Abstract
The mobilization proteins of the broad-host-range plasmid R1162 can initiate conjugative transfer of a plasmid from a 19-bp locus that is partially degenerate in sequence. Such loci are likely to appear by chance in the bacterial chromosome and could act as cryptic sites for transfer of chromosomal DNA when R1162 is present. The R1162-dependent transfer of chromosomal DNA, initiated from one such potential site in Pectobacterium atrosepticum, is shown here. A second active site was identified in Escherichia coli, where it is also shown that large amounts of DNA are transferred. This transfer probably reflects the combined activity of the multiple cryptic origins in the chromosome. Transfer of chromosomal DNA due to the presence of a plasmid in the cytoplasm describes a previously unrecognized potential for the exchange of bacterial DNA.
The discovery over 50 years ago of bacterial mating by Lederberg and Cavalli-Sforza (summarized by Cavalli-Sforza [10]) was a major step in the development of modern microbial genetics. It was later realized that chromosomal DNA transfer was due to the integration of a plasmid, the F factor. This plasmid is able to effect its own transfer and, in the integrated state, chromosomal DNA as well. A unique site on the plasmid, the origin of transfer (oriT), is essential for this process. A complex of plasmid- and host-encoded proteins assemble at oriT (15); the F-encoded relaxase then cleaves one of the DNA strands, forming a covalent, protein-DNA intermediate (27) that is delivered to the type IV secretion apparatus, also encoded by F (24). The possibility of low-level transfer initiating from chromosomal sites was raised early in the history of conjugation (12). However, it is now clear that the exacting architecture of the oriT DNA-protein complex, both for the F factor and related oriTs, results in a very high degree of sequence and structural specificity, and no secondary origins in the chromosome have been identified.
The broad-host-range plasmid R1162 (RSF1010), like the F factor, is also transferred from cell to cell. Although the mechanism of transfer is similar for the two plasmids, the oriTs are structurally very different. In prior studies we have determined that the R1162 oriT is small, structurally simple, and able to accommodate base changes at different positions without a complete loss of function (5, 21). As a result of this relaxed specificity, the R1162 mobilization (Mob) proteins can activate the related but different oriT of another plasmid, pSC101 (28).
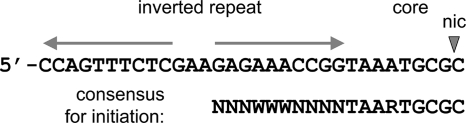
The R1162 oriT is shown in Fig. 1. The R1162 relaxase MobA interacts with the core region, highly conserved in the R1162 Mob family (5), and the adjacent, inner arm of the inverted repeat (23). The protein forms at nic a tyrosyl phosphodiester linkage with the 5′ end of the DNA strand (30), which is then unwound from its complement as the protein-DNA complex is passed into the recipient cell. Circular plasmid DNA is reformed by a reverse of the initial protein-DNA transesterification, with the relaxase now binding to the 3′ end of the transferred DNA. This binding requires the complete inverted repeat, which probably forms a hairpin loop to recreate the double-stranded character of the relaxase binding site on duplex DNA.
FIG. 1.
Base sequence of R1162 oriT on the nicked strand. The relaxase cleaves at nic; subsequent transfer is 5′ to 3′ so that most of oriT is transferred last. Below is the consensus sequence of DNA active for initiation of transfer.
An important feature of the steps in DNA processing at the R1162 oriT is that an inverted repeat is not required for the initiation of transfer or passage of DNA into a new cell. As a result of this and the permissiveness of the relaxase to base changes within oriT, different sites in the chromosome that are not part of a plasmid oriT might nevertheless be capable of initiating transfer when cloned into a plasmid. We previously identified one such site in the chromosome of Erwinia carotovora subsp. atroseptica (now Pectobacterium atrosepticum) (21). In addition, by testing libraries of oriTs with one or more mutations for relaxase-induced nicking, we found that a large population of DNAs with different sequences was potentially active (21). The consensus sequence for activity, derived from these studies, is shown in Fig. 1.
I show here that ectopic sites on the bacterial chromosome, active for the initiation of transfer of plasmid DNA, can also serve for the transfer of chromosomal DNA when the Mob proteins are provided in trans. Thus, the presence of R1162 in the cell can result in a cryptic sexuality in bacteria, resulting in the transfer of chromosomal genes. Although such events are likely to be infrequent, the broad-host-range of R1162 and its close relatives, with their ability to reside stably in the cytoplasms of many different bacteria, could be another source of gene exchange for a variety of different species.
MATERIALS AND METHODS
Bacterial strains.
The strains used in the present study are P. atrosepticum (Erwinia carotovora subsp. atroseptica) (ATCC BAA-672) (6) and the Escherichia coli K-12 strains RM1352, a derivative of C600 (2) resistant to streptomycin (Strr) and nalidixic acid (Nalr), MG1655 (8), and JW2128 and JW2161, which are members of the KEIO collection and derivatives of BW25113 (1).
Plasmids.
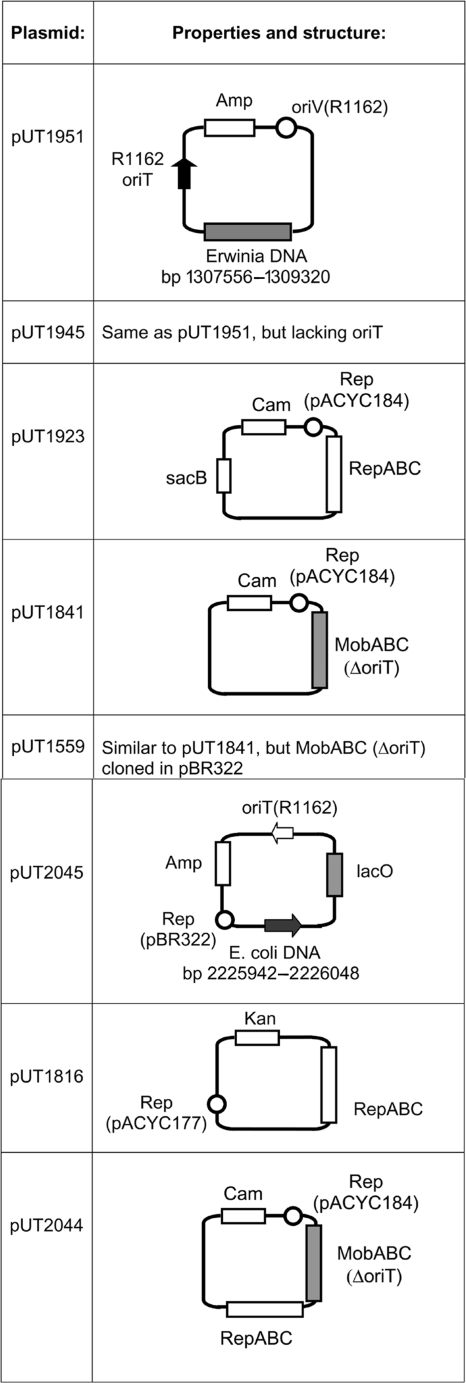
Sketches of plasmids constructed for the present study are given in Fig. 2.
FIG. 2.
Plasmids constructed for the present study. RepABC and MobABC refer, respectively, to the replication and mobilization genes of R1162 (31). Helper plasmids were constructed by using pBR322 (9), pACYC177, or pACYC184 (11); the locations of the replicons are indicated. Abbreviations for antibiotics, here denoting the locations of resistance genes: Amp, ampicillin; Cam, chloramphenicol; Kan, kanamycin.
DNA primers for PCR.
The primers used are listed in Table 1.
TABLE 1.
Primers
| Analysis | Primer | Sequence | Locationa |
|---|---|---|---|
| Confirming that pUT1951 and | 686 | CTCCCGTATCGTAGTTATC | Internal to Ampr gene |
| pUT1945 are inserted into the chromosome (Fig. 3) | 733 | ACATACCCGACACATTG | c bp 1306725-1306741 Erwinia chromosome |
| Identifying plasmids formed | 357 | GGAAATGTTGAATACTCATACTCTTC | Internal to Ampr gene |
| in donors when replication proteins are supplied | 718 | GCGGCCGCAATTGAGCAGAGGTTCGCAGCC | c bp 1309304-1309320 Pectobacterium chromosome |
| Mapping Kanr DNA after transfer from JW2128 and | 791 | CGCCAATCGGCACAAACAATAG | c bp 2228862-2228883 MG1655 chromosome |
| JW2161 | 792 | CAGTCATAGCCGAATAGCCT | Internal to Kanr gene, JW2128 and JW2161 |
| 804 | GCAGGCTGGATGCGTTAC | c bp 2265473-2265490 MG1655 chromosome | |
| Cloning cryptic E. coli oriT | 787 | GGCCGGATCGATCGGCAACGCGACCGGT | c bp 2225942-2225957 MG1655 chromosome |
| 789 | GGCCGGCAATTGAGCAGCTCAACGATGTCGCG | c bp 2226029-2226048 MG1655 chromosome | |
| Replacement of cryptic oriT in E. coli with Camr FRT | 795 | CTTGCCACTTTCGGCAACGCGACCGGTTTAAGCTTCAATGACTTTGTCGTGTAGGCTGGAGCTGCTTC | Amplification of Camr DNA (reverse) |
| fragment | 796 | AGCTCAACGATGTCGCGGGGAAAATGAAAATTATTGAGGCACGGTTAAATGGGAATTAGCCATGGTCC | Amplification of Camr DNA (forward) |
c, complement to the indicated base pairs. Ampr, ampicillin resistance.
Mutagenesis.
E. coli genomic DNA (bp 2,225,979-2,225,997) was deleted and replaced by a chloramphenicol resistance (Camr)-FRT fragment by lambda red-mediated recombination as described by Datsenko and Wanner (13), except that the PCR was done in two steps to increase the size of the arms homologous to the chromosome (25). The primers used are shown in Table 1, and the source of the Cam DNA was pKD3 (13).
Bacterial growth and mating.
For matings involving E. coli, donors were grown overnight in TYE medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl), supplemented with trimethoprim (200 μg/ml), ampicillin (100 μg/ml), and kanamycin sulfate and chloramphenicol (each 25 μg/ml), as appropriate. The recipient RM1352 was grown in TYE containing streptomycin sulfate (50 μg/ml) and nalidixic acid (25 μg/ml). Donors and recipient were diluted 1:10 and 1:5, respectively, in fresh medium lacking drugs and grown for 90 min, and then 0.5 ml each of the donor and recipient were drawn onto a 25-mm-pore-size nitrocellulose filter and washed with 2 ml of medium with gentle suction. The collected cells on the filter were incubated right side up on a TYE plate for 4 h at 37 C and then diluted and plated onto medium containing kanamycin, streptomycin, and nalidixic acid to select for transconjugants. Donor cells were enumerated by plating on medium containing kanamycin.
Pectobacterium strains were grown in TYE containing the concentrations of antibiotics indicated above, but cultures were incubated at 29 C. Donors were grown in medium containing trimethoprim, chloramphenicol, and ampicillin, and the recipient, which contained the plasmid pUT1816 (Fig. 2), was grown in medium containing kanamycin. Matings were also on 25-mm filters, but 0.2 ml of a stationary-phase donor culture, and 0.5 ml of the recipient were drawn onto the filter and then washed with 5 ml of TYE. Filters were incubated on a TYE plate for 24 h at 29°C and then plated on medium containing kanamycin (25 μg/ml) and TurboAmp (100 μg/ml; Stratagene) for selection of transconjugants. Donors were enumerated on medium containing TurboAmp alone. Cells were incubated 2 to 3 days before colonies were counted. All matings were done in quadruplicate, and the results were averaged. There was no large variation in results for mating the same pair of strains.
The initiating activity of a cryptic oriT on a plasmid was tested as described previously (21). In brief, the test plasmid (pUT2045, Fig. 2) contained the cryptic oriT and an R1162 oriT for termination. Initiation at the cryptic site and termination at the R1162 oriT results in deletion of lacO on the plasmid, which is detected by plating for transconjugants on medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and examining the samples for white colonies. Initiation of transfer from the R1162 oriT does not result in the formation of plasmids deleted for lacO, and thus the colonies are blue due to titration of the Lac repressor.
RESULTS
The R1162 Mob proteins can initiate transfer from a cryptic oriT in the chromosome.
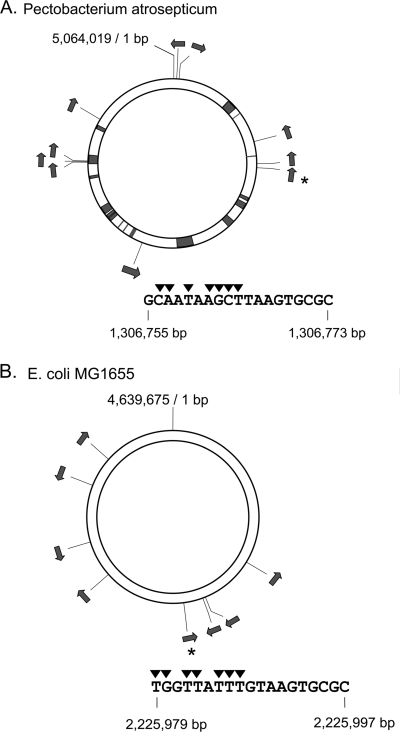
A sketch of the chromosome of P. atrosepticum (6) showing the locations of sites conforming to the consensus for initiation of transfer is in Fig. 3A. We previously showed that one of these sites (bp 1306755 to 1306773, Fig. 2) is active for the initiation of plasmid DNA transfer (21). This DNA is between the genes hupB (HU protein β subunit) and ppiD (peptidyl-prolyl cis-trans isomerase) and is unlikely to be part of an integrated plasmid. A straightforward method to determine whether this site is functional in situ would be to select after mating for a marker downstream from the site in the direction of transfer. However, the frequency of transfer would probably be low, and this approach would require recombination, as well to stabilize a newly acquired marker in the recipient, which would further reduce the frequency of transconjugants. Therefore, I took advantage of the fact that in the R1162 Mob system initiation of transfer at one oriT can be efficiently terminated at another (23), eliminating the need for rescue by homologous recombination. In addition, such an approach would provide a plasmid in the recipient that could be used for the direct characterization of the transferred DNA.
FIG. 3.
Chromosome maps of P. atrosepticum (6) (A) and E. coli K-12 MG1655 (8) (B), showing locations of potential oriTs. Arrows indicate the presumed direction of transfer. For each map, the oriT indicated by the asterisk was selected for study; the sequence of each is shown below. Filled sections of the Pectobacterium map are the positions of likely genomic islands (6).
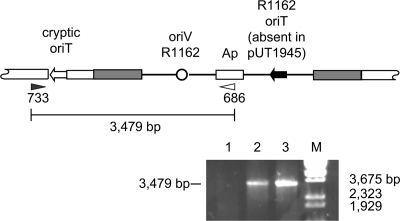
An R1162 oriT with the normal sequence was introduced downstream from the cryptic oriT by homologous recombination. The plasmid pUT1951 (Fig. 2) contains the R1162 oriT, as well as oriV, the origin of replication, but requires the R1162 replication proteins to be provided in trans. In addition, this plasmid contains cloned Pectobacterium DNA downstream from the cryptic oriT. The plasmid pUT1951 was introduced into Pectobacterium and maintained by pUT1923, which contains the R1162 replication (Rep) genes and the B. subtilis gene sacB (Fig. 2). Integration of pUT1951 into the chromosome, as well as loss of the helper plasmid, was selected by plating on medium containing ampicillin and 10% sucrose (17). The absence of pUT1923 was confirmed by sensitivity to chloramphenicol and the lack of plasmid DNA in the cell. Insertion of pUT1951 at the correct site, to result in the structure shown in Fig. 4, was then verified by PCR.
FIG. 4.
(Top) Structure of the recombinant formed by integration of pUT1951 and pUT1945 (not drawn to scale). The shaded DNA was cloned in these plasmids and duplicated by the recombination. This DNA begins 783 bp from the cryptic oriT. (Bottom) Confirmation by PCR of the correct location of the integrated plasmids pUT1951 and pUT1945. The locations of the primers 686 and 733 (Table 1) and the distance (in bp) along the PCR product is indicated in the drawing above. Lane 1, parental strain; lanes 2 and 3, integrated pUT1945 and pUT1951. The marker (M) is BstEII-digested lambda DNA.
R751, a plasmid encoding a type IV secretion system efficiently used by R1162 for transfer (35), and pUT1841, which encodes the R1162 Mob proteins (Fig. 2), were introduced into the strain containing the integrated pUT1951. The resulting cells were then mated with a recipient strain containing pUT1816, which encodes the R1162 Rep proteins and resistance to kanamycin. Under these circumstances, transfer initiated at the cryptic oriT and terminated at the integrated R1162 oriT would reform a plasmid in the recipient, which could then be maintained by the Rep proteins. Cells containing plasmids were selected by plating on medium containing ampicillin and kanamycin. Bacterial matings (done in quadruplicate [see Materials and Methods]) resulted in transconjugants at a frequency of 4.6 × 10−7 per donor. No transconjugants were obtained (<2.5 × 10−9 per donor) when the chromosome contained instead pUT1945, which lacks oriT but is otherwise identical to pUT1951 (Fig. 2), or when donor cells lacked either R751 or pUT1841 (<5.7 × 10−9 and <6.0 × 10−8 transconjugants per donor, respectively).
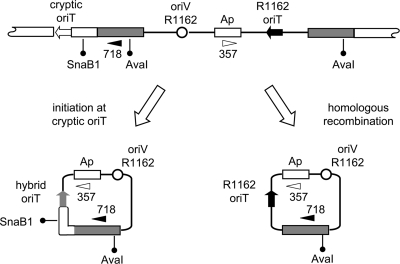
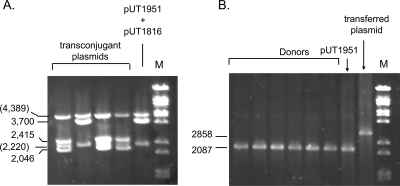
Although the downstream oriT allowed the detection of rare instances of conjugative transfer, its integration into the donor by homologous recombination introduced the possibility that these transfers were the result of excision of the satellite plasmid, also by homologous recombination, immediately followed by transfer into the Rep+ recipient (Fig. 5). Indeed, we have shown previously that plasmids such as pUT1951 can be transformed into donor cells and then mated out without first having to undergo vegetative replication (19). To determine whether excision by recombination were responsible for the transconjugant colonies, plasmid DNA from transconjugant cells was analyzed by digestion with AvaI and SnaBI. If plasmid DNA was reformed by initiation at the cryptic oriT and termination at the R1162 oriT, then it would contain the single AvaI site found in the segment duplicated in the chromosome, and the SnaB1 site in the unduplicated part of the chromosome adjacent to the cryptic oriT (Fig. 5). Digestion with SnaBI and AvaI would then result in 2,415 and 2,046-bp fragments. If the satellite plasmids were generated by homologous recombination of the duplicated segment, to form a plasmid identical to pUT1951, then only the AvaI site would be present, and digestion with these enzymes would result in a single, 3,700-bp fragment. Plasmid DNA that had been transferred in four independent matings was digested with these enzymes. The results (Fig. 6A) show that three of the plasmids were formed by initiation from the cryptic oriT. In other trials, all of the transferred plasmids were due to initiation from the cryptic origin, suggesting that homologous recombination to recreate pUT1951 is only a minor route to the generation of transferring plasmids.
FIG. 5.
Alternate mechanisms for formation of ampicillin resistance plasmids in transconjugant cells. At the top is a map of the integrated plasmid pUT1951, and at the bottom are the structures of the plasmids that would be generated by initiation of transfer at the cryptic oriT (left) or by general homologous recombination within the duplicated Pectobacterium DNA (right). The sites for cleavage by AvaI and SnaBI, used to distinguish the two types of plasmids (Fig. 6A), are indicated. Also shown are the location of the primers (filled and closed horizontal arrowheads) for PCR analysis of excised plasmids in the donor (Fig. 6B).
FIG. 6.
(A) Agarose gel electrophoresis of plasmid DNA, digested with AvaI and SnaBI, from four, independently isolated transconjugant colonies of P. atrosepticum. Also shown for comparison is identically digested plasmid DNA extracted from cells containing pUT1951 and pUT1816. (B) PCR analysis of satellite plasmid DNA formed in six independent Pectobacterium donors containing the R1162 Rep proteins. For comparison, the same PCRs were carried out with pUT1951 DNA and with satellite plasmid from transconjugant cells.
Transfer does not involve prior formation of a plasmid in the donor cell.
The results described above show that DNA transfer can be initiated from a cryptic oriT in the chromosome, with termination at a trailing oriT to form a plasmid, but do not show when this plasmid is being formed. One possibility is that the plasmid is first being generated in the donor, by unwinding of the nicked DNA strand between the two oriTs. This is not a far-fetched possibility: plasmids containing two, directly repeated copies of the R388 oriT (unrelated to the R1162 oriT) spontaneously generate deletions by such a process (26). Similar deletions in the absence of transfer have not been observed for the R1162 Mob system (7), but they could be occurring at a low frequency.
If excision of pUT1951 by strand unwinding in the donor cell is the route to the formation of transconjugants, then the resulting plasmids in the donors should be detected if they are stabilized by providing the R1162 Rep proteins. Pectobacterium cells containing integrated pUT1951 were transformed with the Rep helper plasmid pUT1816 (Fig. 2). Six, independently derived transformants were then analyzed for the presence of satellite plasmid by PCR with the primers 357 and 718 (Table 1) shown in Fig. 5. Only excised, satellite plasmid would be amplified with the primers, since they would be outwardly facing after hybridization to the integrated pUT1951. The results with six, independently derived donor strains are shown in Fig. 6B. In each case the PCR product was identical to that generated with pUT1951 itself, suggesting that the templates were the rare circular molecules generated by excision due homologous recombination. The larger PCR product reflecting a circular molecule due to recombination at the two oriTs was not observed. Taken together, with the results of the previous section, I conclude that formation of the satellite plasmid in the transconjugants and prior generation of satellite plasmids in the donors occur by largely different and independent routes. It would thus appear that the chromosomal DNA is being injected directly into the recipient, as with the integrated F factor.
The E. coli chromosome is conjugatively mobilized by the R1162 Mob system.
Although the results made clear that in Pectobacterium chromosomal DNA could be conjugatively mobilized from a cryptic origin, the presence of a trailing origin, as well meant that only plasmids containing a short segment of the chromosome were detected. To determine whether larger segments of the chromosome, containing many genes, could also be mobilized, I first identified a cryptic oriT in E. coli K-12 strain MG1655. This strain contains eight loci conforming to the oriT consensus for initiation (Fig. 3B). One of these (bp 2225979 to 2225997), located within mdtQ′, a putative multidrug transporter gene inactivated in MG1655 by a frameshifting mutation (8), was selected for further analysis. I first tested this potential oriT for initiation of plasmid transfer in pUT2045, Fig. 2). The activity of the origin (the proportion of white transconjugant colonies; see Materials and Methods) was 0.75%, indicating that this origin is ∼15 times more active than the one in Erwinia (21).
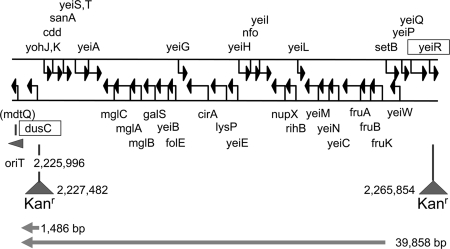
I next determined whether a large segment of chromosomal DNA could be detectably transferred, by using two strains from the KEIO collection (1). JW2128 contains a kanamycin resistance (Kanr) fragment in dusC near the cryptic oriT, 1,486 bp downstream in the direction of transfer (Fig. 7). JW2161 contains the Kanr insertion in yeiR, 39,858 bp from the oriT and thus much more distant (Fig. 7). Donor strains were constructed by introducing R751 and pUT1841 into JW2128 and JW2161. These were then mated with a Nalr Strr derivative of C600, and transconjugants were selected on medium containing kanamycin nalidixic acid and streptomycin. The double selection against the donor was necessary to avoid a background due to spontaneous mutation in the donor. The results of this analysis (Table 2) show that Kanr Nalr Strr colonies appeared when both the Tra+ and the Mob+ functions were provided in the donor.
FIG. 7.
Map of chromosomal DNA of BW 25113 (1) in region containing Kanr insertions in dusC and yeiR to form the KEIO derivatives JW2128 and JW2161, respectively. Distances (in bp) from the cryptic oriT to the Kanr marker are shown in each case. The arrows at bottom indicate the direction of transfer.
TABLE 2.
Mobilization of the E. coli chromosome by R751 and the R1162 Mob systema
| Donor strain | No. of transconjugants per donor with chromosomal marker:
|
||
|---|---|---|---|
| Tra+ Mob+ | Mob+ | Tra+ | |
| dusC::Kan | 5.5 × 10−7 | <7.2 × 10−9 | 1.8 × 10−8 |
| yeiR::Kan | 4.5 × 10−7 | 3.4 × 10−9 | <1.4 × 10−8 |
| [ΔoriT]::Cam yeiR::Kan | 4.3 × 10−7 | NT | 1.5 × 10−9 |
Donors contained R751 and either pUT1841 or pUT1559 (Fig. 2) (Tra+ Mob+), pUT1841 or pUT1559 alone (Mob+), or R751 alone (Tra+). NT, not tested.
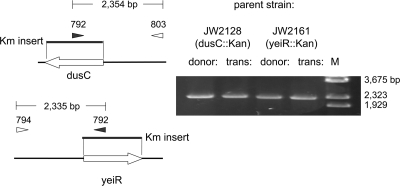
Several tests were performed to determine that the Kanr colonies were true transconjugants, with the Kanr DNA inserted by recombination into the chromosome at the correct location. First, all of the Kanr cells were Lac+, since they formed blue colonies on medium containing X-Gal and IPTG (isopropyl-β-d-thiogalactopyranoside; the donors are white on this medium because of the mutation lacZ4787 [1]). Second, the Kanr marker was recombined at the correct position in the chromosome of the recipient. I isolated genomic DNA from cells of each of the donor strains, as well as the corresponding transconjugants. The DNA was amplified with primers hybridizing to an internal segment of the Kanr gene and to an adjacent region of the chromosome (primers 792 + 793 and primers 792 + 794, Table 2). The sizes of the PCR products were the same for both donor and transconjugant, showing that the Kanr marker remained at its normal location in the chromosome after transfer (Fig. 8).
FIG. 8.
Analysis of Kanr transconjugants by PCR. The locations of the primers (Table 1) and the expected sizes of the PCR products are shown on the left. On the right, PCR results for donor and transconjugant after mating, displayed on 1.2% agarose gel are displayed. The marker is BstEII-digested λ DNA.
The data indicate that there is at least one active, cryptic oriT in the E. coli chromosome and also that at least 40,000 bp of DNA can be transferred. One possibility is that transfer is strongly polar, so that the mobilization frequency of a particular segment is determined by the activity of the nearest cryptic origin. Alternatively, the transfer frequency might reflect the combined contribution of all of the active cryptic origins on the chromosome. To distinguish between these possibilities, bp 2225979 to 2225997 in JW2161 (Fig. 3) were replaced with DNA encoding Camr, and matings were then carried out as before. There was still transfer of the Kanr marker in yeiR; furthermore, many but not all of the transconjugants were Camr (28 of 40 tested). The nearest potential cryptic oriTs that could be responsible for transfer of this DNA, taking into account orientation, are approximately 703,000 and 678,000 bp from the Kan insertion. These results favor a model where there is infrequent transfer initiated from a number of different cryptic origins on the chromosome. However, once transfer is begun, a large fragment of DNA can be mobilized.
DISCUSSION
The R1162 Mob proteins can act in trans to activate cryptic, oriT-like sequences in the chromosome, resulting in the transfer of segments of the chromosome at low but significant frequency. In the example reported here, DNA was rescued by homologous recombination and successful transfer would therefore be limited to closely related organisms, However, there is no reason in principle why DNA fragments capable of nonhomologous recombination cannot also be transferred by R1162 as well. Since R1162 has a very broad host range (29), this system of gene transfer could operate in many different species of bacteria. The mobilizing vector R751 used in these studies also has a broad host range, as do other IncP1 plasmids. These plasmids efficiently transfer R1162 and can be transferred into many species and maintained with various degrees of stability (14, 32). Thus, although the frequency of transfer will probably be low, the potential for such transfer exists in many different bacteria.
In the present study, transfer of the E. coli chromosome cannot be specifically linked to the closest cryptic site. Nevertheless, the eight cryptic sites in MG1655 are probably required, since transfer depends on the R1162 Mob proteins (Table 2). Some bacteria, such as several species of Pseudomonas, have fewer potential cryptic origins in their chromosome, and it will be interesting to see whether indeed the chromosomes of these organisms are poorly transferred by R1162. On the other hand, no strain is permanently immune: R1162-like Mob proteins and oriTs are a very large family (16) and can be acquired on the chromosome as parts of genomic islands, thus introducing one or more potential cryptic sites. In Pectobacterium, three of the ten potential cryptic sites are located as a cluster in a likely genomic island (Fig. 3). The three are related (two have identical sequence) and are intergenic, suggesting that they are derived from true oriTs. The appearance and persistence of such loci, even after the accumulation of mutations, can result in the transfer of genomic island DNA, as well as other DNA when R1162 is then introduced into the cell.
Plasmids encoding a protein related to MobA, and containing a putative oriT recognizable by sequence, are widely distributed in bacteria, with examples found not only in the proteobacteria but also in the firmicutes, actinobacteria, and cyanobacteria. An interesting general question is how many of these diverse mobilization systems also have a relaxed specificity for initiation. It is clear that this property is not an invariable characteristic of the group. The plasmid pSC101 encodes a MobA protein that is similar to the R1162 MobA (28), and the oriT is very similar as well, but very few changes in the sequence of this oriT are tolerated by the pSC101 Mob proteins (21). However, even if the R1162 MobA is unique, its broad host range magnifies its potential: R1162 was isolated from Pseudomonas aeruginosa (4), while virtually identical plasmids were independently isolated from E. coli as RSF1010 (18) and from Salmonella enterica serovar Typhimurium as R300B (4). In addition, there are plasmids, listed in Table 3, with MobAs and oriTs very closely related to those in this group, and thus candidates to have MobAs with relaxed recognition of oriT.
TABLE 3.
Plasmids with MobA and oriT very closely related to those of R1162, RSF1010, and R300B
| Plasmid | MobA identity (%) | oriT identity (bp fraction) | Host | Source or reference | GenBank accession no. |
|---|---|---|---|---|---|
| pCCK381 | 71.6 | 29/32 | Pasteurella multocida | 22 | NC 006994 |
| pIE1130 | 79.2 | 30/32 | Uncultured eubacterium | Unpublished data | AJ271879 |
| pMS260 | 79.2 | 29/32 | Actinobacillus pleuropneumoniae | 20 | AB109805 |
| pIE1115 | 72 | 29/32 | Uncultured eubacterium | Unpublished data | AJ293027 |
| pDN1 | 71.9 | 29/32 | Dichelobacter nodosus | 34 | NC 002636 |
Although the IncP1 plasmids are themselves sufficient to ensure the spread of R1162 into many different strains, type IV secretion systems located on the chromosome can also bring about transfer. For example, the dot-icm virulence system of Legionella pneumophila has been shown to transfer RSF1010 (33). More recently, a pathogenicity island in S. enterica serovar Typhi was reported to mobilize R300B (3). These and other resident, type IV secretion systems could act together with the plasmid Mob proteins to bring about the transfer of chromosomal DNA.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Baba, T., T. Ara, Hasegawa. M, Y. Takai, Y. Okumura, M. Baba, K. Datsenko, M. Tomita, B. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Systems Biol. 20061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. J. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36525-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, S., D. Pickard, S. Whitehead, J. Farrar, and G. Dougan. 2008. Mobilization of the incQ plasmid R300B with a chromosomal system in Salmonella enterica serovar Typhi. J. Bacteriol. 1904084-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, P. T., and N. J. Grinter. 1974. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J. Bacteriol. 120618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, E., and R. Meyer. 2003. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J. Bacteriol. 1853538-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, K., M. Sebaihia, L. Pritchard, M. Holden, L. Hyman, M. Holeva, N. Thomsom, S. Bentley, L. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. Quail, M. Sanders, D. Walker, S. Whitehead, G. Salmond, P. Birch, J. Parkkhill, and I. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 10111105-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharjee, M., X. M. Rao, and R. J. Meyer. 1992. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J. Bacteriol. 1746659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F., I. Plunkett, G., C. Bloch, N. Perna, V. Burland, M. Riley, J. Collado-Vides, J. Glasner, C. Rode, G. Mayhew, J. Gregor, N. Davis, H. Kirkpatrick, M. Goeden, D. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1462. [DOI] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of amplifiable multicopy DNA cloning vehicles II: a multiple cloning system. Gene 295-113. [PubMed] [Google Scholar]
- 10.Cavalli-Sforza, L. 1992. Forty years ago in Genetics: the unorthodox mating behavior of bacteria. Genetics 132635-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clowes, R., and E. Moody. 1966. Chromosomal transfer from “recombination-deficient” strains of Escherichia coli K-12. Genetics 53717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K., and B. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gelder, L., J. Ponciano, P. Joyce, and E. Top. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153452-463. [DOI] [PubMed] [Google Scholar]
- 15.Fekete, R., and L. Frost. 2000. Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J. Bacteriol. 1824022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francia, M., A. Varsaki, M. Garcillan-Barcia, A. Latorre, C. Drainas, and F. de la Cruz. 2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2879-100. [DOI] [PubMed] [Google Scholar]
- 17.Gay, P., D. Le Coq, M. Steinmetz, T. Bekelman, and C. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerry, P., J. van Embden, and S. Falkow. 1974. Molecular nature of two non-conjugative plasmids carrying drug resistance genes. J. Bacteriol. 117619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, D., and R. Meyer. 1999. The MobA-linked primase is the only replication protein of R1162 required for conjugal mobilization. J. Bacteriol. 1812973-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, H., H. Ishi, and M. Akiba. 2004. Analysis of the complete nucleotide sequence of an Actinobacillus pleuropneumoniae streptomycin-sulfonamide resistance plasmid, pMS260. Plasmid 5141-47. [DOI] [PubMed] [Google Scholar]
- 21.Jandle, S., and R. Meyer. 2006. Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J. Bacteriol. 188499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kehrenberg, C., and S. Schwarz. 2005. Plasmid-borne florfenicol resistance in Pasteurella multocida. J. Antimicrob. Chemother. 55773-775. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K., and R. J. Meyer. 1989. Unidirectional transfer of broad host-range plasmid R1162 during conjugative mobilization. Evidence for genetically distinct events at oriT. J. Mol. Biol. 208501-505. [DOI] [PubMed] [Google Scholar]
- 24.Lawley, T., W. Klinke, M. Gubbins, and L. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lets. 2241-15. [DOI] [PubMed] [Google Scholar]
- 25.Lesic, B., and L. Rahme. 2008. Use of lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol. Biol. 920-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llosa, M., S. Bolland, and F. de la Cruz. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J. Bacteriol. 1763210-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matson, S. W., and B. Morton. 1991. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 26616232-16237. [PubMed] [Google Scholar]
- 28.Meyer, R. 2000. Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J. Bacteriol. 1824875-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawlings, D., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherzinger, E., V. Kruft, and S. Otto. 1993. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur. J. Biochem. 217929-938. [DOI] [PubMed] [Google Scholar]
- 31.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75271-288. [DOI] [PubMed] [Google Scholar]
- 32.Thomas, C., and C. Smith. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 4177-101. [DOI] [PubMed] [Google Scholar]
- 33.Vogel, J., H. Andrews, S. Wong, and R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279873-876. [DOI] [PubMed] [Google Scholar]
- 34.Whittle, G., M. Katz, E. Clayton, and B. Cheatham. 2000. Identification and characterization of a native Dichelobacter nodosus plasmid, pDN1. Plasmid 43230-234. [DOI] [PubMed] [Google Scholar]
- 35.Willetts, N., and C. Crowther. 1981. Mobilization of the nonconjugative IncQ plasmid RSF1010. Genet. Res. 37311-316. [DOI] [PubMed] [Google Scholar]