Abstract
The coordinated high-light response of genes encoding subunits of photosystem I (PSI) is achieved by the AT-rich region located just upstream of the core promoter in Synechocystis sp. strain PCC 6803. The upstream element enhances the basal promoter activity under low-light conditions, whereas this positive regulation is lost immediately after the shift to high-light conditions. In this study, we focused on a high-light regulatory 1 (HLR1) sequence included in the upstream element of every PSI gene examined. A gel mobility shift assay revealed that a response regulator RpaB binds to the HLR1 sequence in PSI promoters. Base substitution in the HLR1 sequence or decrease in copy number of the rpaB gene resulted in decrease in the promoter activity of PSI genes under low-light conditions. These observations suggest that RpaB acts as a transcriptional activator for PSI genes. It is likely that RpaB binds to the HLR1 sequence under low-light conditions and works for positive regulation of PSI genes and for negative regulation of high-light-inducible genes depending on the location of the HLR1 sequence within target promoters.
In photosynthesis, light energy is absorbed by the light-harvesting antennae pigments and converted into chemical energy by the reaction centers. However, when the supply of light energy exceeds its consumption, elevated excitation pressure results in excess production of reactive oxygen species, leading to severe damage to many cellular processes (3, 8, 20). Thus, the absorption of excess light energy must be avoided under light-saturated conditions by decreasing the amount of light-harvesting antenna complexes per reaction center or the amount of reaction center complexes itself.
In cyanobacteria, the decrease of photosystem (PS) content, as well as phycobilisome content, is typically observed under high-light (HL) conditions, and the main component to be downregulated is not PSII but PSI (14, 22). The physiological significance of the selective repression of PSI content during HL acclimation has been demonstrated by the characterization of the two mutants of Synechocystis sp. strain PCC 6803, disruptants of pmgA (sll1968) and sll1961, both of which have defect in keeping their PSI content at low level under HL conditions (9, 14). They grew better than the wild-type cells during a short-term exposure (e.g., 24 h) to HL because a higher amount of PSI accelerated the rate of photosynthetic electron transport (14). Under prolonged HL conditions, however, the growth of the mutants was severely inhibited (9, 14, 30), presumably due to the generation of reactive oxygen species at the acceptor side of PSI. These observations strongly suggest that the repression of PSI content is indispensable for growth under continuous HL conditions.
In Synechocystis sp. strain PCC 6803, the PSI complex is comprised of 11 subunits and genes encoding these subunits (PSI genes) are dispersed throughout the genome (11, 17). PSI genes are actively transcribed under low-light (LL) conditions, whereas their transcription is coordinately and strictly downregulated upon the shift to HL conditions preceding the decrease in protein level (13, 15, 16, 23, 31). We conducted the deletion analysis of PSI promoters and found that an AT-rich upstream region from −70 to −46, relative to the transcription start site, is involved in upregulation of the promoter activity in every PSI gene (24, 25). The addition of an AT-rich upstream element to the core promoter region stimulated the promoter activity 5- to 100-fold under LL conditions, whereas this positive regulation was suppressed within 1 h after the shift to HL. This change in the activity of the upstream element was well correlated with changes in PSI transcript levels upon the shift from LL to HL conditions, showing that the upstream element is responsible for the coordinated HL response of PSI genes. However, transcriptional factors involved in the regulation have remained unknown.
In the present study, we found that a response regulator RpaB binds to the AT-rich upstream region of PSI genes. Although RpaB has thus far been reported as a repressor for HL-inducible genes (19, 29), our results suggest the activation of PSI genes by RpaB under LL conditions. It is likely that RpaB is active under LL conditions and works for positive regulation of PSI genes and for negative regulation of HL-inducible genes depending on the location of its binding site within target promoters.
MATERIALS AND METHODS
Strains and culture conditions.
A glucose-tolerant strain of Synechocystis sp. strain PCC 6803 was grown at 31°C on solid BG-11 medium containing 5 mM TES-KOH (pH 8.2), 0.3% (wt/vol) sodium thiosulfate, and 1.5% (wt/vol) agar under continuous illumination provided by fluorescent lamps at 20 μmol of photons m−2 s−1. To maintain the rpaB-disrupted strain and reporter-transformed strains, chloramphenicol (25 μg/ml) and spectinomycin (20 μg/ml) were added, respectively. Cell density was estimated at optical density at 730 nm (OD730) using a spectrophotometer (model UV-160A; Shimadzu).
Escherichia coli and DNA manipulation.
XL1-Blue MRF′ (Stratagene) was the host for all plasmids constructed in the present study. When required, ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), or spectinomycin (20 μg/ml) was added to Terrific broth medium for selection of plasmids in E. coli. The procedures for the growth of E. coli strains and for the manipulation of DNA were as described in Sambrook et al. (28). Sequencing of plasmids was carried out by the dideoxy-chain termination method using a dye terminator cycle sequencing kit (Applied Biosystems).
Overexpression and purification of His-RpaB.
The rpaB coding region was PCR amplified using the primers NdeI-rpaBcod-F (5′-AACATATGGTCGATGACGAGGCC-3′) and BamHpa-rpaBcod-R (5′- AAGGATCCGTTAACTTACGGTTCTTCCCCCGG-3′), cloned into the pT7Blue T-Vector (Novagen), digested with NdeI and BamHI (the sites are underlined), and subcloned into the same restriction sites in pET28a (Novagen) to create pETrpaB for expression of a fusion protein with an N-terminal His tag. The nucleotide sequence was confirmed by DNA sequencing.
E. coli BL21(DE3) harboring pETrpaB was grown overnight to an OD600 of 10 in 500 ml of 2× yeast extract-tryptone medium containing 20 μg of kanamycin/ml. Cells were harvested by centrifugation, resuspended in 20 ml of buffer S (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM dithiothreitol [DTT]), and disrupted by eight rounds of sonication for 30 s each at 4°C. The inclusion body fraction containing the overexpressed protein was pelleted by centrifugation at 12,000 × g for 30 min. The pellet was washed several times with buffer S containing 4% (wt/vol) Triton X-100 and then with distilled water until the supernatant became clarified. The pellet was then solubilized in 8 M urea solution (50 mM Tris-HCl [pH 8.0], 1 mM DTT, 8 M urea) at room temperature for 30 min and centrifuged at 18,000 × g for 30 min to remove insoluble materials. The supernatant was sequentially dialyzed in three steps: first against 50 mM Tris-HCl (pH 8.0) containing 1 mM DTT and 4 M urea for 1 h, then against the same buffer containing 2 M urea for 1 h, and finally overnight against 20 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 10 mM imidazole.
After centrifugation at 18,000 × g for 30 min, the supernatant was applied to a HiTrap chelating HP column (GE Healthcare) that was preequilibrated with 20 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 10 mM imidazole. After a washing step with 20 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 100 mM imidazole, His-RpaB was eluted with 20 mM phosphate buffer (pH 7.4) containing 0.5 M NaCl and 300 mM imidazole. Purified His-RpaB was desalted by using a HiTrap desalting column (GE Healthcare). Protein composition was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by staining with Coomassie brilliant blue R-250.
Gel mobility shift assay.
Probes and competitor DNA fragments for gel mobility shift assays were obtained by PCR amplification using genomic DNA as a template. psaEsub1 and psaEsub2 fragments were obtained using mutated forward primers: PpsaEsub1-F (5′-TAGAACCACTCCCAGGAGCAGGGACCCCTAAAGAATTGTTTT-3′) and PpsaEsub2-F (5′-TAGAACCACTCCCAGGAGCAGGGATATGTAAAGACCCCTTTT-3′), respectively. The 3′ end of the DNA fragment for each probe was labeled with digoxigenin (DIG)-ddUTP by the terminal transferase method according to the manufacturer's instructions (DIG gel shift kit; Roche). Assays were performed by using a DIG gel shift kit as previously described (26).
Generation of luxAB reporter strains.
All plasmids used for the reporter assay were derivatives of pPT6803-1, which is a recombinational plasmid carrying the promoterless luxAB genes, the neutral site sequence of Synechocystis sp. strain PCC 6803 (the downstream region of the ndhB gene) and the spectinomycin resistance cassette (1, 24). Reporter construct E1 containing the region from −70 to + 90 of psaE, E2 containing the region from −45 to +90 of psaE, and A61 containing the region from −69 to +2 of psaA were generated as described in Muramatsu and Hihara (25). The base-substituted constructs, E1sub1 and E1sub2, were generated by using a KOD-Plus mutagenesis kit (Toyobo) according to the manufacturer's instruction. Namely, E1 construct was mutagenized by inverse PCR using the primer set PpsaE-luxSub1 (5′-CGCCCCTAAAGAATTGTTTTGGGAAAG-3′) and luxAB-3 (5′-GCTTTCAATTTCCGCTTT-3′) to generate E1sub1 and the primer set PpsaE-luxSub2 (5′-CGTATGTAAAGACCCCTTTTGGGAAAGTCGGGGGGA-3′) and luxAB-3 to generate E1sub2.
Measurement of bioluminescence from cells harboring luciferase reporter genes.
For in vivo bioluminescence measurements of Synechocystis cells, cells grown on solid BG11 medium were suspended in distilled water. An aliquot (200 μl) was transferred to a reaction tube and set immediately in the luminescence counter (Lumi-counter model 2500; Microtech-Nichion). A total of 100 μl of 0.15% n-decanal (vol/vol) was injected into a reaction tube with a syringe, and bioluminescence from the cells was measured during 120 s after the injection of n-decanal. Specific luciferase activities were calculated as relative units/OD730.
Generation of the rpaB-disrupted strain.
For generation of the rpaB-disrupted strain, a cosmid clone having insertion of a chloramphenicol resistance cassette into the coding region of rpaB (nucleotide 2014496 according to the numbering in CyanoBase) was selected from the transposon-mutagenized cosmid library of Synechocystis sp. strain PCC 6803 (27) and transformed to the wild-type and A61 reporter strains. Transformants were selected by addition of chloramphenicol (25 μg/ml).
Determination of pigment contents.
Cells grown on solid BG11 medium were suspended in distilled water, and in vivo absorption spectra were measured at room temperature by using a spectrophotometer (model 557; Hitachi) with an end-on photomultiplier. Chlorophyll and phycocyanin contents were calculated from the peak heights of absorption spectra using the equations of Arnon et al. (2).
RESULTS
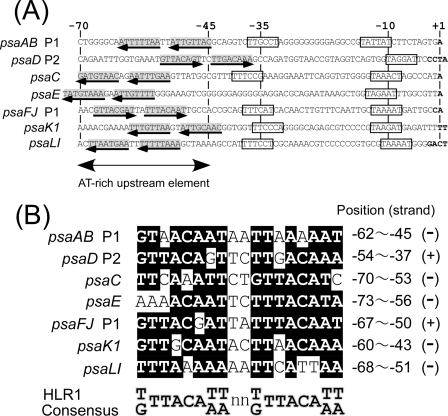
To identify transcriptional regulators that bind to the light responsive AT-rich upstream region of PSI genes, we searched for a common regulatory sequence located in this region. As shown in Fig. 1, there exists an HL regulatory 1 (HLR1) motif, an imperfect direct repeat comprised of two octamers (G/T)TTACA(T/A)(T/A) separated by two nucleotides (18), in the upstream region of every PSI gene. The HLR1 motif was initially identified by Eriksson et al. (7) as a common sequence located upstream of HL-inducible genes such as psbA2, psbA3, hliA, and nblA in Synechocystis sp. strain PCC 6803. Promoter analysis revealed that deletion of the HLR1-containing region resulted in elevated expression of psbA2 in Synechocystis sp. strain PCC 6803 (7) and of hliA in Synechococcus elongatus PCC 7942 (18) under LL conditions. These observations indicate that the HLR1 sequence is recognized by a repressor protein working under LL conditions. Recently, the RpaB (Slr0947, Ycf27, and Rre26) response regulator was reported to bind to the HLR1 sequence of hliB in Synechocystis sp. strain PCC 6803 (19) and of rpoD3 in S. elongatus PCC 7942 (29). Unlike above-mentioned HL-inducible genes, PSI genes are downregulated under HL, and their AT-rich upstream element is likely to be recognized by an activator protein under LL conditions (25). Thus, it is intriguing to see if RpaB, the repressor for HL-inducible genes, can bind to PSI promoters to work for positive regulation under LL conditions.
FIG. 1.
(A) Nucleotide sequences of the core promoter and the light-responsive AT-rich upstream element of PSI genes in Synechocystis sp. strain PCC 6803. The sequences are aligned according to the major transcription start site noted as +1. Putative −35 and −10 hexamers are boxed. The direct repeat of the HLR1 motif is indicated by arrows. (B) Alignment of the HLR1 sequences found in the upstream region of PSI promoters. Residues identical to the HLR1 consensus sequence (18) are shaded in black. The position relative to the transcription start site and the orientation of HLR1 sequence are shown for each PSI gene.
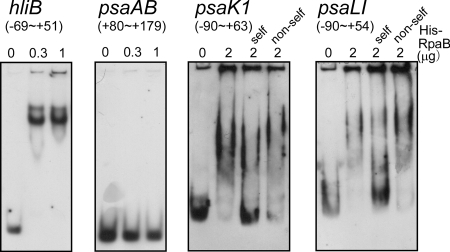
In order to test whether RpaB binds to the upstream region of PSI genes, we overexpressed RpaB protein with N-terminal histidine tag (His-RpaB) in E. coli. His-RpaB exclusively accumulated in the insoluble fraction of the overnight culture without induction with IPTG (isopropyl-β-d-thiogalactopyranoside). The insoluble fraction was repeatedly washed with 4% Triton X-100, and the resulting inclusion body fraction was solubilized by 8 M urea. After centrifugation, the supernatant was renatured by dialysis, and His-RpaB was purified to near homogeneity through nickel affinity chromatography. We first checked the activity of purified His-RpaB by gel mobility shift assay with various promoter fragments (Fig. 2). When the promoter region of hliB, one of the target genes of RpaB (19), was used as a probe, the addition of 0.3 μg of His-RpaB was sufficient for the formation of a shifted complex and the disappearance of the free probe. On the other hand, no shifted complex was observed when the 5′ untranslated region and the coding region of psaAB (+80 to +179) was used as a probe. The HLR1-containing fragments of psaK1 and psaLI promoters formed a shifted complex with His-RpaB, and the specificity of this complex formation was tested by competition assays. Unlabeled probe fragments (“self”) significantly competed for binding with the labeled probe, but unlabeled coding region fragments with the same length as the probe fragments (“non-self”) were not sufficient to compete for binding.
FIG. 2.
Gel mobility shift assay of various DNA fragments with His-RpaB. DIG-labeled upstream DNA fragments of hliB (−69 to +51), psaAB (+80 to +179), psaK1 (−90 to +63), and psaLI (−90 to +54) were incubated for 30 min with His-RpaB added at the indicated concentrations. Samples were separated on a 6% polyacrylamide gel. For the competition assay of psaK1, unlabeled probe fragment (−90 to +63: self) or nonspecific fragment from the coding region (+61 to +213: non-self) was added at a 150-fold excess of the probe concentration. For the competition assay of psaLI, unlabeled probe fragment (−90 to +54: self) or nonspecific fragment from the coding region (+121 to +264: non-self) was added at a 200-fold excess of the probe concentration.
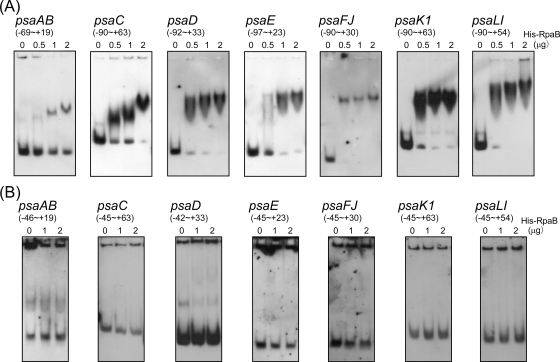
The binding of His-RpaB protein to each PSI promoter then was tested. When the DNA fragment containing the upstream element from positions −70 to −46 was used as a probe, a shifted complex was clearly observed in every PSI gene (Fig. 3A). To obtain the completely shifted band, addition of 0.5 μg of His-RpaB was enough in the case of psaD, psaFJ, psaK1, and psaLI, whereas a greater amount of His-RpaB was required in the case of psaAB, psaC, and psaE. Next, the upstream element was deleted from each probe used in Fig. 3A, and a gel mobility shift assay was performed again. As shown in Fig. 3B, no shifted complex was observed, showing that His-RpaB binds to the HL-responsive upstream element of PSI genes.
FIG. 3.
Gel mobility shift assay of the promoter segments of PSI genes with His-RpaB. (A) A DIG-labeled promoter segment of each PSI gene containing the upstream element from −70 to −46 was incubated for 30 min with His-RpaB added at the indicated concentrations. Samples were separated on a 6% polyacrylamide gel. (B) The upstream element was deleted from each probe used in panel A, and a gel mobility shift assay was performed in the same procedure.
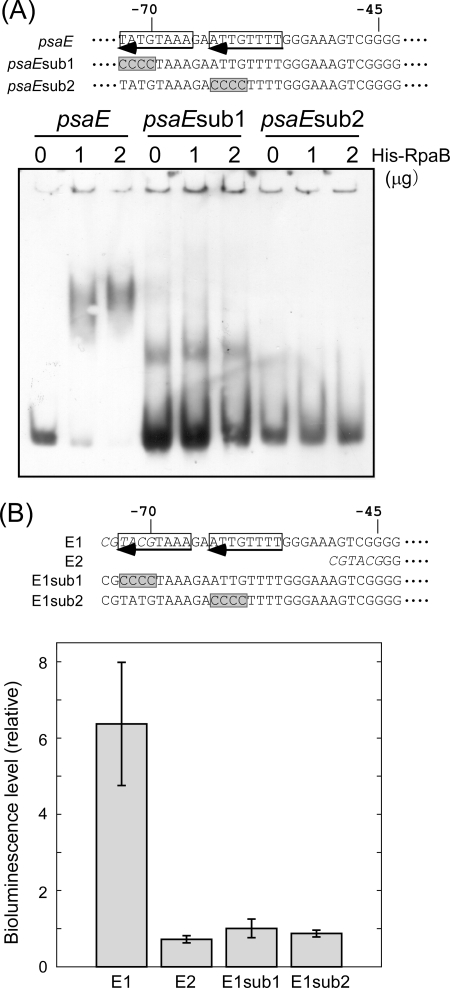
In order to confirm the binding of His-RpaB to the HLR1 sequence, a gel mobility shift experiment was performed with probes having base substitution within the HLR1 sequence. Figure 4A shows the effect of four-base substitutions in each half-site of the HLR1 sequence in the psaE probe (psaEsub1 and psaEsub2). Either substitution resulted in the complete loss of the binding of His-RpaB. This clearly shows that binding of His-RpaB requires the HLR1 sequence. Next, the effect of the same base substitutions on the promoter activity of the psaE gene under LL conditions was examined using luxAB reporter constructs (Fig. 4B). Previously, we reported that E1 strain containing the luxAB genes fused to the region from −70 to +90 of the psaE gene showed much higher reporter activity than E2 strain containing the luxAB genes fused to the region from −45 to +90 (25). In the E1 strain, a part of the HLR1 sequence (−73 to −71) was not contained. Instead, the addition of a BsiWI site (CGTACG) to the 5′ terminus of the promoter fragment resulted in the formation of TACGTAAA (−73 to −66) that seems to be recognized as a half-site of the HLR1 sequence. When this half-site was changed to CCCCTAAA (E1sub1), the promoter activity largely decreased nearly to the level of E2 strain. Similarly, the base substitution in the other half-site (E1sub2) resulted in the decline in the promoter activity. These observations suggest that RpaB binds to the HLR1 sequence of PSI genes and acts as a transcriptional activator under LL conditions.
FIG. 4.
Effect of base substitution within the HLR1 sequence of the psaE promoter. (A) Gel mobility shift assay of the psaE promoter segments with or without base substitution. DIG-labeled psaE promoter fragments (−97 to +23) with or without base substitution were incubated for 30 min with His-RpaB added at the indicated concentrations. Samples were separated on a 6% polyacrylamide gel. Nucleotide sequences of promoter fragments used for the experiment are shown at the top of the panel. The direct repeat of the HLR1 sequence in the psaE promoter is indicated by arrows, and the sites of base substitution in psaEsub1 and psaEsub2 fragments are shaded in gray. (B) Bioluminescence level from low-light grown Synechocystis cells harboring luxAB reporter genes fused to the psaE promoter with or without base substitution. Error bars represent the standard deviation among three independent measurements. Nucleotide sequence of 5′-terminal region of each reporter fusion is shown at the top of the panel. BsiWI sites included in E1 (−70 to +90) and E2 (−45 to +90) are shown in italics. The sites of base substitution in E1sub1 and E1sub2 are shaded in gray.
To examine in vivo role of RpaB, a gene-disrupted mutant was created by the insertion of a chloramphenicol resistance cassette into rpaB. However, as reported by Ashby and Mullineaux (4), rpaB was an essential gene in Synechocystis sp. strain PCC 6803, and only a transformant with a decreased copy number of rpaB was obtained. The transformant formed small yellowish colonies on BG11 agar plates, while its growth rate and pigment contents showed a substantial increase by the addition of 5 mM glucose. In liquid culture, no propagation was observed irrespective of the presence or absence of glucose. During the cultivation on agar plates, large green colonies of suppressor mutants emerged in high frequency. We collected yellowish colonies of the original transformant grown on the agar plate without glucose, measured the absorption spectra, and calculated the cellular pigment contents. The cellular chlorophyll contents (in pg/cell) of the wild type and the mutant were 0.047 ± 0.003 and 0.036 ± 0.004, respectively. Similarly, the cellular phycocyanin contents (in pg/cell) of the wild type and the mutant were 0.282 ± 0.014 and 0.205 ± 0.015, respectively. When the luxAB reporter genes fused to the psaAB promoter fragment from −69 to +2 was introduced, the reporter activity from the mutant cells was only 26% of the wild-type level. Thus, it seemed that decrease of the copy number of rpaB resulted in decrease of psaAB promoter activity, cellular pigment contents, and growth rate under LL conditions.
DISCUSSION
In this report, we show that the response regulator RpaB binds to the HLR1 sequence located in the AT-rich upstream element of PSI genes (Fig. 3 and 4A). Base substitution in the HLR1 sequence (Fig. 4B) or a decrease in copy number of the rpaB gene resulted in the decrease in the promoter activity of PSI genes under LL conditions. Previously, we observed that base substitution of TTTTT (−61 to −57) or TTATT (−54 to −50) but not of GGGGC (−68 to −64) resulted in drastic decrease of the psaAB promoter activity under LL conditions (24). This can be another example that the HLR1 sequence (−62 to −45, in the case of psaAB) is essential for the activation of PSI promoters. Taken together, our observations suggest that RpaB is an activator for PSI genes working under LL conditions.
RpaB has thus far been reported as a negative regulator for HL-inducible genes working under LL conditions (19, 29). Here, we reported for the first time that RpaB can work for positive regulation under the same LL conditions. The regulatory role of RpaB is likely to be determined by the location of the HLR1 sequence in the target promoters. In the case of HL-inducible genes, the HLR1 sequence is located within the core promoter region (7, 18) or within the 5′ untranslated region (29), and it was proposed that binding of RpaB to the HLR1 sequence prevents the interaction between RNA polymerase and the core promoter sequence. On the other hand, we found that the HLR1 sequence is located upstream of the core promoter region in the case of PSI genes (Fig. 1). In various bacterial species, transcriptional activators, including several response regulators, were reported to bind upstream of the core promoter region. They interact with the C-terminal domain of the α subunit of RNA polymerase (α-CTD), leading to an increase in the rate of transcription initiation (5, 6, 10). It is possible that RpaB interacts with α-CTD at the HLR1 sequence of PSI genes to enhance the promoter activity.
Recently, Hanaoka and Tanaka (12) showed by chromatin immunoprecipitation analysis that binding of RpaB to its target promoters, hliA and rpoD3 in S. elongatus PCC 7942, was promptly lost upon the shift to HL. Thus, it is supposed that the coordinated downregulation of PSI genes, as well as upregulation of HL-inducible genes, is accomplished by release of RpaB from the HLR1 sequence under HL conditions in Synechocystis sp. strain PCC 6803. How then is the change in light intensity transmitted to RpaB? Seki et al. (29) observed that overexpression of the truncated RpaB protein harboring only the phosphoreceiver domain resulted in the derepression of hliA and rpoD3 genes under LL conditions in S. elongatus PCC 7942. This phenomenon could be due to a decrease in the phosphorylation level of the native RpaB protein, and these authors proposed that RpaB is phosphorylated by its cognate histidine kinase under LL to work for negative regulation. The prime candidate for the cognate histidine kinase is thought to be NblS (also known as Hik33 or DspA), since the HLR1 sequence is found in the promoter region of genes regulated by NblS (18, 19). However, phosphorylation of RpaB by NblS/Hik33 under LL conditions is still controversial. Based on their extensive studies using DNA microarray technique, Los et al. (21) concluded that Hik33 is inactive under nonstress conditions and is activated in response to the environmental stress. We observed that the effect of disruption of Hik33 is far smaller than that of RpaB under LL conditions (not shown). The mechanism of the perception of the change in light intensity and the following signal transduction to modulate the RpaB activity remain to be elucidated.
Acknowledgments
This study was supported by a Grant-in-Aid for Young Scientists (to Y.H.) from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Aoki, S., T. Kondo, and M. Ishiura. 2002. A promoter-trap vector for clock-controlled genes in the cyanobacterium Synechocystis sp. PCC 6803. J. Microbiol. Methods 49265-274. [DOI] [PubMed] [Google Scholar]
- 2.Arnon, D. I., B. D. McSwain, H. Y. Tsujimoto, and K. Wada. 1974. Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim. Biophys. Acta 357231-245. [DOI] [PubMed] [Google Scholar]
- 3.Asada, K. 1994. Production and action of active oxygen species in photosynthetic tissues, p. 77-104. In C. H. Foyer and P. M. Mullineaux (ed.), Causes of photooxidative stress and amelioration of defense systems in plants. CRC Press, Boca Raton, FL.
- 4.Ashby, M. K., and C. W. Mullineaux. 1999. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181253-260. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, P. E., A. E. Maris, M. S. Yang, and S. Stibitz. 2003. The response regulator BvgA and RNA polymerase alpha subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol. Cell 11163-173. [DOI] [PubMed] [Google Scholar]
- 6.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79743-746. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, J., G. F. Salih, H. Ghebramedhin, and C. Jansson. 2000. Deletion mutagenesis of the 5′ psbA2 region in Synechocystis 6803: identification of a putative cis element involved in photoregulation. Mol. Cell. Biol. Res. Commun. 3292-298. [DOI] [PubMed] [Google Scholar]
- 8.Foyer, C. H., and J. F. Allen. 2003. Lessons from redox signaling in plants. Antioxid. Redox Signal. 53-5. [DOI] [PubMed] [Google Scholar]
- 9.Fujimori, T., M. Higuchi, H. Sato, H. Aiba, M. Muramatsu, Y. Hihara, and K. Sonoike. 2005. The mutant of sll1961, which encodes a putative transcriptional regulator, has a defect in regulation of photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 139408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng, H., Y. Zhu, K. Mullen, C. S. Zuber, and M. M. Nakano. 2007. Characterization of ResDE-dependent fnr transcription in Bacillus subtilis. J. Bacteriol. 1891745-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grotjohann, I., and P. Fromme. 2005. Structure of cyanobacterial photosystem I. Photosynth. Res. 8551-72. [DOI] [PubMed] [Google Scholar]
- 12.Hanaoka, M., and K. Tanaka. 2008. Dynamics of RpaB-promoter interaction during high light stress, revealed by chromatin immunoprecipitation (ChIP) analysis in Synechococcus elongatus PCC 7942. Plant J. 56327-335. [DOI] [PubMed] [Google Scholar]
- 13.Herranen, M., T. Tyystjarvi, and E. M. Aro. 2005. Regulation of photosystem I reaction center genes in Synechocystis sp. strain PCC 6803 during light acclimation. Plant Cell Physiol. 461484-1493. [DOI] [PubMed] [Google Scholar]
- 14.Hihara, Y., K. Sonoike, and M. Ikeuchi. 1998. A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803 in response to high light. Plant Physiol. 1171205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, L., M. P. McCluskey, H. Ni, and R. A. LaRossa. 2002. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 1846845-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3109-136. [DOI] [PubMed] [Google Scholar]
- 18.Kappell, A. D., D. Bhaya, and L. G. van Waasbergen. 2006. Negative control of the high light-inducible hliA gene and implications for the activities of the NblS sensor kinase in the cyanobacterium Synechococcus elongatus strain PCC 7942. Arch. Microbiol. 186403-413. [DOI] [PubMed] [Google Scholar]
- 19.Kappell, A. D., and L. G. van Waasbergen. 2007. The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the cyanobacterium Synechocystis PCC 6803. Arch. Microbiol. 187337-342. [DOI] [PubMed] [Google Scholar]
- 20.Latifi, A., M. Ruiz, and C. C. Zhang. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev., in press. [DOI] [PubMed]
- 21.Los, D. A., I. Suzuki, V. V. Zinchenko, and N. Murata. 2007. Stress responses in Synechocystis: regulated genes and regulatory systems, p. 117-157. In A. Herrero and E. Flores (ed.), Cyanobacteria: molecular biology, genomics, and evolution. Horizon Scientific Press, Norfolk, United Kingdom.
- 22.Murakami, A., and Y. Fujita. 1991. Regulation of photosystem stoichiometry in the photosynthetic system of the cyanophyte Synechocystis PCC 6714 in response to light-intensity. Plant Cell Physiol. 32223-230. [DOI] [PubMed] [Google Scholar]
- 23.Muramatsu, M., and Y. Hihara. 2003. Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp. PCC 6803. Planta 216446-453. [DOI] [PubMed] [Google Scholar]
- 24.Muramatsu, M., and Y. Hihara. 2006. Characterization of high-light-responsive promoters of the psaAB genes in Synechocystis sp. PCC 6803. Plant Cell Physiol. 47878-890. [DOI] [PubMed] [Google Scholar]
- 25.Muramatsu, M., and Y. Hihara. 2007. Coordinated high-light response of genes encoding subunits of photosystem I is achieved by AT-rich upstream sequences in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1892750-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, K., and Y. Hihara. 2006. Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small, redox-responsive, LuxR-type regulator. J. Biol. Chem. 28136758-36766. [DOI] [PubMed] [Google Scholar]
- 27.Ozaki, H., M. Ikeuchi, T. Ogawa, H. Fukuzawa, and K. Sonoike. 2007. Large-scale analysis of chlorophyll fluorescence kinetics in Synechocystis sp. PCC 6803: identification of the factors involved in the modulation of photosystem stoichiometry. Plant Cell Physiol. 48451-458. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Seki, A., M. Hanaoka, Y. Akimoto, S. Masuda, H. Iwasaki, and K. Tanaka. 2007. Induction of a group 2 sigma factor, RPOD3, by high light and the underlying mechanism in Synechococcus elongatus PCC 7942. J. Biol. Chem. 28236887-36894. [DOI] [PubMed] [Google Scholar]
- 30.Sonoike, K., Y. Hihara, and M. Ikeuchi. 2001. Physiological significance of the regulation of photosystem stoichiometry upon high light acclimation of Synechocystis sp. PCC 6803. Plant Cell Physiol. 42379-384. [DOI] [PubMed] [Google Scholar]
- 31.Tu, C. J., J. Shrager, R. L. Burnap, B. L. Postier, and A. R. Grossman. 2004. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803. J. Bacteriol. 1863889-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]