Abstract
FtsH proteins have dual chaperone-protease activities and are involved in protein quality control under stress conditions. Although the functional role of FtsH proteins has been clearly established, the regulatory mechanisms controlling ftsH expression in gram-positive bacteria remain largely unknown. Here we show that ftsH of Lactobacillus plantarum WCFS1 is transiently induced at the transcriptional level upon a temperature upshift. In addition, disruption of ftsH negatively affected the growth of L. plantarum at high temperatures. Sequence analysis and mapping of the ftsH transcriptional start site revealed a potential operator sequence for the CtsR repressor, partially overlapping the −35 sequence of the ftsH promoter. In order to verify whether CtsR is able to recognize and bind the ftsH promoter, CtsR proteins of Bacillus subtilis and L. plantarum were overproduced, purified, and used in DNA binding assays. CtsR from both species bound specifically to the ftsH promoter, generating a single protein-DNA complex, suggesting that CtsR may control the expression of L. plantarum ftsH. In order to confirm this hypothesis, a ΔctsR mutant strain of L. plantarum was generated. Expression of ftsH in the ΔctsR mutant strain was strongly upregulated, indicating that ftsH of L. plantarum is negatively controlled by CtsR. This is the first example of an ftsH gene controlled by the CtsR repressor, and the first of the low-G+C gram-positive bacteria where the regulatory mechanism has been identified.
FtsH proteins are membrane-bound ATP- and Zn2+-dependent metalloproteases belonging to the AAA (ATPase associated with different cellular activities) protein family, a distinct subfamily of the Walker-type ATPases (27). These ATP-dependent proteases also have intrinsic chaperone activity and have therefore been designated charonins (for reviews, see references 22 and 33). Consistent with its dual chaperone-protease function, FtsH plays an important role in the protein quality control network, which not only allows refolding or degradation of denatured and misfolded proteins generated under stress conditions but also enables the temporal control of many cellular processes by regulating the protein stability of specific, critical regulators (18, 19, 22). In Escherichia coli, ftsH is a heat-inducible gene (21), while in Bacillus subtilis, it is transiently induced at the transcriptional level upon osmotic and temperature upshifts (12). Recently, a similar pattern of expression was also observed in lactic acid bacteria (LAB) such as Oenococcus oeni and Lactobacillus plantarum. In O. oeni, ftsH expression was increased by high temperatures and osmotic shock, and its protective role against heat shock was demonstrated by heterologous expression in E. coli (3). In addition, genome-wide microarray-based expression profiling revealed that ftsH is upregulated by bile treatment in L. plantarum WCFS1 (5). Therefore, FtsH seems to be an important component of the stress response machineries developed by several bacteria in order to withstand harsh conditions and sudden environmental changes. Although ftsH is induced by stress in B. subtilis, O. oeni, and L. plantarum, no typical stress-responsive elements of gram-positive bacteria genes have been observed in its promoter. In B. subtilis, a model organism for gram-positive bacteria, in which the stress response has been extensively studied, heat-inducible genes are divided into different subclasses based on their regulatory mechanisms (34). In particular, class I genes encode classical chaperones such as DnaK and GroEL, whose expression involves a highly conserved operator sequence, which is the binding site for the HrcA repressor. Class II genes are regulated by the alternative σB factor. Class III genes, such as clp genes, are controlled by the class III stress gene repressor CtsR (designation from “class three stress gene repressor”), which binds to a specific heptanucleotide direct repeat (RGTCADN NANRGTCADN), referred to as the CtsR box (9). Genes regulated by as yet unknown mechanisms are grouped under class IV; these include ftsH, whose heat shock induction was shown to be CtsR independent in B. subtilis (9, 34).
Lactobacillus plantarum, a facultatively heterofermentative LAB, is one of the most widespread LAB in the environment. A natural inhabitant of the human gastrointestinal tract, it is also found in several food fermentation products for which stress conditions such as heat, cold, and acidity are common. In wine, although L. plantarum is capable of malolactic fermentation, it usually contributes to the production of undesirable products such as biogenic amines and precursors of ethyl carbamate and is therefore generally considered a nuisance. Vinification generates multiple stress conditions, including an acidic pH, ethanol, extreme temperatures, and growth-inhibitory compounds such as fatty acids and tannins. The survival of L. plantarum in this stressful environment indicates that it has developed several tolerance and resistance mechanisms. Study of the stress response of L. plantarum is thus essential in order to understand the high adaptability of this microorganism to stress conditions.
The genome of L. plantarum WCFS1 has been completely sequenced (23). Analysis of the 5′ noncoding region of the L. plantarum ftsH gene allowed us to identify a putative operator sequence highly similar to the CtsR binding site. In this work, we report that the ftsH gene of L. plantarum is heat induced and is under the control of CtsR. To our knowledge, this is the first report identifying ftsH as a novel member of the CtsR stress response regulon, and this is the first ftsH gene in low-G+C gram-positive bacteria for which the regulatory mechanism has been identified.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strains DH10B, TG1, and BL21 λ70 DE3 were used for DNA cloning and overexpression and were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C. L. plantarum WCFS1 (23) was routinely grown in MRS broth (7) (initial pH 6.2) at 28°C without shaking. When required, appropriate antibiotics were added at the following concentrations: ampicillin at 100 μg ml−1 and erythromycin at 200 μg ml−1 for E. coli; erythromycin at 10 μg ml−1 and chloramphenicol at 10 μg ml−1 for L. plantarum.
For heat, salt, and bile stresses, 0.1 ml of stationary-phase L. plantarum cells (optical density at 600 nm [OD600], 2.6) was diluted in 30 ml of fresh MRS broth (pH 6.2), and growth was initiated at 28°C. When the OD600 reached 0.6 (pH 6.1), the cultures were transferred to water baths maintained at 42°C for various times or at 50°C for a short (5-min) heat shock. For salt and bile stresses, L. plantarum cells were harvested by centrifugation (at 4,500 × g for 10 min) and resuspended in 30 ml of fresh MRS broth containing either 0.8 M NaCl or 0.15% (wt/vol) porcine bile extract; stresses were imposed for 10 min and 1 h. Aliquots were removed, and total RNA was extracted and used for quantitative real-time PCR (qRT-PCR) analysis. The control culture was grown at 28°C in MRS medium.
The expression of the ftsH gene in the wild type was monitored over a complete culture cycle (20 h) performed at 28°C in MRS. Growth was monitored by both OD measurement (OD600) and direct plate counting. Total RNA used for qRT-PCR analyses was extracted at the various growth stages. In order to analyze ftsH expression in an L. plantarum ΔctsR mutant, stationary-phase L. plantarum ΔctsR cells were diluted in fresh MRS medium and allowed to grow to mid-exponential phase (OD600, 0.6 to 0.8). Total RNA was then extracted and used for qRT-PCR analysis.
The growth rates of the L. plantarum ftsH mutant and wild-type strains were determined by diluting overnight cultures 1:1,000 in fresh MRS medium and monitoring growth by OD600 measurement and direct plate counting. A complete cycle of growth at 28°C and at 42°C (heat stress condition) was monitored.
DNA manipulation and analysis.
Standard methods were used for DNA manipulations, including isolation, restriction endonuclease analysis, and ligation (32). Taq polymerases, restriction enzymes, alkaline phosphatase, and T4 DNA ligase were purchased from Roche (Milan, Italy), Invitrogen (Milan, Italy), New England Biolabs (Hertfordshire, United Kingdom), Fermentas (Burlington, Ontario, Canada), and Promega (Milan, Italy) and were used as recommended by the suppliers.
Double-stranded plasmid DNA was isolated using QIAprep spin miniprep kits (Qiagen, Milan, Italy). PCR products and DNA restriction fragments were purified with the QIAquick PCR purification and gel extraction kits (Qiagen, Milan, Italy). L. plantarum chromosomal DNA was prepared using a microbial DNA extraction kit (Cabru, Milan, Italy) according to the manufacturer's procedure. For PCR experiments, 20 ng of genomic DNA from L. plantarum was added to a 50-μl PCR mixture and amplified with the Expand Long Template PCR system (Roche, Milan, Italy) by following the manufacturer's instructions. The reaction mixture was cycled through the following temperature profile: 94°C for 5 min, 5 cycles of 94°C for 1 min, 45°C for 1 min, and 68°C for 2 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, and 68°C for 2 min. The PCR was ended by incubation at 72°C for 5 min. The oligonucleotides used in this study are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′-3′) | Application |
|---|---|---|
| FtsHP | CGTTTGTGAGCCGGAATTACCGC | Primer extension analyses |
| ldhDF | ACGCCCAAGCTGATGTTATATC | qRT-PCR |
| ldhDR | AGTGTCCCACGAGCAAAGTT | qRT-PCR |
| gyrAF | CATGCGGTTAGGCGATGAT | qRT-PCR |
| gyrAR | ATCGGCGTTGACGGTTTG | qRT-PCR |
| FtsH1F | TTACGGTCTTGACTATGCAGAACGCTATCGGAACT | Gel mobility shift assay |
| FtsH2F | TCGATGAAGGAGGCACATATGAACAATCGACGCA | Gel mobility shift assay |
| FtsH1R | GCTGGATACTAACGTTTTTAACGTTATTCTTATCTAATTG | Gel mobility shift assay |
| FB1ctsR | ATCTCGAGTTAAAATCCTGCGGTTAGTG | ctsR chromosomal deletion |
| RB1ctsR* | GACTTTGCATGTGCTTCACC | ctsR chromosomal deletion |
| FB2ctsR* | CGGGCCCGGATAATTATCGG | ctsR chromosomal deletion |
| RB2ctsR* | TCATCCGTAATCGTAACCCG | ctsR chromosomal deletion |
| FBctsR* | TGAACCGGCAACAAGGCATG | ctsR chromosomal deletion |
| CtsRrtF | AATTTGGTCGATGATGCTGATG | qRT-PCR |
| CtsRrtR | TAAGTCCCGGTCCGTTAATCC | qRT-PCR |
| CtsR1 | GGTGGTCTCCCATGCAAAGTCAAAATATC | ctsR overexpression |
| CtsR2 | CTCCTCGAGGCTTTCGTAACGCAAGTGGTT | ctsR overexpression |
| CatF | TCAAATACAGCTTTTAGAACTGG | ctsR chromosomal deletion |
| CatR | CCAGTAAATGAAGTCCATGGA | ctsR chromosomal deletion |
| ftsHKOF | ATGGTACCGGACTTATTCGAACAAGCTAAG | ftsH disruption |
| ftsHKOR | TAGGATCCGTAAGCTGCTTGTTGGTTG | ftsH disruption |
| pUCeryF | CCAGGCTTTACACTTTATGC | ftsH disruption |
| pUCeryF | TGGAAAGTTACACGTTACTAAAG | ftsH disruption |
| ftsHrtF | GCAGCTACCTTCGAAGAATCCA | qRT-PCR |
| ftsHrtR | GGGAAACTTGGTTCAGCAACA | qRT-PCR |
| ftsHF | AAAACTGCAGAATCGACGCAATGGAC | ftsH disruption |
| ftsHR | GCTCTAGACGCTCATAACCGAATTAACG | ftsH disruption |
Asterisks indicate 5′-phosphorylated oligonucleotides.
RNA isolation and analysis.
Total RNAs were extracted using the UltraClean microbial isolation kit (Cabru, Milan, Italy) according to the manufacturer's instructions. The quality of the RNA samples was verified by electrophoresis on 1.2% agarose gels, and RNA concentrations were calculated using Quantity One software (Bio-Rad, Milan, Italy). About 1 μg of total RNA was used to synthesize cDNA using Quantitect reverse transcription kit (Qiagen, Milan, Italy), which includes DNase I treatment. The absence of chromosomal DNA contamination was confirmed by real-time PCR on corresponding DNase I-treated RNA.
qRT-PCR was performed on an Applied Biosystems 7300 real-time PCR system using SYBR green I detection. The ldhD and gyrA genes of L. plantarum were used as internal controls for the analysis of ftsH gene expression during abiotic stresses (10, 15). Five microliters of 20-fold-diluted cDNA was added to 15 μl of a real-time PCR mixture containing the Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and 100 nM each primer (Table 1). Cycling conditions included an initial denaturation-enzyme activation at 95°C for 10 min, followed by 35 cycles of 20 s at 95°C, 30 s at 58°C, and 30 s at 72°C. Fluorescence was monitored during each extension phase, and a melting-curve analysis was performed after each run to confirm the amplification of specific transcripts. Data were analyzed using AB 7300 software, by applying the two-standard-curves quantification method. Each assay included triplicate PCR of the samples, negative no-template controls, and standard curves for both the internal-control and target genes, obtained by amplifying serial dilutions (ratio, 1:10) of the cloned target sequence.
Primer extension analysis.
Total RNA was isolated as described previously (30) from L. plantarum cells grown in MRS medium to mid-exponential phase at 30°C or 42°C for 15, 30, or 60 min. Primer extension products of ftsH transcripts were obtained using oligonucleotide FtsHP (Table 1), and primer extension was performed as previously described (30).
Overproduction and purification of CtsR.
B. subtilis CtsR was overexpressed and purified as previously described (9). The L. plantarum ctsR coding sequence was PCR amplified using primer pair CtsR1-CtsR2 (Table 1) and was cloned between the NcoI and XhoI sites of plasmid pET28/16 (6), generating a carboxy-terminal translational fusion with six histidine residues under the control of a T7 bacteriophage promoter. The recombinant pET2816CtsR vector was transformed into E. coli strain BL21 λDE3, in which the T7 RNA polymerase gene is under the control of the inducible lacUV5 promoter (36).
The His-tagged recombinant protein was then overexpressed and purified as previously described (13). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 15% acrylamide gels was performed as described previously (25), and protein concentrations were determined using the Bio-Rad protein assay based on the method developed by Bradford (4).
Gel mobility shift DNA binding assays.
DNA fragments corresponding to the promoter region of the L. plantarum ftsH gene were generated by PCR with biotin-labeled primers (Table 1). Primers FtsH1F, FtsH2F, and FtsH1R were used to amplify two DNA fragments of 343 bp and 202 bp, with or without the predicted CtsR binding site, respectively. Binding of CtsR to DNA was carried out in a 10-μl reaction volume containing 1 μg of poly(dI-dC) (Pharmacia, Milan, Italy), 25 mM Na2HPO4/NaH2PO4 (pH 7), 150 mM NaCl, 0.1 mM EDTA, 2 mM MgSO4, 1 mM dithiothreitol, and 10% glycerol. The DNA binding reaction was initiated by the addition of CtsR, and the mixture was incubated at room temperature for 20 min. Samples were then loaded directly onto a 4% polyacrylamide gel (50 mM Tris, 400 mM glycine, 1.73 mM EDTA) for electrophoresis (14 V cm−1). Electrophoresis was performed for 1 h at room temperature. DNA fragments were transferred by semidry electrotransfer to a nylon membrane (Amersham, Milan, Italy) and revealed with horseradish peroxidase-coupled streptavidin by ECL detection (Pierce, Rockford, IL).
Construction of a chromosomal deletion mutant of the ctsR gene.
The deletion mutant for L. plantarum ctsR was constructed using the Cre-lox-based mutagenesis system (26). DNA fragments corresponding to the chromosomal regions upstream (800-bp fragment; primer pair FB1ctsR-RB1ctsR) and downstream (1,200-bp fragment; primer pair FB2ctsR-RB2ctsR) of ctsR were amplified by PCR using a proofreading DNA polymerase and L. plantarum WCFS1 chromosomal DNA. The amplicons were cloned between the XhoI-SmiI and Ecl136II restriction sites of the suicide vector pNZ5319 (26), and the recombinant mutagenesis vector, pNZ5319CTSR, was introduced into L. plantarum WCFS1 by electroporation. Chloramphenicol-resistant transformants were selected and replica plated to check for erythromycin sensitivity, reflecting loss of the plasmid vector. Candidate double-crossover mutant clones were analyzed by PCR, and correct integration of the lox66-P32-cat-lox71 cassette into the genome was further verified by PCR using primer FBctsR, annealing uniquely to the genomic region, combined with the mutagenesis vector-specific primers (CatF and CatR) (Table 1). In order to excise the P32-cat selectable marker cassette, the cre expression plasmid pNZ5348 (26) was transformed into the ctsR::lox66-P32-cat-lox71 gene replacement mutant. Erythromycin-resistant and chloramphenicol-sensitive colonies were checked by PCR for Cre-mediated recombination and correct excision of the P32-cat cassette by using primers spanning the recombination locus (FBctsR and RB2ctsR) (Table 1).
ctsR deletion was confirmed by genomic DNA sequencing, and the absence of the gene transcript was verified by qRT-PCR (primers CtsRrtF and CtsRrtR) (Table 1).
Disruption of the ftsH gene.
The ftsH gene of L. plantarum WCFS1 was disrupted by single-crossover plasmid integration as reported previously (39). An 870-bp internal ftsH fragment was PCR amplified using primers ftshKOF and ftshKOR and was cloned into pUC18ery between the KpnI and BamHI restriction sites. The resulting recombinant plasmid, pUCFTSH, was transformed into L. plantarum by electroporation, and candidate integrants were obtained on MRS agar plates containing 10 μg erythromycin ml−1. Correct integration of pUCFTSH in the ftsH locus was confirmed by PCR analysis using primers annealing to flanking genomic regions (ftsHF and ftsHR) combined with vector-specific primers (pUCeryF and pUCeryR) (Table 1). A single ftsH disruption mutant was selected and used in subsequent studies. The absence of the ftsH transcript was confirmed by qRT-PCR (primers ftsHrtF and ftsHrtR) (Table 1).
RESULTS
Heat shock induces ftsH expression in L. plantarum.
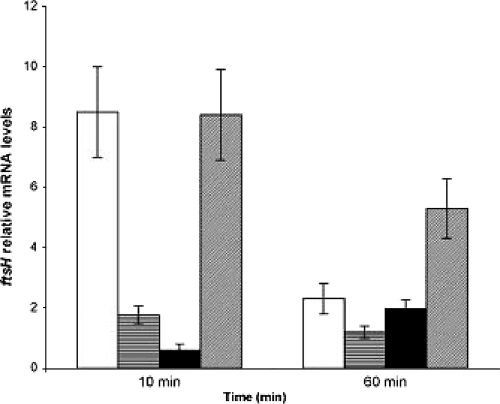
Analysis of the ftsH mRNA level during growth under optimal conditions revealed that this gene is expressed at a basal level throughout growth, with a tendency for the level to increase progressively in the late-stationary phase (data not shown). Expression of ftsH was then analyzed following exposure of cells to a high temperature (42°C), an osmotic stress (0.8 M NaCl), bile (0.15% porcine bile), or heat and osmotic stresses combined (42°C and 0.8 M NaCl), imposed for 10 min and 1 h (Fig. 1). The constitutive ldhD gene was used as an internal control in the qRT-PCR experiments (10, 15). Results were also confirmed by normalizing to gyrA mRNA levels. ftsH transcriptional levels were calculated relative to the mRNA level detected in the control unstressed cells. As reported in Fig. 1, strongly increased ftsH expression was observed 10 min after a temperature upshift to 42°C (eightfold increase). Further induction was observed after 1 h (twofold increase). In contrast, weaker induction was detected in response to osmotic (0.8 M NaCl) and bile (0.15% porcine bile) stresses after a 10-min exposure. Osmotic stress significantly induced expression of the ftsH gene after 10 min (twofold increase), while only a 1.6-fold increase was noted after 1 h for bile stress (Fig. 1). Marked induction (five- to eightfold) was observed in response to the combined heat and osmotic shocks.
FIG. 1.
Relative mRNA levels of L. plantarum ftsH in response to various types of stress as determined by qRT-PCR. mRNA levels were calculated relative to the transcript level detected in corresponding unstressed cultures and were normalized using ldhD as an internal control. Total RNA was extracted and analyzed in the same way 10 and 60 min after exposure to stress. The data presented are averages for three independent experiments; error bars indicate standard deviations. Stress conditions were a heat stress at 42°C (open bars), osmotic stress in 0.8 M NaCl (horizontally striped bars), bile stress (0.15% porcine bile) (filled bars), and combined heat and osmotic stresses (42°C and 0.8 M NaCl) (hatched bars).
ftsH deletion affects growth at a high temperature.
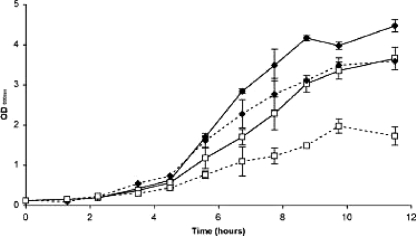
In order to elucidate the importance of FtsH in L. plantarum, a mutant strain was constructed by insertional mutagenesis. qRT-PCR analyses confirmed the absence of ftsH transcripts in the mutant strain (data not shown). The ftsH mutant displayed a lower growth rate than the wild type under optimal growth conditions (Fig. 2). The growth impairment of the mutant strain was much more marked when cells were cultivated under heat stress conditions (42°C) (Fig. 2). This finding suggests that, in comparison to its role in other bacteria, for which no viable null mutant could be obtained (34), FtsH plays a less critical role in L. plantarum. However, FtsH function becomes particularly crucial for coping with harsh conditions, such as high temperatures, indicating the involvement of this protease in heat stress response mechanisms.
FIG. 2.
Growth of L. plantarum wild-type and ftsH mutant strains at an optimal temperature and under heat stress conditions. Cells were cultivated either at 28°C (solid lines) or at a suboptimal temperature of 42°C (dashed lines). The increase in OD600 is shown as a function of time (hours) and was monitored over 12 h for both the wild-type (♦) and ftsH mutant (□) strains. Data shown are means ± standard deviations for one of three independent experiments.
Promoter analysis of ftsH and identification of a putative CtsR binding site.
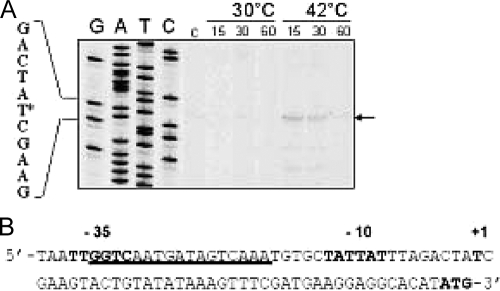
In order to identify regulatory elements controlling ftsH expression, the 5′ mRNA transcriptional start site of ftsH was determined by primer extension analysis on total RNA extracted from exponentially growing L. plantarum cells following incubation at 30°C and 42°C for 15, 30, and 60 min (Fig. 3). Extension products were obtained only from RNA extracted from heat-stressed cells (Fig. 3A). Indeed, a marked band corresponding to the primer extension product appeared after a 10-min temperature upshift, decreased in intensity between 10 and 30 min after the temperature upshift, and became extremely faint after 60 min of heat stress, corroborating the heat induction results obtained by qRT-PCR (see Fig. 1). No extension product was detected for RNA extracted from control cells harvested before stress treatment or from control cells incubated at 30°C for 15, 30, or 60 min. The transcription initiation site was identified at position −41 relative to the translational start codon. −35 (TTGGTC) and −10 (TATAAT) hexamers separated by 18 nucleotides, highly similar to the consensus of L. plantarum sigma A-dependent promoters, were identified at an appropriate distance from the transcriptional start site (Fig. 3B). Sequence analysis of the promoter also revealed a potential binding site for the CtsR repressor, partially overlapping the −35 box and closely resembling the consensus heptad direct-repeat (RGTCADN NAN RGTCADN) CtsR binding site defined in several gram-positive bacteria (9). The potential CtsR box is located between the −35 and −10 boxes, consistent with the role of CtsR as a repressor and in agreement with the locations of previously characterized ctsR operators (9, 34).
FIG. 3.
Mapping and sequence analysis of the L. plantarum ftsH promoter. (A) Primer extension analysis of ftsH mRNA. Total RNA was isolated from L. plantarum WCFS1 cells grown to exponential phase at 30°C (control [C]) and after incubation at either 30°C or 42°C for 15, 30, or 60 min. Lanes G, A, T, and C show DNA sequencing products obtained on genomic DNA with the same primer used for primer extension. The corresponding nucleotide sequence is shown on the left. The transcription start site is indicated by an asterisk. (B) Nucleotide sequence of the ftsH promoter region. −10 and −35 sequences and the transcriptional start site are in boldface. The ctsR binding site is underlined. The translation initiation codon (ATG) is in boldface.
Purified CtsR binds specifically to the ftsH promoter region of L. plantarum.
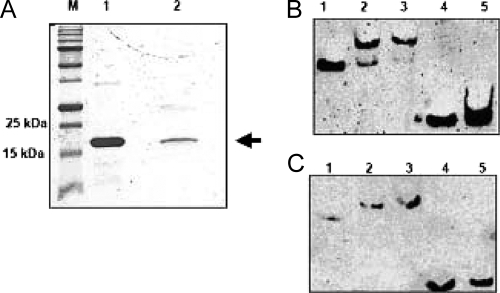
An in vitro approach was used to demonstrate direct interaction of CtsR with its putative target site in the ftsH promoter. To verify whether CtsR was able to recognize and bind the ftsH promoter, a gel mobility DNA binding assay was performed (Fig. 4). The B. subtilis and L. plantarum ctsR genes were cloned and overexpressed in E. coli (see Materials and Methods). The overproduced proteins were purified (Fig. 4A) and used in DNA binding experiments with a biotin-labeled DNA fragment corresponding to the promoter region of L. plantarum ftsH (see Materials and Methods). CtsR from both species was able to bind specifically to the ftsH promoter, generating a single protein-DNA complex, with a gradual displacement of the DNA fragment as the protein concentration increased (Fig. 4B and C). This binding was specific, as shown by the absence of any protein-DNA complex when CtsR proteins were incubated with promoter fragments lacking the CtsR target sequence (Fig. 4B and C). The cross-reactivity between B. subtilis CtsR and the L. plantarum ftsH promoter confirmed the high degree of conservation of this transcriptional regulator and its corresponding target, even among distantly related bacteria. These results indicate that L. plantarum CtsR likely controls the expression of ftsH by interacting directly with its promoter region.
FIG. 4.
Overexpression, purification, and gel mobility shift assays with purified CtsR. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis of purified His-tagged CtsR from E. coli strain BL21 λDE3 carrying the recombinant pET2816CtsR plasmid. Purified recombinant CtsR proteins from B. subtilis (lane 1) and L. plantarum (lane 2) were used. The position of CtsR is indicated by an arrow. M, molecular mass standard. (B and C) Electrophoretic mobility shift assay. Biotin-labeled DNA fragments corresponding to the promoter region of ftsH were incubated with increasing amounts of purified CtsR from B. subtilis (B) and L. plantarum (C). Lanes 1, no CtsR; lanes 2 and 3, 100 and 250 ng of CtsR, respectively; lanes 4 and 5, promoter fragments lacking the CtsR box were preincubated with or without 250 ng of CtsR protein, respectively.
ftsH is derepressed in an L. plantarum ΔctsR mutant.
In order to determine the role of CtsR in controlling the expression of ftsH, a ΔctsR mutant strain of L. plantarum was generated (see Materials and Methods). The expression of ftsH in the ΔctsR mutant strain was analyzed by qRT-PCR and compared to that of the wild type. As reported in Fig. 5, under optimal growth conditions, ftsH was more than threefold derepressed in the mutant strain relative to its expression in the wild-type control (Fig. 5A); the lack of ctsR gene expression in the mutant was confirmed by the absence of any ctsR transcript detectable by PCR (data not shown). This result indicates that ftsH expression is repressed by CtsR, in agreement with the data obtained by gel mobility shift assays. This is the first instance where the ftsH gene of a gram-positive bacterium has been shown to be under CtsR regulation, hence placing it among the class III stress genes.
FIG. 5.
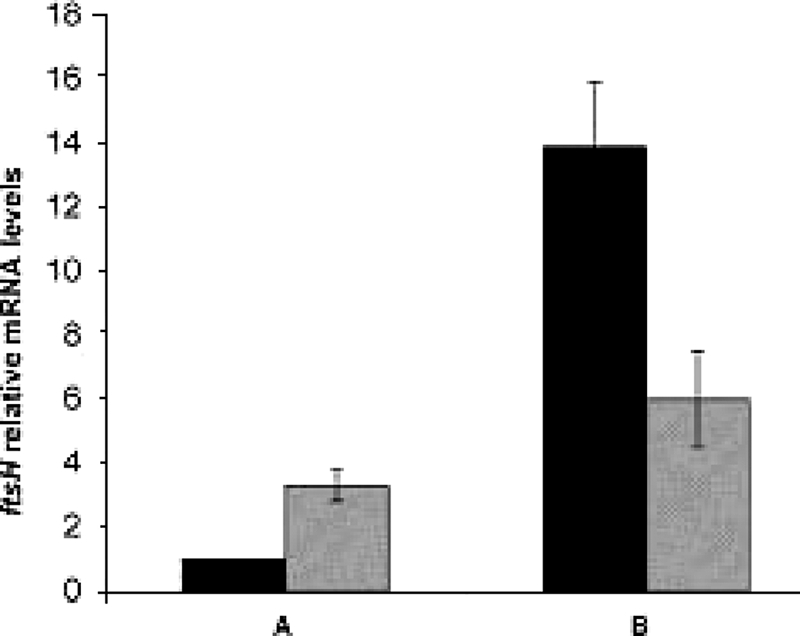
Comparison of ftsH mRNA levels in wild-type and ΔctsR mutant strains of L. plantarum by qRT-PCR. ftsH expression was analyzed in wild-type (filled bars) and ΔctsR mutant (hatched bars) strains under optimal-temperature (30°C) growth conditions (A) and following a heat stress imposed for 5 min at 50°C (B). In all experiments, mRNA levels were normalized to that for the ldhD housekeeping gene. Under unstressed conditions (A), ftsH mRNA levels in the ΔctsR mutant strain were calculated relative to the mRNA level of the wild-type strain, which was assigned a value of 1. Under heat shock conditions (B), ftsH mRNA levels in the wild-type and ΔctsR mutant strains were calculated relative to the corresponding mRNA levels for the unstressed condition (A). Data are averages ± standard deviations of three independent experiments.
When the L. plantarum culture was shifted to 50°C for 10 min, the amounts of ftsH transcript increased further both in the wild type (about 14-fold) and, to a minor extent, in the mutant strain (6-fold) relative to those of the corresponding unstressed control cultures (Fig. 5B). The residual induction observed in the mutant strain could suggest a heat shock control mediated by additional regulators other than CtsR and has been observed previously for many CtsR-dependent genes (6, 8, 9).
DISCUSSION
Functional studies have revealed an important role for FtsH in the bacterial stress response. In several bacteria, including E. coli, B. subtilis, Lactococcus lactis, O. oeni, Helicobacter pylori, and L. plantarum, ftsH expression is induced in response to heat and other stress factors (3, 12, 14, 21). Moreover, in E. coli, FtsH controls the cellular levels of the heat shock sigma factor (σ32) in response to varying temperatures (21, 37). Although the involvement of FtsH in protection against environmental stress has been documented for various bacterial species, little is known about the mechanisms involved in its transcriptional regulation in response to stress. To date, no known typical stress-responsive cis-regulatory element has been found in its promoter that could substantiate its classification as a “stress response gene” (20). Therefore, the ftsH gene has recently been assigned to the heat-inducible class IV genes, whose regulatory mechanisms are still unknown (9, 34). We show that, as has been reported for other bacterial species, the ftsH gene of L. plantarum is significantly induced upon exposure to stress conditions, especially heat stress. The involvement of ftsH in the stress response was also confirmed by the behavior of the mutant strain. Indeed, compared to the wild type, the ftsH mutant strain displayed a significant growth defect when subjected to heat stress.
Interestingly, the consequences of the ftsH mutation are remarkably species specific, ranging from drastic growth impairment (2) to negligible/milder effects on sporulation, development, and the stress response (11, 16, 29). For some bacteria, such as E. coli, L. lactis, and H. pylori, the apparent impossibility of isolating any viable ftsH-null mutant indicates that this protease is essential (1, 17, 31, 38). In contrast, in species such as B. subtilis and Caulobacter crescentus, FtsH seems dispensable for growth under physiological conditions (11, 16). Furthermore, minor effects on normal growth and the cellular stress response were recently observed in a ΔftsH strain of Corynebacterium glutamicum (28). Since we were able to isolate a viable insertional inactivation mutant strain, FtsH is clearly not essential in L. plantarum.
According to our results, the relevance of ftsH function in L. plantarum reflects the situation observed in bacteria such as B. subtilis, C. crescentus, and C. glutamicum but with a marked contribution to growth under heat shock conditions.
Sequence analysis of the ftsH promoter allowed us to predict a potential CtsR binding site. Primer extension experiments indicate that the putative CtsR operator overlaps the promoter and occupies a position consistent with a regulatory function by this transcriptional repressor. By adopting two different experimental approaches, we have shown that L. plantarum ftsH is indeed controlled by CtsR. Purified CtsR from both B. subtilis and L. plantarum recognized and bound specifically to the ftsH promoter. Moreover, using a recently developed Cre-lox-based mutagenesis system (26), we were able to obtain a ΔctsR strain in which the basal transcriptional level of the ftsH gene was significantly derepressed compared to that of the wild type. To our knowledge, this is the first report demonstrating that an ftsH gene is under the control of CtsR. ftsH transcriptional induction was still evident upon heat shock in the ΔctsR mutant strain, suggesting a control mechanism involving CtsR together with one or more additional regulators. Similar results were also obtained for B. subtilis (9, 24) and L. lactis (40), where the expression of some members of the CtsR regulon appears to be controlled by additional regulators.
In conclusion, the ftsH gene of L. plantarum is involved in protection against stress, mainly heat shock, and is controlled by the class III stress gene repressor CtsR. From an evolutionary point of view, CtsR control of ftsH might be either vestigial or a novel and exclusive acquisition of the L. plantarum gene by the CtsR regulon with respect to other, closely related bacterial species. We favor the latter hypothesis. Indeed, although in Oenococcus oeni CtsR acts as a master regulator of most of the known stress response genes, it does not appear to control ftsH expression in this wine bacterium (3, 20). Although little is known about the mechanisms used by L. plantarum to adapt to environmental fluctuations, preliminary analysis of the L. plantarum genome allowed the identification of potential CtsR binding sites upstream of several genes in addition to ftsH, including those encoding small heat shock proteins (hsp18.5) and subunits of the Clp ATP-dependent protease (35; G. Spano and D. Fiocco, unpublished data). This finding strongly suggests that the particular stress conditions encountered by L. plantarum have led to the coordinated CtsR-dependent expression of ftsH with that of other stress response genes, and this possibility will be the subject of further investigation.
Acknowledgments
D. Fiocco was supported by the University of Foggia as a postdoctoral scientist. P. Hols is a research associate at the FNRS. Work in the group of T. Msadek was supported by research funds from the European Commission (grant BACELL Health LSHG-CT-2004-503468), the Centre National de la Recherche Scientifique (CNRS URA 2172), and the Institut Pasteur (GPH 9).
Footnotes
Published ahead of print on 12 December 2008.
REFERENCES
- 1.Akiyama, Y., T. Ogura, and K. Ito. 1994. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention efficiency of a cytoplasmic reporter. J. Biol. Chem. 2695218-5224. [PubMed] [Google Scholar]
- 2.Begg, K. J., T. Tomoyasu, W. D. Donachie, M. Khattar, H. Niki, K. Yamanaka, S. Hiraga, and T. Ogura. 1992. Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J. Bacteriol. 1742416-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourdineaud, J. P., B. Nehme, S. Tesse, and A. Lonvaud-Funel. 2003. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl. Environ. Microbiol. 692512-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bron, P. A., D. Molenaar, W. M. de Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100728-738. [DOI] [PubMed] [Google Scholar]
- 6.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat-shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 471061-1073. [DOI] [PubMed] [Google Scholar]
- 7.De Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23130-135. [Google Scholar]
- 8.Derré, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32581-593. [DOI] [PubMed] [Google Scholar]
- 9.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31117-131. [DOI] [PubMed] [Google Scholar]
- 10.Desroche, N., C. Beltramo, and J. Guzzo. 2005. Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J. Microbiol. Methods 60325-333. [DOI] [PubMed] [Google Scholar]
- 11.Deuerling, E., A. Mogk, C. Richter, M. Purucker, and W. Schumann. 1997. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 23921-933. [DOI] [PubMed] [Google Scholar]
- 12.Deuerling, E., B. Paeslack, and W. Schumann. 1995. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J. Bacteriol. 1774105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 1861175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duwat, P., S. D. Ehrlich, and A. Gruss. 1995. The recA gene of Lactococcus lactis: characterization and involvement in oxidative and thermal stress. Mol. Microbiol. 171121-1131. [DOI] [PubMed] [Google Scholar]
- 15.Fiocco, D., E. Crisetti, V. Capozzi, and G. Spano. 2008. Validation of an internal control gene to apply reverse transcription quantitative PCR to study heat, cold and ethanol stresses in Lactobacillus plantarum. World J. Microbiol. Biotechnol. 24899-902. [Google Scholar]
- 16.Fischer, B., G. Rummel, P. Aldridge, and U. Jenal. 2002. The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 44461-478. [DOI] [PubMed] [Google Scholar]
- 17.Ge, Z., and D. E. Taylor. 1996. Sequencing, expression, and genetic characterization of the Helicobacter pylori ftsH gene encoding a protein homologous to members of a novel putative ATPase family. J. Bacteriol. 1786151-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottesman, S. 1999. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 2142-147. [DOI] [PubMed] [Google Scholar]
- 19.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11815-823. [DOI] [PubMed] [Google Scholar]
- 20.Grandvalet, C., F. Coucheney, C. Beltramo, and J. Guzzo. 2005. CtsR is the master regulator of stress response gene expression in Oenococcus oeni. J. Bacteriol. 1875614-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman, C., D. Thevenet, R. D'Ari, and P. Bouloc. 1995. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. USA 923516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, K., and Y. Akiyama. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59211-231. [DOI] [PubMed] [Google Scholar]
- 23.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 1806681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 731126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langer, T. 2000. AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25247-251. [DOI] [PubMed] [Google Scholar]
- 28.Lüdke, A., R. Kramer, A. Burkovski, D. Schluesener, and A. Poetsch. 2007. A proteomic study of Corynebacterium glutamicum AAA+ protease FtsH. BMC Microbiol. 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lysenko, E., T. Ogura, and S. M. Cutting. 1997. Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143971-978. [DOI] [PubMed] [Google Scholar]
- 30.Muscariello, L., R. Marasco, M. De Felice, and M. Sacco. 2001. The functional ccpA gene is required for carbon catabolite repression in Lactobacillus plantarum. Appl. Environ. Microbiol. 672903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson, D., A. A. Lauridsen, T. Tomoyasu, and T. Ogura. 1994. A Lactococcus lactis gene encodes a membrane protein with putative ATPase activity that is homologous to the essential Escherichia coli ftsH gene product. Microbiology 1402601-2610. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Schumann, W. 1999. FtsH—a single-chain charonin? FEMS Microbiol. Rev. 231-11. [DOI] [PubMed] [Google Scholar]
- 34.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 35.Spano, G., and S. Massa. 2006. Environmental stress response in wine lactic acid bacteria: beyond Bacillus subtilis. Crit. Rev. Microbiol. 3277-86. [DOI] [PubMed] [Google Scholar]
- 36.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 37.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, et al. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma. EMBO J. 142551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomoyasu, T., T. Yuki, S. Morimura, H. Mori, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1993. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J. Bacteriol. 1751344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24387-397. [DOI] [PubMed] [Google Scholar]
- 40.Varmanen, P., H. Ingmer, and F. K. Vogensen. 2000. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology 1461447-1455. [DOI] [PubMed] [Google Scholar]