anaerobic bacterium capable of respiratory growth with different electron acceptors, namely, fumarate, nitrate, and chlorinated ethenes (tetrachloroethene [perchloroethylene (PCE)] and trichloroethene [TCE]) (19). The organism utilizes pyruvate, hydrogen, or formate as an electron donor. When grown with hydrogen or formate as an electron donor and PCE as an electron acceptor, ATP synthesis is coupled to reductive dechlorination via electron transport phosphorylation (dehalorespiration) (8). The key enzyme of PCE utilization that converts PCE via TCE to cis-1,2-dichloroethene (cDCE) is the PCE reductive dehalogenase (PceA). Mature PceA is a monomeric enzyme harboring one norpseudovitamin B12 and two Fe/S clusters as cofactors (12, 15). The cytoplasmic precursor of the PCE dehalogenase (pre-PceA) bears an N-terminal signal peptide of 37 amino acids, including the motif SRRXFXK, which identifies pre-PceA as a substrate of the Tat (twin arginine translocation) protein export pathway (2). Mature PceA is localized in the periplasm, as shown by freeze fracture immunogold labeling techniques (9).
The pce operon comprises pceA, the PCE dehalogenase gene, and pceB, encoding a putative membrane integral protein (16). Genes for transcriptional regulation of the pceAB genes are not found in a 6-kb DNA fragment including the pce operon (GenBank accession number AF022812).
The ability of reductive dechlorination by anaerobic bacteria was found in phylogenetically distant species (8). Transcriptional regulation of dehalogenase gene expression was described for the 3-chloro-4-hydroxy-phenylacetate (Cl-OHPA) dehalogenase (CprA) of the gram-positive Desulfitobacterium dehalogenans. In this organism, the enzyme was strictly regulated by the absence or presence of the substrate. When the cultures were grown with alternative electron acceptors, such as fumarate or nitrate, CprA transcription ceased within the first subcultivation and was induced within less than 30 min (21). A CRP/FNR-like transcriptional activator, CprK, encoded within the cpr operon, was shown to interact with the DNA upstream of the cpr promoter region only when Cl-OHPA was bound to the protein (18). A direct role of CprK in the induction of dehalogenase gene expression in D. dehalogenans was assumed, and a model for transcriptional regulation of cprA expression was developed (13).
While detailed information is available on the control of dehalogenase gene expression in D. dehalogenans grown in the presence of chlorinated aromatic compounds, such as Cl-OHPA, a transcriptional regulation of the PCE dehalogenase genes of Desulfitobacterium species has not been described so far (23). Tsukagoshi and coworkers (22) reported the induction of PCE dehalogenase gene expression in Desulfitobacterium hafniense strain KCB1 as controlled by the presence of PCE.
Our study focused on the maintenance of the PCE-dechlorinating ability of gram-negative S. multivorans in the absence and presence of PCE. Recently, it was reported that the subcellular localization of the PCE dehalogenase of S. multivorans is dependent on the availability of PCE (9). This finding pointed to a unique type of regulation of enzyme targeting by the Tat machinery that might involve proteins encoded by genes not localized in the immediate vicinity of the pceAB operon. It seemed feasible that the expression itself of pceA and pceB is controlled by regulatory genes widely spread in the S. multivorans genome. In this communication, we describe the loss of the PCE dehalogenase in S. multivorans after numerous subcultivation steps on media depleted of chlorinated substrates. Using this subcultivation procedure, a culture of S. multivorans with an inducible PCE dehalogenase was obtained.
MATERIALS AND METHODS
Media and growth conditions.
S. multivorans (DSMZ 12446) (19) was grown under anaerobic conditions at 28°C in a pyruvate (40 mM)- and fumarate (40 mM)-containing medium (14). Rubber-stoppered serum glass bottles were used for cultivation. Unless otherwise stated, anaerobic growth with halogenated ethenes was performed either with formate (40 mM) or pyruvate (40 mM) as an electron donor and PCE (10 mM; see below) as an electron acceptor (19). When the cells were grown with formate, acetate (5 mM) was added as a carbon source. When PCE was provided as an electron acceptor in cultures growing on formate-PCE or pyruvate-PCE, the chlorinated ethenes were dissolved in hexadecane and added to the medium (1 ml of a 0.5 M solution was added to 50 ml culture; the final PCE concentration was 10 mM). Under these conditions, the initial PCE concentration was about 200 μM in the aqueous phase. Hence, subtoxic concentrations of PCE (<300 μM) in the aqueous phase were ensured. Upon PCE consumption, the substrate gradually dissolved in the medium until PCE was completely gone from the hexadecane phase, the gas phase, and the medium. Using this procedure, high concentrations of chlorinated ethenes could be applied without affecting growth (19).
For cultivation of PCE dehalogenase-repressed S. multivorans on solid-medium, 3% agar (grade purified) was added to the pyruvate/fumarate-containing medium and anaerobic roll tubes were prepared. Single colonies were picked under aerobic conditions, transferred to anaerobized pyruvate/fumarate medium, and grown to the late exponential growth phase. These precultures were used for inoculation (10%) of pyruvate-PCE (10 mM in hexadecane).
Quantification of chlorinated ethenes.
PCE, TCE, and cDCE were quantified gas chromatographically by flame ionization detection using a CP 9000 gas chromatograph (Chrompack, Frankfurt, Germany) with a 2-m 10% Ucon LB column (inner diameter, 2 mm; Werner Günther Analysentechnik, Moers, Germany) and N2 as a carrier gas (flow of 28 ml/min at 300 kPa). The following temperatures were applied: oven at 80°C, injector at 150°C, and detector at 250°C. The samples were taken from the aqueous phase of the culture media. The gas sample volume was 0.25 ml, taken from the headspace of heated liquid samples (5 min at 98°C). Nonane was used as an internal standard. The retention times under these conditions were 1.3 min for cDCE, 1.8 min for TCE, 2.8 min for PCE, and 3.4 min for nonane. The detection limit of PCE, TCE, and cDCE was 1 μM.
Preparation of cell extracts and enzyme activity assay.
S. multivorans cells were harvested in the late exponential growth phase by centrifugation (12,000 × g, 15 min at 10°C). The pellet was suspended 1:3 in 50 mM Tris-HCl (pH 7.5), and the cells were disrupted by ultrasonic treatment (15 cycles of 20 s of sonication and 10 s of intermission, 21% amplitude; Bioblock Scientific Vibracell, Illkirch, France). The crude extract was obtained as supernatant after centrifugation (20,800 × g, 5 min at 10°C).
As described earlier, the PCE dehalogenase (EC 1.97.1.8) activity was determined photometrically at 25°C in rubber-stoppered glass cuvettes filled with nitrogen (15). The assay was started by addition of the crude extract. Enzyme activity was measured by the oxidation of reduced methyl viologen with PCE as an electron acceptor. The assay was conducted in Tris-HCl buffer [100 mM, pH 7.5; 0.5 mM methyl viologen (ɛ578 = 9.7 mM−1 cm−1); 4 mM (NH4)2SO4; 0.5 mM PCE]. Before the reaction was started by the addition of extract, methyl viologen was reduced chemically up to an absorption of 3.0 at 578 nm by the addition of titanium(III) citrate solution (25). Protein concentrations were determined in accordance with the method described by Bradford (3) using the Bio-Rad reagent (Bio-Rad Laboratories, München, Germany). Bovine serum albumin was used as a standard.
Immunoblot analysis.
For immunological detection of PceA, cell extracts (1 μg protein per lane) were subjected to denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE). After incubation of a polyvinylidene difluoride (PVDF) membrane and sodium dodecyl sulfate-PAGE in blotting buffer (25 mM Tris, 100 mM glycine, 20% [vol/vol] methanol), the proteins were transferred (15 V, 1 h) using a semidry blot device (Bio-Rad Laboratories, Munich, Germany) (1). The PVDF membrane was subsequently blocked for 1 h in PBST (140 mM NaCl, 10 mM KCl, 6.4 mM Na2HPO4, 2 mM KH2PO4, 0.05% [vol/vol] Tween 20) containing 1% blocking reagent (Roche, Mannheim, Germany). After the PVDF membrane was washed three times with PBST (10 min), PCE dehalogenase antibodies (9) diluted 1:50,000 in PBST were applied at 18°C for 16 h. Subsequently, the membrane was washed twice with PBST (10 min) and incubated for 1 h with the secondary antibody, goat anti-rabbit antibody-alkaline phosphatase conjugate (Bio-Rad Laboratories, Munich, Germany), and diluted 1:3,000 in PBST. After washing of the PVDF membrane three times with PBST (10 min), the reaction mixture for the alkaline phosphatase reaction, 0.34 mg nitroblue tetrazolium and 0.175 mg 5-bromo-4-chloro-3-indolyl phosphate per ml developing buffer (100 mM Tris-HCl, pH 9.5; 100 mM NaCl; 50 mM MgCl2), was added. The reaction was stopped after 10 min with 10 mM Tris-HCl, pH 7.4, containing 1 mM EDTA.
Amplification of the pceA gene.
The presence of pceA was analyzed by colony PCR amplifying a 1.6-kb fragment that covered the entire pceA gene and a significant part of the putative operator region (starting at a position −110 nucleotides upstream from ATG in the pceA sequence) including the transcription start (16). Single clones of S. multivorans that showed no PceA formation were repeatedly cultured on pyruvate plus fumarate. The cells were suspended in sterile water (protein concentration of 20 ng/μl), and the sample was used as genomic template DNA. The oligonucleotides (5′→3′) 3162AAC ACA TTA AAA AAT AAA TAA CTG TAC TTG GGG3195 and 4777TCA TGA TTT TTT AAC CCT ATC CTT TAA AGC4746 (GenBank accession no. AF022812) were used for amplification. PCR mixtures (25 μl) contained 100 ng of total cell protein, 25 pmol of each primer, 2.5 U Taq polymerase (segenetic; Borken, Germany), 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 0.01% (vol/vol) Tween 20, 1.5 mM MgCl2, and 20 mM dNTPs. PCR was performed with an Eppendorf thermocycler (Mastercycler personal; Wesseling-Berzdorf, Germany). The PCR program started with initial denaturing (2.5 min, 96°C) and was followed by 35 cycles of polymerization (0.5 min, 50°C; 1 min, 72°C; 0.5 min, 96°C) and a final cycle with prolonged annealing and polymerization times (1 min, 45°C; 14 min, 72°C).
RT-PCR analysis of pceA transcripts.
Total RNA of S. multivorans (1 × 109 cells) was isolated according to the manufacturer's instructions using the RNeasy mini kit (Qiagen, Hilden, Germany). The removal of DNA contaminations was done with DNase I, RNase free, from Roche (Grenzach-Wyhlen, Germany) at 37°C for 1 h. DNase was inactivated at 75°C for 15 min. Reverse transcriptase (RT) reaction was performed with 1 μg total RNA using the OneStep RT-PCR kit (Qiagen, Hilden, Germany) at 50°C for 1 h. The subsequent PCR was started by addition of 1 U Taq polymerase (segenetic; Borken, Germany). The following primers were used for the RT-PCR: (5′→3′) 3162AAC ACA TTA AAA AAT AAA TAA CTG TAC TTG GGG3195 and 3501TGA GTA AAC GCT GTT CGT ACT TCA GC3476 (GenBank accession no. AF022812). The PCR program was run as follows: initial denaturing (15 min, 96°C) and 29 cycles of polymerization (1 min, 96°C; 0.5 min, 50°C; 2 min, 72°C).
RESULTS
PCE deprivation affected PCE dehalogenase biosynthesis.
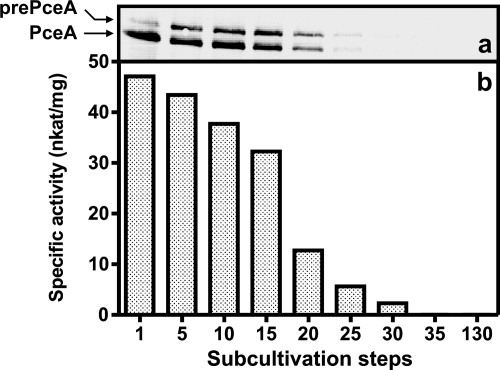
Previous observations that the catalytically active PCE dehalogenase (PceA) was found in S. multivorans grown a few passages in the absence of PCE led to the assumption that the pceAB operon is constitutively expressed in this organism (24). To prove this hypothesis, S. multivorans was repeatedly subcultivated with pyruvate and fumarate as energy substrates in the absence of chlorinated ethenes. The media used for the experiment were prepared under aerobic conditions, filled into serum glass bottles that were subsequently capped with butyl rubber stoppers, and anaerobized by alternate evacuation and flushing with N2. Using this procedure, contaminations of the culture headspace with traces of chlorinated ethenes were excluded. The preculture of the subcultivation experiment was grown on formate plus PCE as sole energy sources to ensure a maximum level of active PCE dehalogenase. In the course of the experiment, cells of S. multivorans grown to the late exponential growth phase on pyruvate plus fumarate without PCE (optical density at 578 nm [OD578] of 0.7, corresponding to 3.5 × 108 cells per ml) were repeatedly transferred to fresh pyruvate/fumarate medium (10% inoculum). Upon long-term subcultivation in the absence of PCE, the level of PceA was monitored by Western blot analysis and enzyme activity measurements before each transfer. During subcultivation, the level of PceA detected immunologically using PceA-directed antibodies gradually decreased (Fig. 1a). After 35 subcultivation steps (approximately 105 generations), the PCE dehalogenase was no longer detectable by immunological analysis in crude extracts. Concomitantly, the specific activity of the enzyme decreased (Fig. 1b). After ≥35 subcultivation steps, a nondechlorinating S. multivorans culture was obtained. This finding was reproducible in more than three independent experiments (data not shown).
FIG. 1.
Loss of the PCE dehalogenase produced by S. multivorans subcultured in medium containing pyruvate (40 mM)/fumarate (40 mM) in the absence of PCE. Each 50-ml subculture was inoculated with 5 ml of a grown culture. (a) Immunological analysis of PceA in crude extracts (1 μg protein per lane) after separation by PAGE under denaturing conditions and subsequent blotting to a PVDF membrane; (b) corresponding reductive PCE dehalogenase activity of PceA. For experimental detail, see Materials and Methods. pre-PceA, 55.9-kDa unprocessed form bearing the N-terminal Tat signal peptide; PceA, 52.1-kDa, processed, exported form without the signal peptide.
The two bands of PceA detected in the immunological analysis of crude extracts separated by denaturing PAGE (Fig. 1a) corresponds to the enzyme with and without the N-terminal PCE dehalogenase Tat signal peptide (pre-PceA, 55.9 kDa; PceA, 52.1 kDa). Since the preculture was grown on formate in the presence of PCE, almost 100% of the PCE dehalogenase was found in the processed, exported form. During the subcultivation, the ratio of PceA to pre-PceA decreased. This result was coincident with previous observations that in cells grown in the absence of PCE a significant portion of the enzyme is localized in the cytoplasm or at the cytoplasmic face of the cell membrane (9). Long-term subcultured cells, which had lost PceA, also lacked the norpseudovitamin B12 cofactor as detected by corrinoid extraction (data not shown) (12).
pceA gene expression is downregulated.
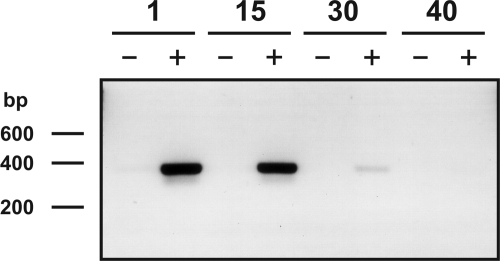
S. multivorans from different subcultivation steps (1, 15, 30, and 40) in the absence of PCE was grown to the late exponential growth phase (OD578 of 0.6) on pyruvate/fumarate-containing medium. To investigate the influence of PCE deprivation on pceA gene expression, total RNA was isolated and analyzed by RT-PCR for the presence of the pceA transcript. Figure 2 shows the gradual decrease of the pceA mRNA level in the course of the subcultivation experiment. When the PceA activity was reduced to 75% after 15 passages in the absence of PCE (Fig. 1b), the level of mRNA derived from pceA transcription was accordingly reduced (Fig. 2). In coincidence with the drastic decrease of PceA activity after 30 subcultivation steps (Fig. 1b), traces of RT-PCR product showed the presence of a minimal level of pceA transcript left (Fig. 2). When S. multivorans was subcultivated 40 passages on pyruvate/fumarate, the PceA protein was totally absent (Fig. 1a) and no pceA transcript was detected (Fig. 2). This result displayed pceA transcriptional regulation in S. multivorans for the first time.
FIG. 2.
pceA transcript level detected by RT-PCR upon subcultivation in the absence of PCE. Total RNA (1 μg) isolated from S. multivorans grown on pyruvate/fumarate-containing medium for 1, 15, 30, and 40 subcultivations (10% inoculum) was used as a template. To each lane an identical volume of 10 μl of the samples was applied to a 1% agarose gel. The gel was stained with ethidium bromide. −, without RT; +, with RT.
To exclude the possibility of a loss of the pceA gene during subcultivation, colony PCR was performed using primers derived from a locus upstream of the transcription start of pceA and from the 3′ terminus of the gene. More than 70 colonies (after 130 subcultivation steps) were tested, and all exhibited the persistence of the pceA gene in the S. multivorans genome during the subcultivation procedure (data not shown).
The pceA gene is still functional.
The colony PCR pointed to a preserved pceA gene upon subcultivation; however, a slight modification of the gene, e.g., by point mutations resulting in nonfunctional pceA transcription, cannot be excluded by this experiment. To test for the presence of a functional pceA gene after 168 subcultivations in the absence of PCE (approximately 500 generations), PceA-depleted S. multivorans was plated on solid medium containing pyruvate and fumarate as energy sources. Twenty colonies were picked and transferred to liquid medium with pyruvate and fumarate. From that preculture, 10% inoculum was transferred to liquid medium containing pyruvate plus PCE as energy sources. The experiment was conducted in three parallel experiments. All 20 clones were able to utilize PCE within 1 or 2 weeks after inoculation (data not shown). Parallel to the initiation of PCE reductive dechlorination, significant growth of the clones with pyruvate and PCE was detected, as found for non-PceA-depleted S. multivorans (data not shown). This result argues against loss of the pceA gene function due to point mutations and points to a long-term downregulation of gene expression in the absence of its substrate, PCE. Repeated subcultivation of S. multivorans with pyruvate alone was not possible, since the presence of an electron acceptor is indispensable for growth on pyruvate. Hence, downregulation of pceA expression could not be determined in the absence of an external alternate electron acceptor.
Induction of pceA expression with chlorinated ethenes.
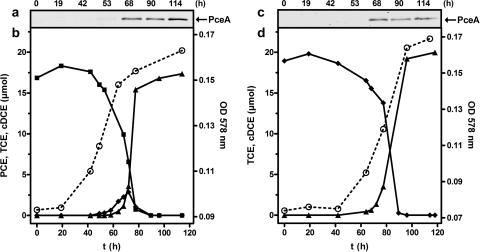
Using a PceA-depleted culture, induction of PCE dehalogenase gene expression in the presence of chlorinated ethenes was studied. Culture medium containing formate as an electron donor and PCE or TCE as an electron acceptor was used. Under these conditions the energy metabolism of S. multivorans strictly depends on reductive dechlorination of PCE or TCE for ATP synthesis via electron transport phosphorylation (dehalorespiration). The chlorinated ethenes (about 20 μmol; Fig. 3) were added directly to the medium (50 ml in a 100-ml serum bottle), and the medium was agitated with the halogenated compound for 3 days prior to inoculation. The induction experiment was started by inoculation with 10% of an S. multivorans preculture that lost the PCE dehalogenase after 80 subcultivation steps in the absence of PCE. The formation of PceA in the presence of PCE or TCE was monitored immunologically. The recovery of the ability to dechlorinate PCE or TCE was examined by detection of cDCE accumulating in the culture media using gas chromatography as described in Materials and Methods. After more than 50 h (approximately three generations) of cultivation, a slight band of the mature PceA protein was detected immunologically (Fig. 2a and c). Almost exclusively, the band of the processed PceA (PCE dehalogenase without the Tat signal peptide) was found. This is in accordance with earlier findings that in the presence of PCE or TCE the PCE dehalogenase is predominately localized in the periplasm (9). The increase of the PceA protein level in the cells was in coincidence with a decrease of the PCE (Fig. 2b) or TCE (Fig. 2d) concentration in the medium almost at the same time. After about 100 h of cultivation, the growth of S. multivorans reached the stationary phase, and PCE or TCE was quantitatively converted to cDCE. This result was reproducible in three independent experiments. When the induction experiment was conducted with pyruvate-PCE instead of a formate-PCE-containing medium, the lag phase of PCE dechlorination was slightly shortened because no adaptation of the cells to a different electron donor and/or carbon source was necessary (data not shown). The concentrations of the chlorinated ethenes were measured in the liquid phase. The total amount of PCE, TCE, and cDCE present in the culture bottle (50-ml liquid phase plus 50-ml gas phase) was calculated considering Henry's constant ([chlorinated ethene]gas/[chlorinated ethene]liquid). Henry's constant for PCE, TCE, and cDCE was 0.92, 0.5, and 0.19, respectively (19).
FIG. 3.
Induction of PceA formation and PCE dechlorination by PceA-depleted S. multivorans grown on formate plus PCE (a and b) or TCE (c and d). PceA was detected by Western blot analysis (1 μg protein/lane) performed with PceA-specific antibodies. Sampling times are given above each lane. PCE, TCE, and cDCE were determined by gas chromatography in the aqueous phase (see Materials and Methods), and the total amount of these compounds was calculated. ▪, PCE; ⧫ TCE; ▴, cDCE, ○, growth (OD578).
To examine the minimal concentration of PCE or TCE required for the induction of dechlorination, increasing initial concentrations of PCE or TCE were added to PceA-depleted cultures of S. multivorans containing formate as an electron donor. Every day, a sample was taken from the cultures, the cells were harvested, and an immunoblot analysis of the crude extracts was performed. To determine the minimal inducer concentration, ethanolic solutions of PCE and TCE were directly added in concentrations ≥1 μM to the medium. The concentration of the chlorinated compound was determined after inoculation by gas chromatography. The concentrations required for induction of PceA formation within 5 days were about 3 μM for PCE and 7 μM for TCE. A concentration of 1 μM PCE or 5 μM TCE did not lead to a detectable PceA biosynthesis within 7 days (data not shown); however, a later induction cannot be excluded.
DISCUSSION
In this communication the reversible loss of reductive dehalogenase gene expression during repeated subcultivation of the gram-negative anaerobe S. multivorans in the absence of chlorinated ethenes is described. After numerous subcultivation steps, a culture was obtained that was unable to synthesize PceA. In such cells, the PCE dehalogenase was induced either by PCE or TCE. The loss and induction have been overlooked in previous studies, since S. multivorans cells were usually grown at most for five subcultivations with pyruvate and fumarate in the absence of PCE (15, 19).
A contamination of the subcultures with small amounts of PCE or TCE can be excluded, since the equipment used for the experiments has never been exposed to chlorinated ethenes. In addition, it seems unlikely that the gradual loss of the enzyme is reproducible if accidental contamination of the laboratory equipment would occur.
When S. multivorans was cultivated on formate-PCE (10 mM in hexadecane), PCE and TCE were completely consumed during growth; the remaining concentrations of both substrates were <1 μM. With respect to the dilution factor of 10−1 (10% inoculum) with each transfer upon subcultivation in the absence of PCE, after five passages (PceA activity still at almost 100%), a maximal concentration of 10 pM PCE or TCE can be present in the aqueous phase, assuming that none of the transferred PCE or TCE has been consumed and/or diffused into the gas phase. The minimal inductor concentration of PCE (3 μM) or TCE (7 μM) was greater by a factor of >100,000. Cells of S. multivorans grown to the late exponential growth phase with PCE (10 mM in hexadecane) accumulated at most 5 μM PCE or TCE. We determined this in an independent experiment. Again, after five subcultivations with still an almost 100% level of PceA activity remaining, a dilution of 10−5 resulted in maximal concentrations of these substrates, which are by far lower than the concentrations sufficient to induce PCE dehalogenase activity.
From these data and assuming a cell concentration of a grown culture of 4 × 1011 cells per liter, it can be calculated that after 10 subcultivation steps, when 80% of the maximal PCE dehalogenase activity was still retained, at most 1 out of 1,000 cells can contain one molecule of PCE or TCE (assuming no consumption or diffusion). From this calculation it is not feasible that contaminant PCE or TCE induced pceA gene expression.
A long-term repression within 35 subcultivations (approximately 105 generations) is very surprising and points to a retentive “memory” of the cells with respect to PCE dehalogenase gene expression. This effect seems to go far beyond the transgenerational inheritance known as “epigenetics.” Specific methylation patterns of the DNA are believed to be responsible for such epigenetic effects in microorganisms (for a recent review, see reference 4). Epigenetic control generally concerns structural components rather than catabolic enzymes (for an example, see reference 7). In addition, the inheritance of traits via epigenetic control usually comprises at most a few generations. A long-term control of catabolic enzyme formation as described in our report for the PCE dehalogenase of S. multivorans appears to be unique and might involve a novel type of regulatory mechanism.
The ability to dehalogenate chlorinated aliphatic compounds is found in bacteria belonging to quite different genera (8). Therefore, the regulation regime for PCE dehalogenase expression might be diverse as well. In the gram-positive PCE-dechlorinating Desulfitobacterium hafniense strain Y51, the pce operon is located on a transposable element (5). In this organism the reductive dehalogenase was not inducible after loss of the transposon. In contrast, colony PCR performed with subcultivated and PceA-depleted S. multivorans indicated that the pceA gene was still present in 100% of the colonies tested. In addition, in S. multivorans, neither IS elements nor transposase genes have been identified upstream or downstream of the pce operon. These results argue for a different molecular basis for the loss of the enzyme in the two organisms. Furthermore, this assumption is supported by the fact that after numerous subcultivation steps in the absence of PCE, a functional PCE dehalogenase can still be induced in S. multivorans.
In D. dehalogenans and D. hafniense strains DCB-2, the gene expressions of the reductive o-chlorophenol dehalogenases (CprA) are inducible as well (6, 17, 21). The gene cprK or cprK1, located within the cpr operon, encodes a transcriptional activator of the CRP/FNR-type (for a review of CRP/FNR-dependent regulation, see reference 11). Within the PCE dehalogenase operon in Desulfitobacterium sp. strain KBC1, a gene encoding a transcriptional regulator, PrdK, of the CAP family activators was detected (22). The PCE dehalogenase (PrdA) gene transcription of this organism is induced by addition of PCE. All mechanisms for the transcriptional regulation of reductive dehalogenase gene expression described so far are short term and obviously different from the regulation in S. multivorans. Based on genome sequence analysis of Dehalococcoides ethenogenes, a model for the regulation of reductive dehalogenase gene expression in this organism was published recently (20). Genes encoding two-component signal transduction systems were found close to reductive dehalogenase genes, but a functional characterization is still lacking.
The induction of PceA-depleted S. multivorans cells was observed with the enzyme substrates PCE and TCE at “threshold” inductor concentrations lower than 10 μM for the induction within 7 days of incubation. The range of minimal inductor concentrations is consistent with the data obtained by Johnson and coworkers (10) for the fast induction of the TCE reductive dehalogenase (TceA) of a Dehalococcoides-containing enrichment culture. In this culture, the dechlorination activity was lost within 48 h of incubation and restored within 24 h.
All regulation mechanisms described so far for the reductive dehalogenation were short-term and seemingly according to conventional models. The molecular basis for the unique type of long-term regulation of dehalorespiration in S. multivorans described in this communication is not yet known and is currently studied in detail in our laboratory.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SCHM 2144/3-1) and from the Helmholtz Society (VH-VI-155). We thank Peggy Brand for skillful technical assistance.
Footnotes
Published ahead of print on 19 December 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., New York, NY.
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22393-404. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Casadesús, J., and D. Low. 2006. Epigenetic regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70830-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Futagami, T., Y. Tsuboi, A. Suyama, M. Goto, and K. Furukawa. 2006. Emergence of two types of nondechlorinating variants in the tetrachloroethene-halorespiring Desulfitobacterium sp. strain Y51. Appl. Microbiol. Biotechnol. 70720-728. [DOI] [PubMed] [Google Scholar]
- 6.Gábor, K., C. S. Veríssimo, B. C. Cyran, P. ter Horst, N. P. Meijer, H. Smidt, W. M. de Vos, and J. van der Oost. 2006. Characterization of CprK1, a CRP/FNR-type transcriptional regulator of halorespiration from Desulfitobacterium hafniense. J. Bacteriol. 1882604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guespin-Michel, J., and M. Kaufman. 2001. Positive feedback circuits and adaptive regulations in bacteria. Acta Biotheor. 49207-218. [DOI] [PubMed] [Google Scholar]
- 8.Holliger, C., C. Regeard, and G. Diekert. 2003. Dehalogenation by anaerobic bacteria, p. 115-158. In M. M. Häggblom and I. D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Publishers, Norwell, MA.
- 9.John, M., R. P. H. Schmitz, M. Westermann, W. Richter, and G. Diekert. 2006. Growth substrate dependent localization of the tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Arch. Microbiol. 18699-106. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, D. R., P. K. Lee, V. F. Holmes, A. C. Fortin, and L. Alvarez-Cohen. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 717145-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 12.Kräutler, B., W. Fieber, S. Ostermann, M. Fasching, K. H. Ongania, K. Gruber, C. Kratky, C. Mikl, A. Siebert, and G. Diekert. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helv. Chim. Acta 863698-3716. [Google Scholar]
- 13.Levy, C., K. Pike, D. J. Heyes, M. G. Joyce, K. Gabor, H. Smidt, J. van der Oost, and D. Leys. 2008. Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol. Microbiol. 70151-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann, A., H. Scholz-Muramatsu, and G. Diekert. 1994. Tetrachloroethene metabolism of Dehalospirillum multivorans. Arch. Microbiol. 162295-301. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 27116515-16519. [DOI] [PubMed] [Google Scholar]
- 16.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 1804140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pop, S. M., N. Gupta, A. S. Raza, and S. W. Ragsdale. 2006. Transcriptional activation of dehalorespiration. Identification of redox-active cysteines regulating dimerization and DNA binding. J. Biol. Chem. 28126382-26390. [DOI] [PubMed] [Google Scholar]
- 18.Pop, S. M., R. J. Kolarik, and S. W. Ragsdale. 2004. Regulation of anaerobic dehalorespiration by the transcriptional activator CprK. J. Biol. Chem. 27949910-49918. [DOI] [PubMed] [Google Scholar]
- 19.Scholz-Muramatsu, H., A. Neumann, M. Meßmer, E. Moore, and G. Diekert. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov. sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 16348-56. [Google Scholar]
- 20.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307105-108. [DOI] [PubMed] [Google Scholar]
- 21.Smidt H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 1825683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsukagoshi, N., S. Ezaki, T. Uenaka, N. Suzuki, and R. Kurane. 2006. Isolation and transcriptional analysis of novel tetrachloroethene reductive dehalogenase gene from Desulfitobacterium sp. strain KBC1. Appl. Microbiol. Biotechnol. 69543-553. [DOI] [PubMed] [Google Scholar]
- 23.Villemur, R., M. Lanthier, R. Beaudet, and F. Lépine. 2006. The Desulfitobacterium genus. FEMS Microbiol. Rev. 30706-733. [DOI] [PubMed] [Google Scholar]
- 24.Wohlfarth, G., and G. Diekert. 1999. Reductive dehalogenases, p. 871-893. In R. Banerjee (ed.), Chemistry and biochemistry of B12. John Wiley & Sons, New York, NY.
- 25.Zehnder, A. J., and K. Wuhrmann. 1976. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 1941165-1166. [DOI] [PubMed] [Google Scholar]