Abstract
Iron-sulfur clusters may have been the earliest catalytic cofactors on earth, and most modern organisms use them extensively. Although members of the Archaea produce numerous iron-sulfur proteins, the major cluster assembly proteins found in the Bacteria and Eukarya are not universally conserved in archaea. Free-living archaea do have homologs of the bacterial apbC and eukaryotic NBP35 genes that encode iron-sulfur cluster carrier proteins. This study exploits the genetic system of Salmonella enterica to examine the in vivo functionality of apbC/NBP35 homologs from three archaea: Methanococcus maripaludis, Methanocaldococcus jannaschii, and Sulfolobus solfataricus. All three archaeal homologs could correct the tricarballylate growth defect of an S. enterica apbC mutant. Additional genetic studies showed that the conserved Walker box serine and the Cys-X-X-Cys motif of the M. maripaludis MMP0704 protein were both required for function in vivo but that the amino-terminal ferredoxin domain was not. MMP0704 protein and an MMP0704 variant protein missing the N-terminal ferredoxin domain were purified, and the Fe-S clusters were chemically reconstituted. Both proteins bound equimolar concentrations of Fe and S and had UV-visible spectra similar to those of known [4Fe-4S] cluster-containing proteins. This family of dimeric iron-sulfur carrier proteins evolved before the archaeal and eukaryal lineages diverged, representing an ancient mode of cluster assembly.
Members of the Archaea produce many proteins that require iron-sulfur cluster cofactors, including redox proteins, aconitase-like dehydratases, radical S-adenosylmethionine enzymes, and RNA polymerase (9, 13, 18, 32). Methanogenic archaea are obligate anaerobes, and many heterotrophic archaea grow anaerobically, indicating that oxidative stress has not limited the proliferation of iron-sulfur proteins in these lineages. Archaea must have a mechanism to assemble Fe-S clusters, but many members lack homologs of the known bacterial and eukaryotic Nif or Isc systems, suggesting that an alternative system is present (see Table S1 in the supplemental material) (14, 15).
Some euryarchaea have homologs of the bacterial genes iscS (encoding cysteine desulfurase) and iscU (encoding a scaffold protein). Yet many other archaea, including the euryarchaea Pyrococcus furiosus, Methanocaldococcus jannaschii, and Methanococcus maripaludis, plus most crenarchaea, either lack a homologous cysteine desulfurase gene or have no homologs of A-type or U-type scaffold genes. Due to their sulfide-rich environments, it is reasonable that the anaerobic archaea may use an inorganic sulfur source to assemble Fe-S clusters. Most archaeal genome sequences do carry homologs of the sufBC genes that are part of the alternative Suf system for Fe-S cluster biosynthesis (36). Biochemical studies have shown that SufC is an ABC-type ATPase and that SufB is a persulfide acceptor that may act as a site for Fe-S cluster assembly (20). The SufB and SufC proteins interact, and SufB stimulates the ATPase activity of SufC. We hypothesize that the Archaea share a common mechanism for Fe-S cluster biosynthesis, supplemented with genes acquired by horizontal gene transfer in some lineages.
A screen for Salmonella enterica bacteria defective in thiamine biosynthesis identified lesions in the apbC locus (28) that compromised Fe-S metabolism (33). An abpC mutant cannot grow with tricarballylate as a carbon and energy source, which may be due to a defect in assembling or repairing [4Fe-4S] clusters in the membrane-bound TcuB protein (21, 22). ApbC is a 40-kDa cytoplasmic protein with Walker A and B nucleotide-binding domains and two conserved carboxy-terminal cysteine residues separated by two amino acids (Cys-X-X-Cys). Mutational analyses have shown that ApbC proteins with directed changes in the Cys-X-X-Cys or Walker A motifs are not active in vivo (6). Suppressor analysis allowed the conclusion that a degree of functional redundancy between ApbC and the Fe-S scaffold protein IscU exists (4, 38). Although purified ApbC does not contain iron or sulfur, biochemical studies showed that ApbC can bind an Fe-S cluster and rapidly transfer it to an apoprotein (5).
It is thought that in eukaryotes, Fe-S clusters are assembled by the mitochondrial iron-sulfur cluster (ISC) system (23). The clusters are transported into the cytosol and delivered by the cytosolic iron-sulfur protein assembly system. Two components of this system, Nbp35 and Cfd1, are homologs of bacterial ApbC (Fig. 1) and act as intermediate Fe-S cluster-trafficking proteins in the cytosol (16, 27, 30). Electron paramagnetic resonance, Mössbauer, and absorbance spectra of the Saccharomyces cerevisiae, human, and Arabidopsis Nbp35 holoproteins suggest that these holoproteins form dimers with stable amino-terminal [4Fe-4S] clusters and a shared carboxy-terminal [4Fe-4S] cluster (10, 34).
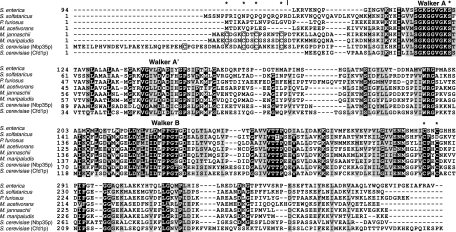
FIG. 1.
A protein sequence alignment of bacterial, archaeal, and eukaryotic ApbC/Nbp35 homologs was constructed using the ClustalW program (version 1.83) (37). The sequence of the S. enterica serovar Typhimurium protein (ApbC; RefSeq accession no. NP_461098.1) is shown without the amino-terminal domain that is not homologous to the amino-terminal domains of the archaeal and eukaryotic proteins. The archaeal homologs are from S. solfataricus (SSO0460; accession no. NP_341994.1), P. furiosus (PF1145; accession no. NP_578874.1), Methanosarcina acetivorans (MA4246; accession no. NP_619111.1), M. jannaschii (MJ0283; accession no. NP_247256.1), and M. maripaludis (MMP0704; accession no. NP_987824.1). The two paralogs from S. cerevisiae are Nbp35 (accession no. NP_011424.1) and Cfd1 (accession no. NP_012263.1). Conserved amino acid residues are shown in white on a black background. Similar residues are shown in black on a gray background. The four conserved amino-terminal cysteine residues shared by the MMP0704 and Nbp35p proteins are boxed. Asterisks above the sequences indicate MMP0704 residues replaced by mutagenesis in this study. A vertical bar indicates the N termini of the truncated proteins MJ0283(19-290) and MMP0704(20-289).
Archaeal homologs of bacterial ApbC and eukaryotic Nbp35 are underannotated as nucleotide-binding proteins or misannotated as cobyrinic acid a,c-diamide synthases in sequence databases. The hallmarks of the Nbp35 sequences are an amino-terminal ferredoxin-like domain, an ATP-binding motif, and two conserved carboxy-terminal cysteine residues that are believed to bind an Fe-S cluster. The amino-terminal ferredoxin-like domain is absent in the ApbC family of proteins. The ApbC and Nbp35 proteins belong to a large superfamily of P-loop-containing nucleoside triphosphate hydrolases that also includes the bacterial MinD and CooC proteins. The M. maripaludis MMP0704 protein shows approximately 40% amino acid identity to both the S. enterica ApbC and S. cerevisiae Nbp35 proteins (Fig. 1). However, the MMP0704 protein also shows 30% sequence identity to two paralogous proteins from M. maripaludis. The genome sequence of M. maripaludis encodes at least nine paralogs, although only the MMP0704 protein contains the conserved cysteine residues found in most ApbC/Nbp35 proteins.
The experiments described herein identified the first archaeal proteins that form functional Fe-S carrier proteins. The apbC/NBP35 homologs from M. maripaludis (MMP0704), M. jannaschii (MJ0283), and Sulfolobus solfataricus (SSO0460) allowed an S. enterica strain with an apbC null mutation to grow on tricarballylate. Genetic studies showed that the Walker A box and at least one cysteine residue from the Cys-X-X-Cys motif were required for in vivo functionality. The unique amino-terminal ferredoxin-like domains of the MMP0704 and MJ0283 proteins were not required. Purified MMP0704 proteins bound Fe-S clusters. Orthologs of ApbC/Npb35 proteins were found in all of the available genomes of free-living archaea, identifying this protein family as an ancient part of the Fe-S assembly system that evolved before the divergence of Archaea and Eukarya.
MATERIALS AND METHODS
Strains, media, and DNA.
Chromosomal DNA was purified from M. jannaschii JAL-1, M. maripaludis S900, and S. solfataricus P2 cells (Table 1). S. enterica strain DM10300 was derived from S. enterica LT2, and the genotype is listed in Table 1. No-carbon essential salts medium supplemented with 1 mM MgSO4, 10 nM thiamine, and trace minerals was prepared as described previously. Tricarballylate or glucose added at a concentration of 11 mM was the sole carbon source (4).
TABLE 1.
List of microorganisms and plasmids
| Strain or plasmid (parent plasmid) | Description and/or partial genotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | Expression strain with T7 RNA polymerase gene | Novagen |
| DH5α | General cloning host | Invitrogen |
| M. jannaschii JAL-1 | Wild type | DSM 2261 |
| M. maripaludis S900 | S2 Δhpt | 25 |
| S. enterica DM10300 | ara-9 apbC55::Tn10d(tet) | 4 |
| S. solfataricus P2 | Wild type | DSM 1617 |
| Plasmids | ||
| pDG499 (pET-11a) | MJ0283 (wild type) | This work |
| pDG530 (pET-11a) | Δ1-18 MJ0283; encodes MJ0283(19-290) protein | This work |
| pDG547 (pET-20b) | Δ1-19 MMP0704; encodes MMP0704(20-289)-His6 protein | This work |
| pDG549 (pET-20b) | MMP0704 (wild type); encodes MMP0704-His6 protein | This work |
| pDG592 (pET-20b) | MMP0704(Cys5,9,12,18Ala) | This work |
| pET-11a | Expression vector | Novagen |
| pET-20b | Expression vector | Novagen |
| pJMB100 (pSU18) | S. enterica apbC (wild type) | This work |
| pJMB102 (pSU18) | MJ0283 (wild type) | This work |
| pJMB103 (pSU18) | Δ1-18 MJ0283 | This work |
| pJMB104 (pSU18) | MMP0704 (wild type) | This work |
| pJMB105 (pSU18) | Δ1-19 MMP0704 | This work |
| pJMB107 (pSU18) | MMP0704(Ser55Ala) | This work |
| pJMB108 (pSU18) | MMP0704(Cys218Ala) | This work |
| pJMB109 (pSU18) | MMP0704(Cys5,9,12,18Ala) | This work |
| pJMB110 (pSU18) | Δ1-19 MMP0704(Ser55Ala) | This work |
| pJMB111 (pSU18) | Δ1-19 MMP0704(Cys218Ala) | This work |
| pJMB112 (pSU18) | MMP0704(Cys220Ala) | This work |
| pJMB113 (pSU18) | MMP0704(Cys218,220Ala) | This work |
| pJMB114 (pSU18) | Δ1-19 MMP0704(Cys220Ala) | This work |
| pJMB115 (pSU18) | Δ1-19 MMP0704(Cys218,220Ala) | This work |
| pJMB116 (pSU18) | SSO0460 | This work |
| pSU18 | Complementation vector | 2 |
Cloning and mutagenesis.
The M. jannaschii gene at locus MJ0283 (RefSeq accession no. NP_247256.1) was amplified by PCR using oligonucleotide primers 5MJ0283BN and 3MJ0283B (all primer sequences are listed in Table 2). The product was ligated between the NdeI and BamHI sites of vector pET-11a to produce plasmid pDG499 (Table 1). The same PCR product was ligated into the HincII site of vector pSU18 to produce plasmid pJMB102. A PCR product obtained using primers 5MJ0283BN2 and 3MJ0283B lacked codons 1 to 18, which encode the amino-terminal ferredoxin-like domain of the MJ0283 protein. The ligation of this DNA fragment (Δ1-18 MJ0283) between the NdeI and BamHI sites of vector pET-11a produced pDG530, and ligation into the HincII site of pSU18 produced pJMB103.
TABLE 2.
Oligodeoxyribonucleotide primers
| Primer name | Sequencea |
|---|---|
| 5MJ0283BN | CGAGGATCCCATATGGCTGAGTGTGATGGAAAATG |
| 3MJ0283B | GCAGGATCCTTATTCTTTTTTACCTTCGACC |
| 5MJ0283BN2 | CGCCATATGGATACAAAGAAACTCTTAGC |
| 5MMP0704N | GCAGCATATGGCAGAAGAATGCTCTGGAAATTG |
| 3MMP0704X | GCAGTTACTCGAGTTCTTTTTTAATTCTTTCAAGAACTGTATTAAC |
| 5MMP0704N2 | GCAGCATATGGATACTAAAAAAATGATGGAACAGCAGAACG |
| 5MMP0704c | ATGGCAGAAGAAGCCTCTGGAAATGCCGATAGCGCCGGTTCAAG |
| 5MMP0704e | GCAGCATATGGCAGAAGAAGCC |
| 3MMP0704d | CATCATTTTTTTAGTATCTGAAGCGTCGGAACTTGAACCGGCGCTA |
| C218A | TGTTGAAAACATGGGCGGATTTGTAGCTCCTGAATGCGACAAA |
| C218A-rev | TTTGTCGCATTCAGGAGCTACAAATCCGCCCATGTTTTCAACA |
| S55A | GGTAAAGGAGGAGTTGGTAAAGCCACAGTTACCG |
| S55A-rev | CGGTAACTGTGGCTTTACCAACTCCTCCTTTACC |
| C220A | AACATGGGCGGATTTGTATGTCCTGAAGCCGACAAACTAATTG |
| C220A-rev | CAATTACTTTGTCGGCTTCAGGACATACAAATCCGCCCATGTT |
| C218AC220A | AACATGGGCGGATTTGTAGCTCCTGAAGCCGACAAAGTAATTTG |
| C218AC220A rev | CAATTACTTTGTCGGCTTCAAGGAGCTACAAATCCGCCCATGTT |
| 5SSO0460N | GCAGCATATGAGCAGTAATCCTTTTAG |
| 3SSO0460B | GCAGGATCCTTATTGATTGGATTCAACAATTT |
| T7-Promoter | GTAATACGACTCACTATAGGG |
| T7-Term | GCTAGTTATTGCTCAGCGG |
| −40 FORWARD | GTTTTCCCAGTCACGAC |
| −48 REVERSE | GCGGATAACAATTTCACACAGGA |
Restriction sites are underlined, and the initiator codon is shown in boldface.
The M. maripaludis gene at locus MMP0704 (RefSeq accession no. NP_987824.1) was amplified using primers 5MMP0704N and 3MMP0704X. The PCR product was ligated between the NdeI and XhoI sites of vector pET-20b to produce plasmid pDG549. The ligation of the same product into the HincII site of vector pSU18 afforded plasmid pJMB104. Primers 5MMP0704N2 and 3MMP0704X were used to amplify a truncated sequence that lacked codons 1 to 19 (Δ1-19 MMP0704). The ligation of this PCR product into pET-20b formed plasmid pDG547, and ligation into pSU18 formed pJMB105.
Primers C218A and C218A-rev were used to perform site-directed mutagenesis on plasmid pJMB104 with the QuikChange kit (Stratagene), producing plasmid pJMB108. Primers S55A and S55A-rev were used to convert plasmid pJMB104 to plasmid pJMB107. Primers C220A and C220A-rev were used to perform site-directed mutagenesis on plasmid pJMB104, producing plasmid pJMB114. Primers C218AC200A and C218AC220A-rev were used to perform site-directed mutagenesis on plasmid pJMB108, producing plasmid pJMB114. Plasmids pJMB110, pJMB111, pJMB114, and pJMB115 were created by the amplification of Δ1-19 MMP0704 mutant sequences from the full-length constructs with the primers 5MMP0704N2 and 3MMP0704X and subsequent subcloning into pSU18. To replace the four amino-terminal cysteine residues in the MMP0704 protein with alanine residues, primers 5MMP0704c and 3MMP0704e were annealed to each other and extended by Deep VentR DNA polymerase (New England Biolabs) to produce a 77-bp product. Together with primer 5MMP0704e, this product was used to amplify the MMP0704 gene from pDG549. The resulting PCR product was ligated into the NdeI and XhoI sites of pET-20b to produce plasmid pDG592. The same PCR product was ligated into pSU18 to form plasmid pJMB109. The DNA sequences of all plasmid inserts and mutations were confirmed by sequencing (at the University of Wisconsin Biotechnology Center or the Institute for Cellular and Molecular Biology DNA facility at the University of Texas at Austin). Primers −40 FORWARD and −48 REVERSE were used to sequence pSU18 inserts.
Phenotypic analysis.
Nutritional requirements were assessed by the quantification of growth on solid and liquid media (4). Liquid growth experiments were performed with 96-well plates, and growth was monitored with an ELx808 high-throughput spectrophotometer (Bio-Tek Instruments). The optical density at 650 nm (OD650) was recorded every 30 min for 48 h, and the incubation chamber was maintained at 37°C. The starting OD650 was routinely between 0.03 and 0.08, with a final OD650 between 0.5 and 1.1. Each culture had at least three replicates. Growth on solid medium was scored after replica printing onto relevant medium and incubation at 37°C for 24 to 48 h.
Protein expression and purification.
Escherichia coli BL21(DE3) cells were transformed with expression vectors by electroporation. Grown aerobically at 37°C in Luria-Bertani Miller medium supplemented with ampicillin (100 μg ml−1), these cultures were induced in mid-logarithmic-growth phase with 0.1% d-lactose. Cells were harvested and lysed as described previously, and the MMP0704 proteins were purified by nickel affinity chromatography using standard methods (13). Fractions containing the MMP0704-His6 protein were combined in dialysis tubing (SpectraPor4; 14,000-molecular-weight cutoff) and dialyzed for 15 h in 2 liters of buffer containing 50 mM KCl and 20 mM Tris-HCl (pH 8.0) at 4°C. Dialysis was continued for 5 h in fresh buffer before the protein inside the dialysis tubing was concentrated by dehydration with 20,000-Da polyethylene glycol. The MJ0283 proteins were purified by heat treatment of cell extracts prepared in a mixture of 50 mM KCl and 20 mM Tris-HCl (pH 8.0) (13). Total protein concentrations were determined by the Bradford dye-binding method (Thermo-Pierce) using bovine serum albumin as a standard.
Reconstitution of protein iron-sulfur centers and determination of iron-sulfur content.
The purified apoproteins were diluted to a final concentration of 2.5 mg ml−1 {for the His-tagged MMP0704 protein comprising residues 20 to 289 [MMP0704(20-289)-His6]} or 1.0 mg ml−1 (for full-length MMP0704-His6) in 625-μl reaction mixtures sealed under argon gas (13). The reconstitution buffer contained 200 mM KCl, 3 mM dithiothreitol, 0.64 mM ATP, 1.3 mM MgCl2, and 50 mM Tris-HCl (pH 8.0). FeCl3 was added to the stirred reaction mixtures at 0°C to obtain a concentration of 500 μM. After 10 min, Na2S was added dropwise to reach a concentration of 500 μM, and the mixture was stirred under argon for 2 h to reconstitute clusters in holoproteins. The MMP0704 holoproteins were desalted using a PD-10 column (GE Healthcare) equilibrated with an anoxic solution of 50 mM Tris-HCl (pH 8.0) (12). Iron and sulfide analyses were performed using standard methods (3, 41). The standard errors for each mean value were calculated by propagating errors from each analysis.
Phylogenetic analysis.
An alignment of seven amino acid sequences from eukaryotic Nbp35 proteins was combined with an alignment of the eight euryarchaeal proteins that have ferredoxin-like domains. The remaining sequences were added to this alignment. All alignments were constructed using the ClustalW program (version 1.83) (37). The full alignment of 47 sequences was manually edited using the Jalview program (version 2.3) (11) to remove positions that were aligned with low confidence, resulting in an average of 303 amino acids per sequence. This alignment was analyzed using the proml program from the Phylip package (version 3.67; J. Felsenstein, University of Washington) to infer a maximum-likelihood phylogeny from this alignment, with the Jones-Taylor-Thornton model of amino acid changes and a γ-distribution of rates (α = 2.4) approximated by three states. Bootstrap analysis was performed with 100 replicates. The full organism names and accession numbers corresponding to the aligned sequences are listed in the supplemental material.
RESULTS
Identification of ApbC/Nbp35 homologs in the Archaea.
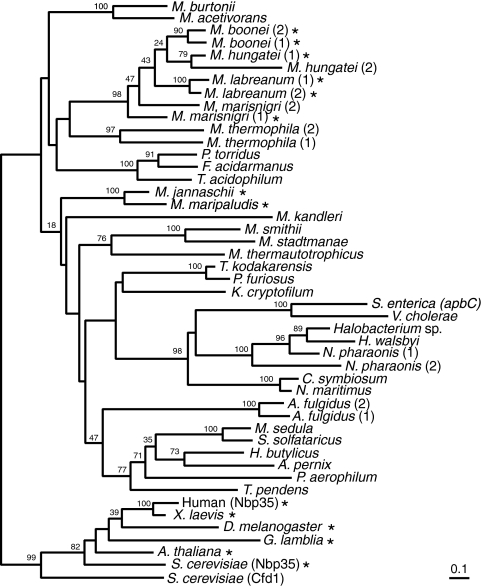
M. maripaludis has numerous paralogs of genes from the P-loop-containing nucleoside triphosphate hydrolase family that includes the apbC and NBP35 genes, but only the MMP0704 sequence encodes a protein conserving the key cysteine residues of this subfamily. To date, all of the known genome sequences of free-living archaea encode homologs of the MMP0704 protein (Fig. 2). Although the phylogeny of these ApbC/Nbp35 homologs does not recapitulate organismal phylogenies inferred from small-subunit rRNA or ribosomal protein sequences, most major archaeal groups are conserved (8). Notable exceptions include the grouping of the low-temperature crenarchaeal homologs with the haloarchaeal homologs and the gammaproteobacterial ApbC homologs. These exceptions may be explained by horizontal gene transfer events, but the direction and frequency of this transfer are obscure.
FIG. 2.
The ApbC/Nbp35 family of iron-sulfur proteins evolved before the Archaea and Eukarya diverged. This phylogeny was inferred using a protein maximum-likelihood method, although a neighbor-joining distance method produced a similar tree. Bootstrap values from the neighbor-joining consensus tree are shown near branches supported by a plurality of 100 trees. Numbers in parentheses after some species names differentiate paralogs from the same genome. Asterisks indicate sequences with amino-terminal ferredoxin-like domains. The complete organism names and accession numbers are listed in the supplemental material. Bar, 0.1 amino acid substitution expected per site.
Most crenarchaeal homologs, including the S. solfataricus SSO0460 protein represented in Fig. 1, are missing the cysteine residue that corresponds to MMP0704 Cys220. In these sequences, an acidic amino acid replaces cysteine. Three crenarchaeal sequences, from the deeply branching organisms Thermofilum pendens, “Cenarchaeum symbiosum,” and “Candidatus Nitrosopumilus maritimus,” do contain Cys220. From these data, we conclude that the archaeal ancestor contained a homolog of the MMP0704 gene (encoding both conserved Cys218 and Cys220 residues) and that this gene has been vertically inherited in most archaeal lineages.
The M. maripaludis apbC/NBP35 homolog complements an S. enterica apbC mutation.
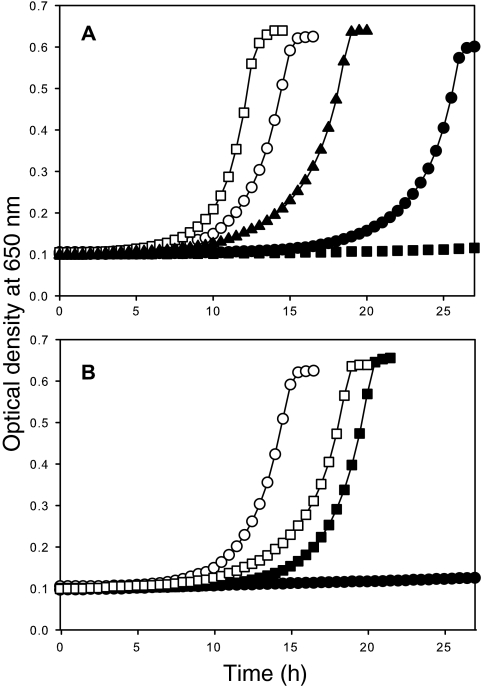
An S. enterica apbC mutant will not grow with tricarballylate as the sole carbon and energy source (4). This growth defect was used to examine the in vivo functionality of methanogen apbC/NBP35 homologs. The M. maripaludis MMP0704 locus and the M. jannaschii MJ0283 locus were cloned into a multicopy vector compatible with S. enterica (pSU18). S. enterica strain DM10300 was transformed with the resulting plasmids, and the abilities of these genes to complement an apbC null mutant was examined. As shown in Fig. 3, strain DM10300 that had pJMB100 (apbC) grew on tricarballylate whereas strain DM10300 that had pSU18 (empty vector) did not. Strain DM10300(pJMB102), expressing MJ0283, did not grow on tricarballylate medium but did grow on glucose medium (Table 3). However, strain DM10300(pJMB104), expressing MMP0704, did grow on tricarballylate despite a significant lag before the start of exponential growth (Fig. 3). The doubling time for this strain was nearly twice as long as that for strain DM10300(pJMB100). Although the MMP0704 and MJ0283 proteins show 73% amino acid identity and 91% similarity, these data indicate that only the MMP0704 protein can replace ApbC for tricarballylate metabolism.
FIG. 3.
M. maripaludis MMP0704 constructs can complement an S. enterica apbC mutant. Strains were grown at 37°C in no-carbon essential salts medium supplemented with thiamine, with tricarballylate as the sole carbon and energy source. (A) The growth of strain DM10300 with pJMB100 (apbC) (open squares), pJMB105 (Δ1-19 MMP0704) (open circles), pJMB109 [MMP0704(Cys5,9,12,18Ala)] (filled triangles), pJMB104 (MMP0704) (filled circles), and empty vector (pSU18) (filled squares) was determined by measuring the OD650. (B) Mutational analysis of conserved cysteine residues in the Δ1-19 MMP0704 gene product. The growth of strain DM10300 with pJMB105 (Δ1-19 MMP0704) (open circles), pJMB111 [Δ1-19 MMP0704(Cys218Ala)] (closed squares), pJMB114 [Δ1-19 MMP0704(Cys220Ala)] (open squares), and pJMB115 [Δ1-19 MMP0704(Cys218,220Ala)] (closed circles) was monitored as described in the legend to panel A.
TABLE 3.
Archaeal ApbC/Npb35 homologs can function in place of S. enterica ApbC in vivoa
| Vector | Vector insert | Doubling timeb (h) with:
|
|
|---|---|---|---|
| Glucose | Tricarballylate | ||
| pJMB100 | apbC | 1.1 ± 0.0 | 1.6 ± 0.1 |
| pJMB102 | MJ0283 | 1.1 ± 0.0 | NGc |
| pJMB103 | Δ1-18 MJ0283 | 1.1 ± 0.0 | 3.1 ± 0.2 |
| pJMB104 | MMP0704 | 1.1 ± 0.1 | 3.0 ± 0.3 |
| pJMB107 | MMP0704(Ser55Ala) | 1.2 ± 0.0 | NG |
| pJMB108 | MMP0704(Cys218Ala) | 1.1 ± 0.1 | NG |
| pJMB112 | MMP0704(Cys220Ala) | 1.1 ± 0.0 | NG |
| pJMB114 | MMP0704(Cys218,220Ala) | 1.0 ± 0.1 | NG |
| pJMB109 | MMPO704(Cys5,9,12,18Ala) | 1.1 ± 0.0 | 2.7 ± 0.3 |
| pJMB105 | Δ1-19 MMP0704 | 1.1 ± 0.0 | 2.2 ± 0.0 |
| pJMB110 | Δ1-19 MMP0704(Ser55Ala) | 1.1 ± 0.0 | NG |
| pJMB111 | Δ1-19 MMP0704(Cys218Ala) | 1.1 ± 0.0 | 2.2 ± 0.0 |
| pJMB114 | Δ1-19 MMP0704(Cys220Ala) | 1.1 ± 0.0 | 2.6 ± 0.1 |
| pJMB115 | Δ1-19 MMP0704(Cys218,220Ala) | 1.1 ± 0.0 | NG |
| pJMB116 | SSO0460 | 1.2 ± 0.0 | 2.7 ± 0.1 |
| pJMB106 | None | 1.1 ± 0.0 | NG |
Complementation was conducted as outlined in Materials and Methods.
Doubling times were calculated using the following formulas: μ = ln(X/Xo)/T, where μ is the growth rate, X is the OD650 value at a given time point, X0 is the OD650 value at time zero, and T is the time between readings X and Xo, and doubling time (g) = (ln 2)/μ (26). The numbers shown represent the averages of results for three independent cultures. Minimal media with the indicated carbon sources were used. The media were supplemented with thiamine.
NG, no growth.
The amino-terminal domain of MMP0704 protein is not required for activity.
Eukaryotic Nbp35 protein copurifies with an oxygen-stable Fe-S cluster thought to be coordinated by the four cysteine residues in the N-terminal ferredoxin domain. These cysteines are absent in the ApbC protein but appear to be conserved in some archaeal sequences (Fig. 1). Most archaea have truncated forms of these proteins (data not shown). To examine the necessity of the cysteine-rich ferredoxin-like domain of MMP0704 protein, an MMP0704 mutant gene with a deletion removing the first 19 codons (Δ1-19 MMP0704) was constructed. An MJ0283 gene with a similar deletion removing the first 18 codons (Δ1-18 MJ0283) was also produced (Fig. 1 and Table 1). Strain DM10300 with either pJMB103 (Δ1-18 MJ0283) or pJMB105 (Δ1-19 MMP0704) grew on tricarballylate with doubling times slightly longer than that of the apbC control (Fig. 3 and Table 3). Additionally, the long lag before exponential growth seen with strain DM10300(pJMB104) was not seen with cells carrying either pJMB103 or pJMB105. We conclude that the truncated forms of MMP0704 and MJ0283 complemented an apbC mutation more efficiently than their full-length counterparts.
To determine whether the ferredoxin-like domain of wild-type MMP0704 protein reduced the effectiveness of the protein in complementation, a mutant protein in which four Cys residues were replaced by Ala was constructed. Figure 3 shows that strain DM10300(pJMB109), expressing the MMP0704(Cys5,9,12,18Ala) variant, was able to grow with tricarballylate. The lag before logarithmic growth and the doubling time of DM10300(pJMB109) were intermediate relative to those of strains with the full-length and truncated constructs.
Mutational analysis of the Walker A box and Cys-X-X-Cys domain of MMP0704.
To determine whether nucleotide binding and hydrolysis are required for in vivo function, the Ser55 residue in the Walker A motif of MMP0704 was replaced with Ala. The corresponding ApbC variant is not able to function in vivo or hydrolyze ATP in vitro (6), and the crystal structure model of the homologous MinD cell division protein shows that the corresponding Thr17 hydroxyl group coordinates an Mg2+ ion required for the hydrolysis of ATP (17). S. enterica DM10300 strains with pJMB107 [MMP0704(Ser55Ala)] or pJMB110 [Δ1-19 MMP0704(Ser55Ala)] could not grow with tricarballylate, but both strains grew with glucose (Table 3).
Roy et al. reported that the replacement of yeast Cfd1 cysteine residues in the Cys-X-X-Cys motif with serine prevented protein function in vivo (30). These residues are predicted to be ligands for a carboxy-terminal Fe-S cluster. To test whether the corresponding cysteine residues in MMP0704 protein are essential, Cys218Ala and Cys220Ala variants were constructed. DM10300 strains with both pJMB111 [Δ1-19 MMP0704(Cys218Ala)] and pJMB112 [Δ1-19 MMP0704(Cys220Ala)] grew in tricarballylate medium (Fig. 3 and Table 3), but strain DM10300 with pJMB113 [Δ1-19 MMP0704(Cys218,220Ala)] could not. Variant full-length MMP0704 proteins in which both Cys218 and Cys220 were changed to Ala could not compensate for the loss of ApbC in vivo (Fig. 3 and Table 3).
A crenarchaeal ApbC/Nbp35 homolog complements an S. enterica apbC mutation.
Like other crenarchaeal homologs, the S. solfataricus SSO0460 protein is missing the four amino-terminal Cys residues present in MMP0704 and MJ0283 proteins, and the second Cys residue of the Cys-X-X-Cys motif is replaced with Asp (Fig. 1). Additionally, the S. solfataricus homolog has a glutamine- and proline-rich amino-terminal domain that is not found in euryarchaeal homologs. These differences raised doubts about the functional similarities between ApbC and the crenarchaeal homologs. Strain DM10300 with pJMB116 (SSO0460) grew on tricarballylate, and cells doubled every 2.7 ± 0.1 h and displayed a lag before logarithmic growth that was similar to that of strain DM10300 with pJMB105 (Δ1-19 MMP0704). These complementation data indicate that the S. solfataricus SSO0460 protein has Fe-S carrier activity, supporting our observation that one of the cysteine residues of the Cys-X-X-Cys motif in the MMP0704 protein is dispensable.
Purification of methanogen ApbC/Nbp35 proteins and reconstitution of the protein iron-sulfur centers.
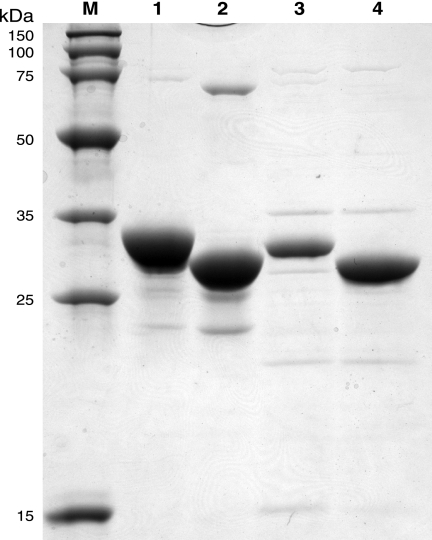
High levels of MMP0704-His6, truncated MMP0704(20-289)-His6, untagged MJ0283, and truncated MJ0283(19-290) proteins were produced in E. coli BL21(DE3) cells as soluble proteins. From a 500-ml culture of BL21(DE3)(pDG547) cells, 7 mg of MMP0704(20-289)-His6 protein was purified by nickel affinity chromatography (Fig. 4). Heating cell extracts containing either the full-length MJ0283 or truncated MJ0283(19-290) protein at 70°C for 10 min produced a substantially pure protein solution after centrifugation.
FIG. 4.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of an overloaded, Coomassie blue-stained gel shows the purities and solubilities of heterologously expressed MMP0704 and MJ0283 proteins (10 μg each). Marker proteins, shown in lane M, have the molecular masses indicated to the left. Lane 1 contains affinity-purified MMP0704-His6 protein, with an apparent molecular mass of 33 kDa (calculated mass, 32.1 kDa). Lane 2 contains affinity-purified MMP0704(20-289)-His6 protein, with an apparent molecular mass of 29 kDa (calculated mass, 30.4 kDa). Lane 3 contains heat-stable, purified MJ0283 protein, with an apparent molecular mass of 31 kDa (calculated mass, 31.2 kDa). Lane 4 contains heat-stable, purified MJ0283(19-290) protein, with an apparent molecular mass of 30 kDa (calculated mass, 29.5 kDa).
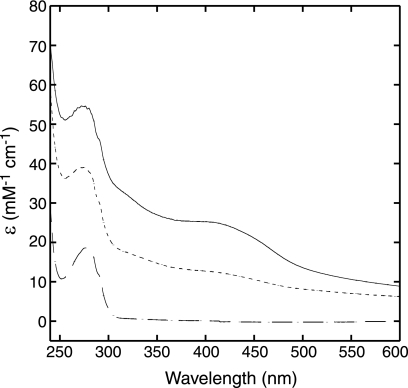
The iron-sulfur centers of the MMP0704 apoproteins were reconstituted under anaerobic conditions. Without ATP in the reconstitution mixture, the proteins aggregated to form high-mass complexes. However, when a fivefold excess of Mg-ATP was added, the proteins remained soluble. The aerobically purified MMP0704 apoproteins were colorless, with no significant UV-visible absorption peaks in the 400-nm region that are characteristic of major S→Fe charge transfer bands. The UV-visible absorption spectra of chemically reconstituted MMP0704(20-289)-His6 and full-length MMP0706-His6 holoproteins had a broad peak at 400 nm, a typical feature for proteins with [4Fe-4S]2+ clusters, and a slight shoulder at 325 nm (Fig. 5). The reconstituted MMP0706-His6 protein had an absorption coefficient at 400 nm of 25 mM−1 cm−1 and an A400/A280 ratio of 0.47. This full-length MMP0704-His6 protein bound 7.5 ± 1.2 equivalents of iron and 7.7 ± 1.6 equivalents of sulfide per monomer. The truncated MMP0704(20-289)-His6 protein had an absorption coefficient at 400 nm of 13 mM−1 cm−1 and an A400/A280 ratio of 0.33. This protein bound 1.7 ± 1 equivalents of iron and 2.2 ± 0.3 equivalents of sulfide per monomer. These values are consistent with either a monomeric [2Fe-2S] center or a dimeric [4Fe-4S] center.
FIG. 5.
UV-visible absorbance spectra of reconstituted MMP0704-His6 and MMP0704(20-289)-His6 holoproteins. The MMP0704-His6 apoprotein (broken line, bottom spectrum) was treated with ferric chloride, sulfide, and Mg-ATP and then desalted to reconstitute the holoprotein (solid line, top spectrum). The MMP0704(20-289)-His6 apoprotein had an absorbance spectrum identical to that of the full-length apoprotein (data not shown), while the truncated holoprotein had an intermediate absorbance spectrum (dashed line, middle spectrum). The millimolar absorption coefficient (ɛ) was calculated from measured absorbance and protein concentration values.
DISCUSSION
The eukaryotic ISC biosynthesis system has a clear proteobacterial origin (35, 39), and it is too sporadically distributed in the Archaea to be an ancestral pathway for Fe-S cluster biosynthesis (see Table S1 in the supplemental material). Those archaea with Isc or Nif systems, such as the Methanosarcinales and Methanomicrobiales lineages, probably acquired them by horizontal gene transfer. Alternatively, most archaea have homologs of the bacterial SufBC proteins, but it is not clear how this system could function without other Fe-S cluster biosynthetic proteins. The archaeal ApbC/Nbp35 protein may be the intermediate carrier protein that receives an Fe-S cluster from the SufBC complex and delivers it to an apoprotein.
The archaeal Fe-S carrier proteins described herein unify an ancient family of cluster-binding proteins that evolved before the divergence of the Archaea and Eukarya. Despite the significant sequence similarity among the gammaproteobacterial ApbC proteins, the eukaryotic Nbp35 proteins, and the archaeal homologs, it was not clear that archaea shared an ancestral Fe-S carrier prior to this work. All members of the large nucleoside triphosphate hydrolase superfamily that includes these proteins adopt highly conserved P-loop folds, making it difficult to identify signature motifs or distinguish orthologs at the primary sequence level.
A remarkable number of archaea have acquired paralogous genes through recent duplications of the MMP0704 ortholog (Fig. 2). Most members of the Methanomicrobiales have duplicate copies that evolved independently in many genera. Similar duplications occurred in the haloarchaeal strain Natronomonas pharaonis and in the sulfate reducer Archaeoglobus fulgidus. In Methanoculleus marisnigri and Methanospirillum hungatei, one copy lost the sequence encoding the amino-terminal ferredoxin-like domain, reminiscent of the Cfd1 gene in yeast. It remains to be determined whether these paralogous genes encode proteins that interact, as in yeast (27), or are differentially expressed to activate different proteins.
The eukaryotic Nbp35 homologs share an amino-terminal ferredoxin-like domain, an ATP-binding motif, and two conserved carboxy-terminal cysteine residues that are believed to bind an Fe-S cluster. The S. cerevisiae Npb35 protein contains an oxygen-stable [4Fe-4S] cluster when overexpressed in and purified from E. coli (34). A truncated Nbp35 variant missing the N-terminal ferredoxin-like domain is not purified with Fe and S (16). Comparative sequence analyses show that all bacterial homologs and most archaeal homologs lack the ferredoxin-like domain present in MMP0704 protein. S. enterica mutants expressing the truncated form of the MMP0704 protein grew faster than cells producing wild-type MMP0704 protein during tricarballylate-dependent growth assays, demonstrating that the amino-terminal domain is dispensable. To test whether this difference in growth rate was due to decreased gene expression or thiol-mediated complexes of the full-length protein, all four amino-terminal cysteines were replaced by alanine. Cells producing this variant protein grew faster than cells with wild-type protein, suggesting that interactions due to the cysteine thiols caused reduced activity. Therefore, the amino-terminal domain is not required for MMP0704 activity in S. enterica, although genetic analysis of M. maripaludis will be required to determine whether this domain is essential in the native host.
The ATPase activities of full-length and truncated MMP0704 proteins are necessary for function in vivo. Both the truncated and full-length MMP0704 proteins require Mg-ATP for efficient in vitro chemical reconstitution of their Fe-S clusters. Key residues in the ATP-binding motif of ApbC are also essential for function in vivo but not for in vitro Fe-S cluster binding (5, 6). In the presence of Mg-ATP, the full-length MMP0704 holoprotein formed a mixture of high-mass species (analyzed by size exclusion chromatography), while the MMP0704(20-289)-His6 holoprotein formed primarily dimers (R. Drevland, unpublished data). Our interpretation of these data is that ATP binding or hydrolysis is required for cluster transfer to MMP0704 protein or cluster construction on MMP0704 protein.
Chemically reconstituted, truncated MMP0704 protein bound approximately 2 mol of Fe and S per mol of protein, formed dimers, and had an absorption spectrum similar to those of proteins known to have [4Fe-4S] clusters. These data are consistent with the hypothesis that truncated MMP0704 protein binds a [4Fe-4S] cluster that forms the interface of two subunits. The ligands for the essential Fe-S cluster in ApbC/Nbp35 proteins were assumed to be two conserved cysteine thiols, Cys218 and Cys220, near the carboxy terminus. A preliminary model of the A. fulgidus AF2382 homolog (Protein Data Bank accession no. 2PH1) shows that the conserved cysteine residues are found in a β turn. The two cysteine sulfur atoms are 4.3 Å apart, coordinating a Zn2+ ion at the dimer interface (F. Forouhar and L. Tong, unpublished data). In most crenarchaeal homologs, an Asp or Glu residue replaces Cys218, yet the S. solfataricus homolog complemented the Salmonella apbC mutation. Aspartate is also a ligand for the [4Fe-4S] cluster of the P. furiosus ferredoxin, where it slightly increases the cluster's reduction potential compared to a cysteine ligand substituent (7). Yet both the Δ1-19 MMP0704(Cys220Ala) and Δ1-19 MMP0704(Cys218Ala) alleles complemented an apbC mutant, suggesting that individual cysteine residues in the Cys-X-X-Cys motif are dispensable. Amino acid insertions in this region of the plant Nbp35 homologs also indicate variable modes of coordination (10). A similar result was reported for variants of the Azotobacter vinelandii NifU Fe-S scaffold protein, where mutations in conserved cysteine codons reduced, but did not abolish, diazotrophic growth (1). Additional electron paramagnetic resonance and Mössbauer studies will be required to determine which residues are ligands for an Fe-S cluster.
Almost all modern organisms have Fe-S proteins, and these cofactors represent the interface of biological chemistry with the complex marine geochemistry of iron sulfides (29). FeS minerals, particularly mackinawite, greigite, and pyrite, feature prominently in several hypotheses for the chemical origin of life, as Lewis acid catalysts or primitive inorganic membranes (19, 24, 31, 40). As iron became less bioavailable, cells' iron acquisition systems became sophisticated and a variety of Fe-S cluster assembly systems evolved. Among archaea with completely sequenced genomes, only the obligate parasite “Nanoarchaeum equitans” lacks an apbC/NBP35 gene. The loss of most iron-sulfur-dependent metabolic proteins from N. equitans may be associated with this gene loss, since the remaining iron-sulfur proteins must be assembled using host-derived clusters. This report represents the first genetic and biochemical study examining Fe-S cluster biosynthesis in archaea. The data herein support the hypothesis that archaeal ApbC/Nbp35 homologs function as Fe-S cluster carrier proteins.
Supplementary Material
Acknowledgments
This work was supported in part by Public Health Service grant AI064444 from the National Institute of Allergy and Infectious Diseases (D.E.G.), National Science Foundation grant MCB-0817903 (D.E.G.), and competitive grant GM47296 from the NIH (D.M.D.). R.M.D. was partially supported by Lewis and Banks graduate fellowships. J.M.B. was supported by NIH Ruth Kirschstein postdoctoral training fellowship 144PZ38.
We thank Farhad Forouhar and Liang Tong for helpful discussions about the A. fulgidus AF2382 protein structure model.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Agar, J. N., P. Yuvaniyama, R. F. Jack, V. L. Cash, A. D. Smith, D. R. Dean, and M. K. Johnson. 2000. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem. 5167-177. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 10275-78. [DOI] [PubMed] [Google Scholar]
- 3.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 131373-378. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, J. M., J. A. Lewis, J. C. Escalante-Semerena, and D. M. Downs. 2008. Salmonella enterica requires ApbC function for growth on tricarballylate: evidence of functional redundancy between ApbC and IscU. J. Bacteriol. 1904596-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, J. M., A. J. Pierik, D. J. A. Netz, R. Lill, and D. M. Downs. 2008. Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry 478195-8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, J. M., J. L. Sondelski, and D. M. Downs. 2009. Bacterial ApbC protein has two biochemical activities that are required for in vivo function. J. Biol. Chem. 284110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brereton, P. S., M. F. J. M. Verhagen, Z. H. Zhou, and M. W. W. Adams. 1998. Effect of iron-sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus. Biochemistry 377351-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brochier-Armanet, C., B. Boussau, S. Gribaldo, and P. Forterre. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6245-252. [DOI] [PubMed] [Google Scholar]
- 9.Buckel, W., and B. T. Golding. 2006. Radical enzymes in anaerobes. Annu. Rev. Microbiol. 6027-49. [DOI] [PubMed] [Google Scholar]
- 10.Bych, K., D. J. A. Netz, G. Vigani, E. Bill, R. Lill, A. J. Pierik, and J. Balk. 2008. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 28335797-35804. [DOI] [PubMed] [Google Scholar]
- 11.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20426-427. [DOI] [PubMed] [Google Scholar]
- 12.Drevland, R. M., Y. Jia, D. R. J. Palmer, and D. E. Graham. 2008. Methanogen homoaconitase catalyzes both hydrolyase reactions in coenzyme B biosynthesis. J. Biol. Chem. 28328888-28896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drevland, R. M., A. Waheed, and D. E. Graham. 2007. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J. Bacteriol. 1894391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontecave, M., and S. Ollagnier-de-Choudens. 2008. Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474226-237. [DOI] [PubMed] [Google Scholar]
- 15.Frazzon, J., and D. R. Dean. 2003. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr. Opin. Chem. Biol. 7166-173. [DOI] [PubMed] [Google Scholar]
- 16.Hausmann, A., D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Mühlenhoff, and R. Lill. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 1023266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 201819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata, A., B. J. Klein, and K. S. Murakami. 2008. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451851-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, C., and G. Wächtershäuser. 1997. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science 276245-247. [DOI] [PubMed] [Google Scholar]
- 20.Layer, G., S. A. Gaddam, C. N. Ayala-Castro, S. Ollagnier-de Choudens, D. Lascoux, M. Fontecave, and F. W. Outten. 2007. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 28213342-13350. [DOI] [PubMed] [Google Scholar]
- 21.Lewis, J. A., and J. C. Escalante-Semerena. 2007. Tricarballylate catabolism in Salmonella enterica. The TcuB protein uses 4Fe-4S clusters and heme to transfer electrons from FADH2 in the tricarballylate dehydrogenase (TcuA) enzyme to electron acceptors in the cell membrane. Biochemistry 469107-9115. [DOI] [PubMed] [Google Scholar]
- 22.Lewis, J. A., A. R. Horswill, B. E. Schwem, and J. C. Escalante-Semerena. 2004. The tricarballylate utilization (tcuRABC) genes of Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 1861629-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lill, R., and U. Mühlenhoff. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77669-700. [DOI] [PubMed] [Google Scholar]
- 24.Martin, W., and M. J. Russell. 2003. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos. Trans. R. Soc. Lond. B 35859-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neidhardt, F. C., J. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, MA.
- 27.Netz, D. J. A., A. J. Pierik, M. Stumpfig, U. Muhlenhoff, and R. Lill. 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3278-286. [DOI] [PubMed] [Google Scholar]
- 28.Petersen, L., and D. M. Downs. 1996. Mutations in apbC (mrp) prevent function of the alternative pyrimidine biosynthetic pathway in Salmonella typhimurium. J. Bacteriol. 1785676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rickard, D., and G. W. Luther III. 2007. Chemistry of iron sulfides. Chem. Rev. 107514-562. [DOI] [PubMed] [Google Scholar]
- 30.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 224826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell, M. J., and A. J. Hall. 2006. The onset and early evolution of life, p. 1-32. In S. E. Kesler and H. Ohmoto (ed.), Memoir 198. Evolution of early earth's atmosphere, hydrosphere, and biosphere—constraints from ore deposits. Geological Society of America, Boulder, CO.
- 32.Schäfer, G., M. Engelhard, and V. Müller. 1999. Bioenergetics of the Archaea. Microbiol. Mol. Biol. Rev. 63570-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 18598-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stehling, O., D. J. A. Netz, B. Niggemeyer, R. Rösser, R. S. Eisenstein, H. Puccio, A. J. Pierik, and R. Lill. 2008. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 285517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachezy, J., L. B. Sánchez, and M. Müller. 2001. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol. Biol. Evol. 181919-1928. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 27728380-28383. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unciuleac, M.-C., K. Chandramouli, S. Naik, S. Mayer, B. H. Huynh, M. K. Johnson, and D. R. Dean. 2007. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 466812-6821. [DOI] [PubMed] [Google Scholar]
- 39.van der Giezen, M., S. Cox, and J. Tovar. 2004. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol. Biol. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wächtershäuser, G. 1998. Origin of life in an iron-sulfur world, p. 206-218. In A. Brack (ed.), The molecular origins of life: assembling pieces of the puzzle. Cambridge University Press, Cambridge, United Kingdom.
- 41.Wu, S.-P., G. Wu, K. K. Surerus, and J. A. Cowan. 2002. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry 418876-8885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.