Abstract
mTOR, the mammalian target of rapamycin, is a critical node for control of cell growth and survival and has widely been implicated in cancer survival signals. mTOR exists in two complexes: mTORC1 and mTORC2. Phospholipase D (PLD) and its metabolite phosphatidic acid (PA) have been implicated in the regulation of mTOR; however, their role has been controversial. We report here that suppression of PLD prevents phosphorylation of the mTORC1 substrate S6 kinase (S6K) at Thr389 and the mTORC2 substrate Akt at Ser473. Suppression of PLD also blocked insulin-stimulated Akt phosphorylation at Ser473 and the mTORC2-dependent phosphorylation of PRAS40. Importantly, PA was required for the association of mTOR with Raptor to form mTORC1 and that of mTOR with Rictor to form mTORC2. The effect of PA was competitive with rapamycin—with much higher concentrations of rapamycin needed to compete with the PA-mTORC2 interaction than with PA-mTORC1. Suppressing PA production substantially increased the sensitivity of mTORC2 to rapamycin. Data provided here demonstrate a PA requirement for the stabilization of both mTORC1 and mTORC2 complexes and reveal a mechanism for the inhibitory effect of rapamycin on mTOR. This study also suggests that by suppressing PLD activity, mTORC2 could be targeted therapeutically with rapamycin.
It has become apparent during the past decade that a critical aspect of tumor progression is the suppression of default apoptotic programs that constitute what is likely the most important protection against cancer. Cellular signals that suppress apoptosis have come to be known as “survival signals.” A common node for survival signals is mTOR, the mammalian target of rapamycin (5, 13, 14, 25). mTOR exists in two distinct complexes, mTORC1 and mTORC2 (21), that differ in their subunit composition and sensitivity to rapamycin. mTORC1 consists of a complex that includes mTOR and a protein known as Raptor (regulatory associated protein of mTOR), whereas mTORC2 consists of a complex that includes mTOR and a protein known as Rictor (rapamycin-insensitive companion of mTOR) (13, 14). There are also mTORC2 complexes that can be distinguished by association with different isoforms of mSin1 (9). While much is known about the regulation of mTORC1 (21), very little is known about the regulation of mTORC2.
mTORC1 is highly sensitive to rapamycin, whereas mTORC2 is relatively insensitive to rapamycin (21). However, it was recently reported that long-term exposure to rapamycin prevented the formation of mTORC2 complexes and blocked the phosphorylation of the mTORC2 substrate Akt at Ser473 (24, 38). Rapamycin, in association with FK506 binding protein 12 (FKBP12), has been reported to interact with mTOR in a manner that is competitive with phosphatidic acid (PA), the metabolic product of phospholipase D (PLD) (2, 4). PLD, like mTOR, has been implicated in survival signals in several human cancer cell lines (1, 10, 11, 27, 32, 39). Since rapamycin-FKBP12 competes with PA for binding to mTOR, the sensitivity of mTORC2 complex formation to rapamycin suggests that PA facilitates the assembly of mTORC2—and ultimately the activation of mTORC2. We report here that the assembly of both mTORC1 and mTORC2 complexes is dependent upon PLD and its metabolite PA. The study also provides mechanistic insight into how rapamycin impacts on mTOR-mediated signals and how PLD regulates mTOR by facilitating the formation of mTOR complexes.
MATERIALS AND METHODS
Cells, cell culture conditions, and transfection.
786-O and MDA-MB-231 cells were obtained from the American Type Culture Collection. All cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Transfections were performed with Lipofectamine LTX for plasmid transfection or Lipofectamine RNAiMAX for small interfering RNA (siRNA) according to the vendor's instructions. Transfection efficiency was determined by transfection of pEGFP-C1, which expresses green fluorescent protein. The percentage of green cells was determined microscopically and was routinely in excess of 90%. For transfection of DNA plasmids, cells were plated at 5 × 105/60-mm plate, and for siRNAs, cells were plated at a lower density of 2 × 105/60-mm plate to allow the turnover of existing proteins that are suppressed by this approach.
Materials.
Reagents were obtained from the following sources. Protein G-Sepharose beads were purchased from Amersham; antibodies against mTOR, Rictor, and Raptor were obtained from Santa Cruz Biotechnology; and antibodies against Akt, P-Akt (Ser473), P-Akt (Thr308), S6 kinase, P-S6 kinase (Thr389), PRAS40, and P-PRAS40 (Thr246) were obtained from Cell Signaling. siRNAs targeting Raptor, Rictor, mTOR, PLD1, and PLD2 were obtained from Sigma-Aldrich. The transfection reagents Lipofectamine LTX and Lipofectamine RNAiMAX were obtained from Invitrogen. Rapamycin and FK506 were purchased from Calbiochem. PA (1-palmitoyl 2-oleoyl) in chloroform was purchased from Avanti-Polaris Lipids. Insulin was obtained from Sigma-Aldrich.
Plasmids.
The pcDNA3.1 control plasmid was obtained from Invitrogen. The green fluorescent protein-expressing pEGFP plasmid was obtained from Clontech. The plasmid expression vector for catalytically inactive mutant PLD1 and PLD2 proteins (pCGN-PLD1-K898R and pCGN-PLD2-K758R) (29, 30) was a generous gift from Michael Frohman (State University of New York, Stony Brook).
Western blot analysis.
Extraction of proteins from cultured cells and Western blot analysis of extracted proteins were performed with the ECL system (Amersham) as described previously (26, 32).
Immunoprecipitation.
Cells were grown in 10-cm-diameter plates. Immediately before lysing, culture plates were rinsed once with cold phosphate-buffered saline and lysed on ice for 20 min in 500 μl of ice-cold 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) immunoprecipitation buffer (40 mM HEPES [pH 7.5], 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 0.5 mM orthovanadate, EDTA-free protease inhibitors [Roche]) containing 0.3% CHAPS. A 500-μg sample of protein was then incubated with appropriate antibodies, and the immunoprecipitates were recovered 16 h later with protein G-Sepharose. The immunoprecipitates were then subjected to Western blot analysis along with 40 μg of total cell lysate.
Cross-linking assay.
Cells were prepared as described for immunoprecipitation. Immediately prior to lysing, the culture plates were rinsed once with cold phosphate-buffered saline and lysed on ice for 30 min with 500 μl of CHAPS immunoprecipitation buffer in the presence of 25 mg/ml dithiobis(succinimidyl)propionate (DSP; Sigma-Aldrich). After a 30-min incubation on ice, 100 μl of 1 M Tris-HCl (pH 7.4) was added to quench the cross-linking and the lysate was left on ice for an additional 30 min. A 500-μg sample of protein was then incubated with mTOR antibody overnight, and the immunoprecipitates were recovered and subjected to Western blot analysis along with 40 μg of total cell lysate.
siRNA.
Cells were plated on 12-well plates at 2 × 105/60-mm plate in medium containing 10% serum without antibiotics. After 1 day, cells were transfected with siRNA with Lipofectamine RNAiMAX according to the manufacturer's directions. After 24 h, the medium was changed to fresh medium containing 10% serum and 2 days later cells were lysed and analyzed by Western blotting.
Assay for PLD activity.
Cells were labeled with [3H]myristic acid (60 Ci/mmol; 1.5 μCi/ml; Perkin-Elmer) for 4 h. 1-Butanol (1-BtOH; 0.8% or as indicated) was added 20 min before lipids were collected. Cells were washed twice in ice-cold phosphate-buffered saline (pH 7.4), after which 500 μl of ice-cold methanol-6N HCl (50:2) was added. The cells were scraped and placed in the first extraction tube containing 155 μl of 1 M NaCl and 500 μl of ice-cold chloroform. These tube are then vortexed for 30 s and centrifuged at 4°C for 3 min at 13,000 rpm. The upper aqueous layer was discarded, and the lower organic phase was transferred to the second extraction tube containing 350 μl of water, 115 μl of methanol, and 115 μl of 1 N NaCl, which was then vortexed and centrifuged as described above. Most of the upper aqueous phase was removed, leaving just enough to cover the lower organic phase. The radioactivity in the organic layer from each sample was then determined by scintillation counting, and 5 × 105 cpm from each sample was dried with nitrogen gas for 10 min. Samples were resuspended in 30 μl of spotting solution (chloroform-methanol, 9:1). The samples were then spotted (10 μl at a time) onto Silica Gel G 60A thin-layer chromatography (TLC) plates (Whatman) and placed in a chamber containing 100 ml of a mobile-phase solution (ethyl acetate-isooctane-glacial acetic acid-H2O, 88:40:16:80) for 2 h. The plates were allowed to air dry for 20 min, sprayed with En3Hancer (Perkin-Elmer), and sealed in a polyethylene bag to protect the silica gel. Plates are then placed in an exposing cassette with X-ray film and developed after 3 to 5 days at −80°C.
Alcohol trap assay for suppression of PA production.
PLD preferentially utilizes primary alcohols over H2O in the transphosphatidylation reaction and generated a phosphatidylalcohol at the expense of PA. This occurs because the primary alcohol can position its hydrophobic tail in the membrane with its hydroxyl group adjacent to the phosphate group on the phospholipid, where the nucleophilic attack can take place with more efficiency than the hydrolysis reaction—in spite of the high concentration H2O. Tertiary alcohols such as tert-BtOH do not work because they do not insert themselves into the membrane in a manner that positions the hydroxyl group adjacent to the phosphate and therefore can be used as negative controls.
Alcohol treatments were performed in 60-mm plates in which the appropriate amounts of 1-BtOH or tert-BtOH were added to Dulbecco's modified Eagle's medium to obtain the desired concentration with vigorous mixing. Due to the high volatility of BtOH, the plates were sealed with Parafilm or small culture flasks were tightly sealed for the duration of the treatment in order to prevent alcohol evaporation. Nontreated controls were sealed as well in order to preserve the consistency of the experiment's conditions. The sealing of plates for up to 8 h did not affect the phenotypes observed.
Preparation of PA.
Right before use, the appropriate amount of PA was dried under N2 and resuspended by vortexing for 2 min in 150 mM NaCl-10 mM Tris-Cl, pH 8.0. The resulting PA suspension was immediately added to the cell culture to a final concentration of 100 μM. Due to the short half-life of PA, this process was repeated every 45 min throughout treatment.
RESULTS
Activation of mTORC1 and mTORC2 is dependent upon PLD and PA.
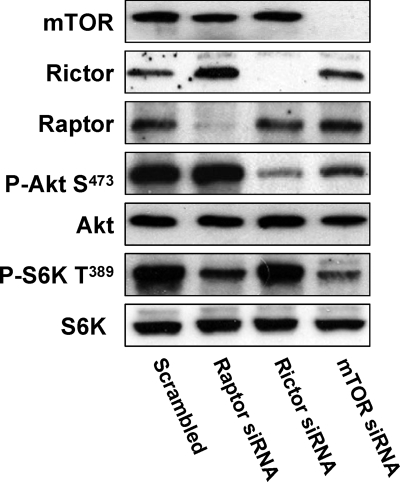
We previously reported that the expression of HIF2α in 786-O cells is dependent upon PLD (32) and that HIF2α is dependent on mTORC2 (33). These studies suggested a link between PLD and mTORC2. PLD, through its metabolite PA, has been implicated in the activation of rapamycin-sensitive mTORC1 in a manner whereby PA is competitive with rapamycin for binding to the FKBP12-rapamycin binding (FRB) domain of mTOR (see reference 7 for a review). Suppression of PLD activity reduces phosphorylation on the mTORC1 substrate S6 kinase at Thr389 (2), indicating a PLD dependence for mTORC1. Recently, Sabatini and coworkers reported that rapamycin suppressed the assembly of mTORC2 complexes (24, 38). This finding suggested that if rapamycin competes with PA for mTOR, then PA could also be required for the activation of mTORC2. mTORC2 phosphorylates Akt at Ser473 (16, 23) and can be used as an indicator of mTORC2 activity. To verify this in 786-O cells, we examined the dependence of Akt phosphorylation at Ser473 on mTOR and Rictor—a critical component of mTORC2. We also examined the dependence of S6 kinase phosphorylation on mTOR and Raptor—a critical component of mTORC1. As shown in Fig. 1, the phosphorylation of Akt at Ser473 in 786-O cells is suppressed by siRNAs for both mTOR and Rictor, but not Raptor, and the phosphorylation of S6 kinase at Thr389 was suppressed by siRNAs for mTOR and Raptor, but not Rictor. These data establish that the phosphorylation of Akt at Ser473 is dependent on mTORC2, which is dependent on Rictor, and that S6 kinase phosphorylation is dependent on mTORC1, which is dependent on Raptor.
FIG. 1.
Phosphorylation of Akt at Ser473 in 786-O cells is dependent on mTORC2. 786-O cells were plated at 2 × 105/60-mm plate. Twenty-four hours later, the cells were transfected with siRNA for Raptor, Rictor, or mTOR or a scrambled siRNA as described in Materials and Methods. Twenty-four hours later, the cells were treated with fresh medium containing 10% serum for an additional 48 h. The cells were then harvested and analyzed for levels of mTOR, Raptor, Rictor, Akt phosphorylated at Ser473 (P-Akt S473), Akt, S6 kinase phosphorylated at T398 (P-S6K T389), and S6 kinase (S6K) by Western blot analysis. All of the data shown are representative of at least three independent experiments.
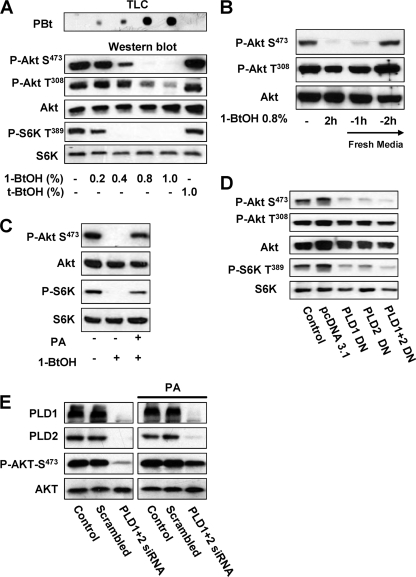
To investigate whether mTORC2 is dependent on PLD activity, we examined whether the phosphorylation of Akt at the mTORC2-dependent Ser473 site was dependent upon PLD activity with the “alcohol trap” transphosphatidylation reaction whereby primary alcohols generate phosphatidylalcohols at the expense of PA production (26). Tertiary alcohols are not utilized by PLD in the transphosphatidylation reaction and serve as negative controls. As shown in Fig. 2A (top), increasing concentrations of 1-BtOH led to increasing concentrations of the transphosphatidylation product phosphatidyl-BtOH (PBt) whereas tert-BtOH did not generate detectable PBt. The increasing levels of PBt indicate that 1-BtOH suppressed PA production in a dose-dependent manner. The effect of increasing concentrations of 1-BtOH on the phosphorylation of Akt at Ser473 and S6 kinase at Thr389 was investigated, and as shown in the lower part of Fig. 2A, 1-BtOH suppressed the phosphorylation of Akt at Ser473 beginning at 0.4% and complete inhibition was seen at 0.8% 1-BtOH. Significantly, S6 kinase phosphorylation at Thr389 was more sensitive to 1-BtOH than was Akt phosphorylation. 1-BtOH had much less of an effect on the phosphorylation of Akt at Thr308—a site that is not phosphorylated by mTORC2 (13). We have found that 1-BtOH can suppress the expression of PDK1 (phosphoinositide-dependent kinase 1), the kinase that phosphorylates Akt at T308 (our unpublished results), which likely explains the slight reduction in the level of Akt phosphorylation at this site caused by high concentrations of 1-BtOH. tert-BtOH at 1% had no effect upon the phosphorylation of the substrates examined here, indicating that the effects were due to the suppression of PA production by PLD. The effect of 1-BtOH was reversible, and phosphorylation of Akt at Ser473 was restored within 2 h after fresh medium lacking 1-BtOH was provided (Fig. 2B). If PA was added to cell cultures treated with 1-BtOH, the suppression of Akt phosphorylation at Ser473 and S6 kinase phosphorylation at Thr389 was reversed (Fig. 2C). This observation indicates that it is PA generated by PLD that stimulates Akt phosphorylation. To further establish that the effect of 1-BtOH on Akt phosphorylation was due to an effect on PLD, we transiently introduced catalytically inactive mutant forms of PLD1 and PLD2 (29, 30), which have been shown to act effectively as dominant negative mutant PLDs (26, 32). As shown in Fig. 2D, the presence of the mutant PLDs strongly suppressed the phosphorylation of Akt at Ser473 while having much less of an effect on the phosphorylation at Thr308. The mutant PLDs also suppressed S6 kinase phosphorylation. We wanted to examine whether the effect of the dominant negative mutant PLDs could be overcome by PA, but this proved problematic due to the toxic effects of exogenous PA vesicles on cells recently treated with Lipofectamine. We therefore used siRNAs for PLD1 and PLD2, which take several days to suppress PLD expression (32). As in Fig. 2D, the PLD siRNAs substantially reduced the levels of Akt phosphorylation at Ser473, and importantly, this effect was reversed by PA (Fig. 2E). The data in Fig. 2 indicate that the mTORC2-dependent phosphorylation of Akt at Ser473 and the mTORC1-dependent phosphorylation of S6 kinase at Thr389 are dependent upon PLD and its metabolite PA.
FIG. 2.
Activation of mTORC2 is dependent on PLD and PA. (A) 786-O cells were plated at 5 × 105/60-mm plate for 24 h and then shifted to medium containing 0.5% serum. 1-BtOH or tert-BtOH was then added where indicated at the concentrations shown. After 2 h, the cells were harvested and analyzed for levels of the transphosphatidylation product PBt by TLC as described in Materials and Methods. The PBt band of the TLC plate is shown at the top. The cells were also evaluated for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt phosphorylated at Thr308 (P-Akt T308), Akt, S6 kinase phosphorylated at T398 (P-S6K T389), and S6 kinase (S6K) by Western blot analysis. (B) 786-O cells were plated as in panel A. 1-BtOH (0.8%) was then added where indicated. After 2 h, the cells were either harvested or placed in fresh medium for 1 or 2 h as indicated, at which time the cells were harvested and analyzed for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt phosphorylated at Thr308 (P-Akt T308), and Akt as in panel A. (C) 786-O cells were prepared and treated with 0.8% 1-BtOH as in panel B. PA (100 μM) was prepared as described in Materials and Methods and was added along with 1-BtOH where indicated. After 2 h, the cells were harvested and analyzed for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt, S6 kinase phosphorylated at Thr389 (P-S6K T389), and S6 kinase as in panel A. (D) 786-O cells were plated at 5 × 105/60-mm plate. Twenty-four hours later, the cells were transfected with vectors expressing catalytically inactive dominant negative (DN) mutant forms of PLD1 or PLD2 as indicated. The parental vector pcDNA3.1 was used as a control. Twenty-four hours later, the cells were treated with fresh medium containing 10% serum for an additional 24 h. Control cells were treated with transfection medium but without transfection. The cells were then harvested and analyzed for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt phosphorylated at Thr308 (P-Akt T308), Akt, S6 kinase phosphorylated at Thr389 (P-S6K T389), and S6 kinase as in panel A. (E) 786-O cells were plated at 2 × 105/60-mm plate. Twenty-four hours later, the cells were transfected with siRNAs for PLD1 and PLD2 or a scrambled siRNA as indicated. Control cells were treated with transfection medium but without the transfection reagent. Forty-eight hours later, the cells were treated with fresh medium containing 10% serum. The cells were then treated with fresh medium containing 10% serum for an additional 24 h and placed in medium containing 0.5% medium for 24 h. Where indicated, cells were treated with 100 μM PA for 3 h. The cells were then harvested and analyzed for levels of PLD1, PLD2, Akt phosphorylated at Ser473 (P-Akt S473), and Akt as in panel A. All of the data shown are representative of at least two independent experiments.
PLD requirement for the formation of mTOR complexes.
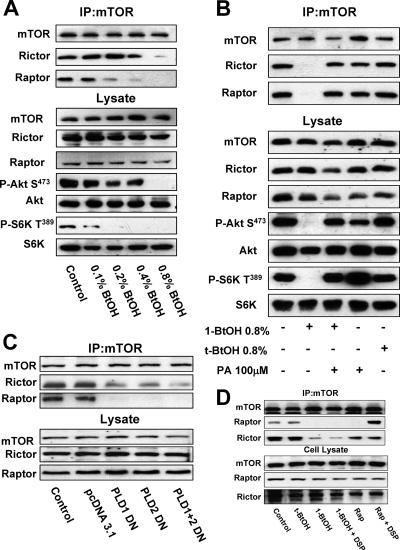
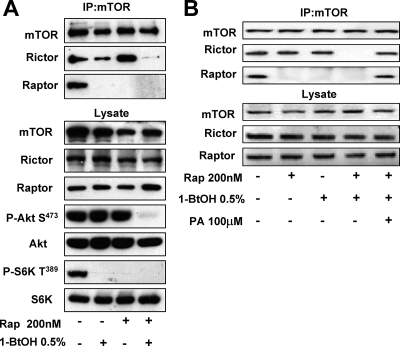
Sabatini and coworkers reported that prolonged rapamycin treatment prevented mTORC2 complex formation (24, 38), suggesting that PA, which is competitive with rapamycin for mTOR binding (2, 4), might be required for the assembly of the mTORC2 complex. To address this question, we examined the impact of PLD activity on the ability of mTOR to coimmunoprecipitate with Rictor and Raptor. 786-O cells were treated with increasing concentrations of 1-BtOH, and the levels of Rictor and Raptor that coimmunoprecipitated with mTOR were evaluated. As shown in Fig. 3A, treatment of the 786-O cells with increasing concentrations of 1-BtOH suppressed the levels of both Rictor and Raptor that coimmunoprecipitated with mTOR. Importantly, the level of 1-BtOH required to suppress Akt phosphorylation at Ser473 correlated with the level required to suppress coimmunoprecipitation of mTOR with Rictor and the level of 1-BtOH required to suppress S6 kinase phosphorylation correlated with the level required to suppress coimmunoprecipitation of mTOR with Raptor (Fig. 3A). If PA was added, the effect of 1-BtOH on the association between mTOR and both Raptor and Rictor was reversed (Fig. 3B). We also examined the effect of the dominant negative mutant PLDs on the association between Raptor and Rictor. As shown in Fig. 3C, both PLD1 and PLD2 suppressed the association between mTOR and both Raptor and Rictor. Of significance, the association between mTOR and Raptor was more sensitive to the mutant PLDs, which is consistent with the greater sensitivity of this association to 1-BtOH, as shown in Fig. 3A. Sabatini and colleagues reported previously that rapamycin disrupted the interaction between mTOR and Raptor, but if a cross-linking reagent was added to the lysates during immunoprecipitation, Raptor was found in the mTOR immunoprecipitates (17). This suggested that rapamycin did not completely disassemble the mTOR complex. We therefore examined whether reducing PA levels with 1-BtOH has a similar effect when a cross-linker is employed. As shown in Fig. 3D, the rapamycin-induced dissociation of mTOR was reversed by the cross-linking reagent DSP. However, when PA levels were reduced with 1-BtOH, DSP did not restore coimmunoprecipitation of mTOR with either Raptor or Rictor. This observation suggests that, in the absence of PA, the mTOR complexes are broken down more completely than when rapamycin displaces PA. Collectively, the data in Fig. 3 suggest that the PLD/PA requirement for mTOR activity is to facilitate or stabilize the assembly of mTOR complexes. The data are consistent with the reported competition between PA and rapamycin-FKBP12 (2, 4) and the observation that rapamycin suppresses the formation of the mTORC2 complex (24, 38). The lack of PA apparently breaks down the complexes more completely than rapamycin.
FIG. 3.
PLD is required for the formation of mTOR complexes. (A) 786-O cells were plated at 5 × 105/60-mm plate for 24 h and then shifted to medium containing 0.5% serum. 1-BtOH was then added at the indicated concentrations. After 3 h, lysates were prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight, and then mTOR immunoprecipitates (IP:mTOR), along with the lysates, were subjected to Western blot analysis for Rictor, Raptor, and mTOR. The lysates were also analyzed for the levels of Akt, Akt phosphorylated at Ser473 (P-Akt S473), S6 kinase, and S6 kinase phosphorylated at Thr389 (P-S6K T389). (B) 786-O cells were prepared as in panel A and then treated with 1-BtOH, tert-BtOH, and PA as indicated. Lysates were prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight, and then the mTOR immunoprecipitates and lysates were subjected to Western blot analysis as in panel A. (C) 786-O cells were plated at 5 × 105/60-mm plate. Twenty-four hours later, the cells were transfected with vectors expressing catalytically inactive dominant negative (DN) mutant forms of PLD1 and PLD2 or the parental vector pcDNA3.1 as indicated. Twenty-four hours later, the cells were treated with fresh medium containing 10% serum for an additional 24 h. Control cells were treated with transfection medium but without transfection. Lysates were then prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight, and then mTOR immunoprecipitates (IP:mTOR), along with the lysates, were subjected to Western blot analysis for Rictor, Raptor, and mTOR. (D) Cells were prepared as in panel A and placed on medium containing 0.5% serum overnight. Cells were treated with the indicated alcohols (0.8%) or 200 nM rapamycin (Rap) for 2 h. The cells were then lysed in the absence or presence of DSP as described in Materials and Methods. Lysates were subjected to immunoprecipitation with anti-mTOR antibody overnight, and then the mTOR immunoprecipitates were subjected, along with the whole-cell lysate, to Western blot analysis for mTOR, Raptor, and Rictor as indicated. All of the data shown are representative of at least two independent experiments.
Differential sensitivity of mTORC1 and mTORC2 to rapamycin.
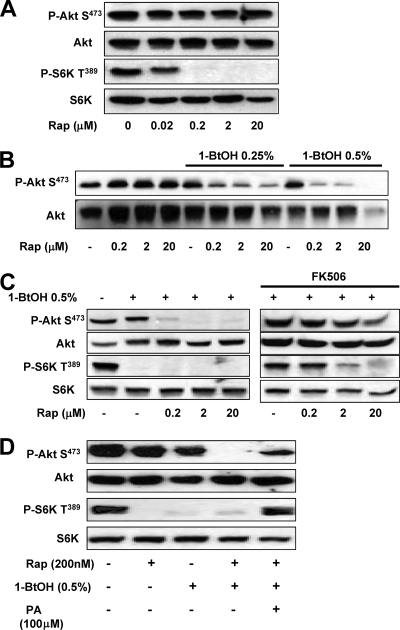
We previously demonstrated that elevated PLD activity conferred rapamycin resistance on breast cancer cells (2). If there was higher PLD activity, higher doses of rapamycin were required to suppress both cell proliferation and S6 kinase phosphorylation (2). Intriguingly, higher concentrations of rapamycin were required to suppress cell proliferation than were required to suppress S6 kinase phosphorylation. These data could be explained by a differential sensitivity of mTORC1 and mTORC2 to rapamycin. To test this hypothesis, we examined the effect of increasing concentrations of rapamycin on the phosphorylation of Akt at Ser473 and S6 kinase at Thr389. As shown in Fig. 4A, S6 kinase phosphorylation was sensitive to concentrations of rapamycin in the low nanomolar range, with a 50% inhibitory concentration of around 20 nM. In contrast, Akt phosphorylation at Ser473 was resistant to rapamycin concentrations of up to 20 μM.
FIG. 4.
Differential sensitivity of mTORC1 and mTORC2 activities to rapamycin and 1-BtOH. (A) 786-O cells were plated at 5 × 105/60-mm plate in medium containing 10% serum and incubated for 24 h. Cells were then shifted to medium containing 0.5% serum overnight. Rapamycin (Rap) was added at the indicated concentrations. The cells were harvested 8 h later, and the levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt, S6 kinase phosphorylated at Thr389 (P-S6K T389), and S6 kinase (S6K) were determined by Western blot analysis. (B) 786-O cells were plated as in panel A. Rapamycin was added at the indicated concentrations in the absence or presence of either 0.25 or 0.5% 1-BtOH as indicated. Eight hours later, the cells were harvested and the levels of Akt phosphorylated at Ser473 (P-Akt S473) and Akt were determined by Western blot analysis. (C) 786-O cells were plated as in panel A. Rapamycin was added at the indicated concentrations in the absence or presence of 0.5% 1-BtOH as indicated. FK506 (10 μM) was added along with rapamycin and 1-BtOH where indicated. Eight hours later, the cells were harvested and the levels of Akt phosphorylated at Ser473 (P-Akt S473) and Akt were determined by Western blot analysis. (D) 786-O cells were prepared and treated with 0.5% 1-BtOH and 200 nM rapamycin as shown. PA (100 μM) was added along with 1-BtOH where indicated. After 8 h, the cells were harvested and analyzed for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt, S6 kinase phosphorylated at Thr389 (P-S6K T389), and S6 kinase as in panel A. All of the data shown are representative of at least two independent experiments.
The data in Fig. 2A revealed that suppression of Akt phosphorylation at Ser473 (mTORC2) required higher concentrations of 1-BtOH than did the suppression of S6 kinase phosphorylation (mTORC1). This finding indicates that the concentration of PA must be substantially lower before mTORC2 activity is reduced—indicating that PA interacts more strongly with mTORC2 than with mTORC1 (see Discussion). The observation also suggests that rapamycin-FKBP12 would disrupt the weaker interaction between PA and mTORC1 at lower concentrations than would be needed to disrupt the stronger interaction between PA and mTORC2. If this is true, then lowering the PA concentration should allow rapamycin to suppress mTORC2. To investigate this, we examined the effect of rapamycin on Akt phosphorylation at Ser473 in the presence of increasing concentrations of 1-BtOH, which reduces the level of PA. As shown in Fig. 4B, the presence of either 0.25 or 0.5% 1-BtOH had little or no effect on Akt phosphorylation at Ser473. However, in the presence of 1-BtOH, Akt phosphorylation was sensitive to 200 nM rapamycin. To verify that the effect of rapamycin on Akt phosphorylation was due to an effect on mTOR, we examined the effect of rapamycin on Akt phosphorylation in the presence of FK506, which competes with rapamycin for binding to FKBP12. As shown in Fig. 4C, FK506 reversed the inhibitory effect of rapamycin on both Akt and S6 kinase. We also examined whether the effect could be reversed by PA, and as shown in Fig. 4D, the phosphorylation of both Akt at Ser473 and S6 kinase at Thr389 was restored by PA.
The sensitivity of Akt phosphorylation at Ser473 to 0.5% 1-BtOH and 200 nM rapamycin suggests that the effect is due to an effect on mTORC2. We therefore examined the sensitivity of the mTOR association with Rictor to rapamycin in the presence of 1-BtOH. As shown in Fig. 5A, the association between mTOR and Rictor was sensitive to 200 nM rapamycin in 0.5% 1-BtOH. As with Akt phosphorylation, the effect of rapamycin and 1-BtOH on the association between mTORC2 and Rictor was reversed by PA—indicating that the effect of 1-BtOH was on PLD-generated PA. These data support the hypotheses that the formation of mTOR complexes is mediated by PA and that rapamycin-FKBP12 interferes with the interaction between PA and mTOR.
FIG. 5.
Differential sensitivity of mTORC1 and mTORC2 complex formation to rapamycin and 1-BtOH. (A) 786-O cells were plated at 5 × 105/60-mm plate, incubated for 24 h, and then shifted to medium containing 0.5% serum. 1-BtOH (0.25%) and the indicated concentrations of rapamycin (Rap) were added as indicated. After 6 h, lysates were prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight, and then the mTOR immunoprecipitate (IP:mTOR) was subjected, along with the lysates, to Western blot analysis for Rictor and Raptor. The lysates were also analyzed for levels of Akt phosphorylated at Ser473 (P-Akt S473), Akt, S6 kinase phosphorylated at Thr389 (P-S6K T389), and S6 kinase (S6K). (B) 786-O cells were prepared as in panel A and treated with 0.5% 1-BtOH and 200 nM rapamycin as indicated. PA (100 μM) was added along with 1-BtOH where indicated. After 6 h, lysates were prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight and then the mTOR immunoprecipitate was subjected, along with the lysates, to Western blot analysis for Rictor. The data shown are representative of three independent experiments.
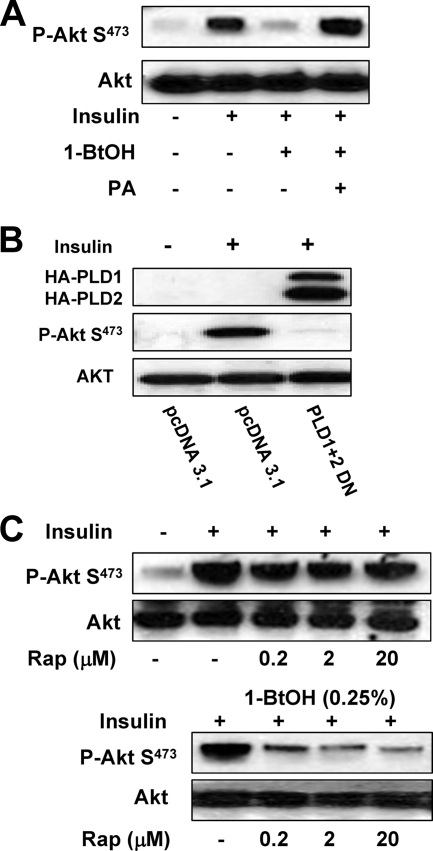
Insulin-stimulated Akt phosphorylation at Ser473 is dependent on PLD activity.
Akt phosphorylation at Ser473 can be stimulated by insulin (9, 16). We therefore wanted to examine the PLD dependence of insulin-stimulated Akt phosphorylation. The 786-O cells used in the experiments described above have constitutively elevated levels of Akt phosphorylation at Ser473. We therefore used MDA-MB-231 cells, which have low levels of basal Akt phosphorylation at this site (1). As shown in Fig. 6A, insulin strongly induced an increase in Akt phosphorylation at Ser473. This increase was suppressed by 0.8% 1-BtOH; however, if PA was added, the effect of 1-BtOH was reversed—indicating a PA requirement for the insulin-induced increase in Akt phosphorylation at Ser473. The insulin-induced increase in Akt phosphorylation at Ser473 was also suppressed in MDA-MB-231 cells if they expressed dominant negative mutant proteins PLD1 and PLD2. We also examined the sensitivity of the insulin-induced increase in Akt phosphorylation to rapamycin. As shown in Fig. 6C, the insulin-induced increase in Akt phosphorylation was resistant to a rapamycin concentration of 20 μM. However, in the presence of 0.25% 1-BtOH, which had little effect on Akt phosphorylation, rapamycin was able to suppress insulin-induced Akt phosphorylation at 200 nM (Fig. 6C). These data further support the hypothesis that mTORC2 can be made rapamycin sensitive by reducing the concentration of PLD-generated PA.
FIG. 6.
Insulin-stimulated Akt phosphorylation at Ser473 is dependent on PLD activity. (A) MDA-MB-231 cells were plated at 5 × 105/60-mm plate in medium containing 10% serum and incubated for 24 h. Cells were then shifted to medium containing 0.5% serum overnight. Insulin (100 nM), 1-BtOH (0.8%), and PA (100 μM) were then added as indicated. The cells were harvested 2 h later, and the levels of Akt phosphorylated at Ser473 (P-Akt S473) and Akt were determined by Western blot analysis. (B) MDA-MB-231 cells were plated at 5 × 105/60-mm plate as in Fig. 2. Twenty-four hours later, the cells were transfected with vectors expressing the catalytically inactive dominant negative (DN) mutant forms of PLD1 and PLD2 or the parental vector pcDNA3.1 as indicated. Twenty-four hours later, the cells were treated with fresh medium containing 10% serum for an additional 24 h. The cells were then harvested and analyzed for the levels of Akt phosphorylated at Ser473 (P-Akt S473) and Akt as in panel A. (C) MDA-MB-231 cells were plated as in panel A. Rapamycin (Rap) was added at the indicated concentrations in the absence or presence of 0.25% 1-BtOH as indicated. Six hours later, the cells were harvested and the levels of phosphorylated Akt and Akt were determined as in panel A. The data shown are representative of two independent experiments.
Suppression of PLD increases the association between mTOR and the mTOR inhibitory protein PRAS40.
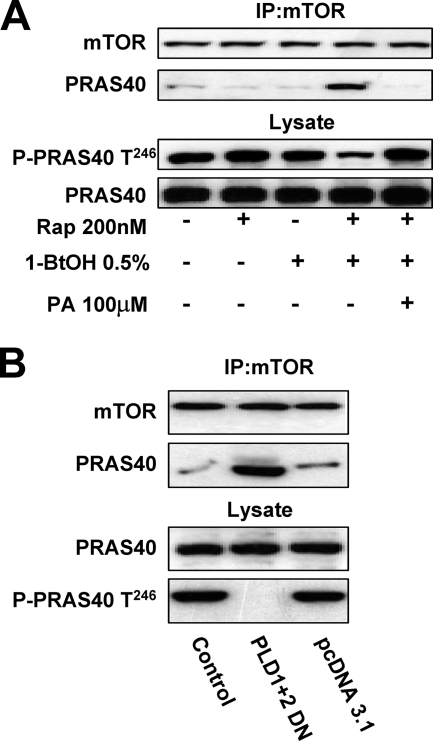
PRAS40 (proline-rich Akt substrate of 40 kDa) is an inhibitor of mTORC1 that is a substrate of mTORC2 (22, 34, 37). The phosphorylation of PRAS40 prevents association with mTOR. We therefore examined the effects of 1-BtOH and rapamycin on the association of PRAS40 with mTOR and the phosphorylation state of PRAS40. As shown in Fig. 7A (top), neither 0.5% 1-BtOH nor 200 nM rapamycin had an impact on the ability of PRAS40 to coimmunoprecipitate with mTOR. However, the combination of 0.5% 1-BtOH and 200 nM rapamycin strongly increased the association between mTOR and PRAS40. This combination of 1-BtOH and rapamycin was that needed to suppress mTORC2 and Akt phosphorylation at Ser473 (Fig. 4B). This combination of 1-BtOH and rapamycin also suppressed the phosphorylation of PRAS40 at the Akt site of Thr246 (Fig. 7A, bottom). The effect of 1-BtOH on the association between mTOR and PRAS40, as well as the effect on PRAS40 phosphorylation, was overcome by PA (Fig. 7A). We also examined the effect of the dominant negative mutant PLDs on the association between mTOR and PRAS40, and as shown in Fig. 7B, the dominant negative mutant PLDs suppressed PRAS40 phosphorylation and increased association with mTOR. These data further support a role for PLD in the regulation of mTORC2 and the positive feedback on mTORC1 through suppression of PRAS40 inhibition.
FIG. 7.
Suppression of PLD increases the association between mTOR and the mTOR inhibitory protein PRAS40. (A) 786-O cells were plated at 5 × 105/60-mm plate, incubated for 24 h, and then shifted to medium containing 0.5% serum. 1-BtOH (0.5%) and rapamycin (200 nM) were added as indicated. After 6 h, lysates were prepared and subjected to immunoprecipitation with anti-mTOR antibody overnight and then the mTOR immunoprecipitate (IP:mTOR) was subjected, along with the lysates, to Western blot analysis for mTOR, PRAS40, and PRAS40 phosphorylated at Thr246 (P-PRAS40). (B) 786-O cells were plated as in panel A. Twenty-four hours later, the cells were transfected with vectors expressing catalytically inactive dominant negative (DN) mutant forms of PLD1 and PLD2 or the parental vector pcDNA3.1 as indicated. Twenty-four hours later, the cells were treated with fresh medium containing 10% serum for an additional 24 h. The cells were then harvested and analyzed for mTOR and PRAS40 in the mTOR immunoprecipitates and PRAS40 and PRAS40 phosphorylated at Thr246 (P-PRAS40) in the lysates as in panel A. The data shown are representative of two independent experiments.
DISCUSSION
Elevated PLD activity has been observed in a large number of human cancers and human cancer cells (8). PLD activity has also been implicated in survival signaling in cancer cells (5, 6). The PLD metabolite PA has been implicated in the activation of mTOR (7), which has also been widely implicated in cancer survival signals (13, 14). However, a role for PLD in the survival signals mediated by mTOR has not been widely accepted. The data provided here reveal that PLD and its metabolite PA are critical for the formation of both mTORC1 and mTORC2 complexes. This study reinforces the concepts that rapamycin suppresses mTOR by interfering with the interaction between mTOR and PA and that mTOR can become more sensitive to rapamycin by reducing PA levels. Data presented here demonstrate that the mTORC2-dependent phosphorylation of Akt at Ser473 requires PLD activity and PA. Akt, like mTOR, is a critical node for cancer survival signals, and phosphorylation of Akt at Ser473 has been strongly correlated with elevated Akt activity (13, 19). The dependence of Akt phosphorylation at Ser 473 on PLD implicates PA as a critical regulator of Akt-mediated survival signals. The PLD dependence was observed in renal cancer cells, where there is elevated basal Akt phosphorylation, and also in insulin-stimulated increases in Akt phosphorylation in a breast cancer cell line, where there are reduced levels of Akt phosphorylation. Data were also presented demonstrating that the suppression of mTORC1 by PRAS40 is reversed when PLD activity is suppressed. Collectively, these data firmly establish a role for PLD-generated PA in the regulation of both mTORC1 and mTORC2 in human cancer cells and suggest that targeting PLD signaling represents a means for suppressing mTOR-dependent survival signals and enhancing the efficacy of rapamycin-based therapeutic strategies in cancers where PLD and mTOR suppress default apoptotic programs that protect against cancer.
While the specificity of rapamycin for mTOR has been known for some time, the mechanism of action has not been established. Jie Chen's group reported that PA interacted with mTOR in a manner that was competitive with rapamycin but did not have any apparent impact on the kinase activity of mTOR (4). Our group subsequently demonstrated that elevated PLD activity increased the concentration of rapamycin needed to suppress S6 kinase phosphorylation and cell proliferation (2). Recent structural studies have revealed that PA interacts with the FRB domain of mTOR and causes structural changes similar to those observed when rapamycin-FKBP12 binds to the FRB domain (35). Sabatini's group reported recently that rapamycin could prevent the association of mTOR with other components of the mTORC2 complex. Our finding that PLD is required for the stability of complexes between mTOR and Rictor and between mTOR and Raptor is consistent with the observations that rapamycin prevents mTOR2 complex formation and that PA interacts with mTOR in a manner that is competitive with rapamycin (2, 4, 35). While rapamycin disrupts the interaction between mTOR and Raptor, the presence of a cross-linker in mTOR immunoprecipitations restored the association (17). In contrast, reducing PA levels with 1-BtOH apparently disrupted the mTOR complexes such that cross-linking did not restore the association between mTOR and either Raptor or Rictor—implying that the lack of PA disrupts the mTOR complexes more than rapamycin does. The major implications of this study are that PLD and PA regulate mTOR signaling by facilitating the formation of mTOR complexes and that rapamycin inhibits mTOR by interfering with the PA-mTOR interaction.
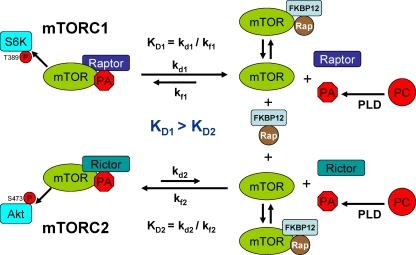
Our previous study demonstrating that elevated PLD activity increased the dose of rapamycin needed to suppress S6 kinase phosphorylation and cell proliferation (2) revealed something that was confusing. The amount of rapamycin needed to suppress cell proliferation was greater than that needed to suppress S6 kinase phosphorylation. The effect of high-dose rapamycin needed to suppress proliferation and induce apoptosis in MDA-MB-231 cells was reversed when rapamycin-resistant mutant mTOR was used (1)—indicating that the effect of the high doses of rapamycin involved mTOR. Data provided in Fig. 3 reveal that higher concentrations of 1-BtOH are required to suppress Akt phosphorylation at Ser 473 than are needed to suppress S6 kinase phosphorylation. This suggests that higher concentrations of 1-BtOH are required to suppress mTORC2 than are required to suppress mTORC1. The implications of this finding are that mTORC2 binds PA more strongly than mTORC1 and that lower concentrations of PA in the cell are needed when PA dissociates in order for PA to stay dissociated from mTORC2. This observation also suggests why higher concentrations of rapamycin are needed to compete for binding to mTORC2 than to compete for binding to mTORC1. The data provided in Fig. 4 reveal a differential rapamycin dose response for S6 kinase phosphorylation and Akt phosphorylation at Ser473. The higher sensitivity of S6 kinase phosphorylation to rapamycin is consistent with a requirement for mTORC1, which has an apparently lower affinity for PA than does mTORC2. The weaker association of PA with mTORC1 means that PA and mTORC1 will dissociate more often, and thus, lower concentrations of rapamycin would be needed to replace PA on mTORC1 when there is a dissociation. In contrast, the higher concentration of rapamycin needed to suppress Akt phosphorylation at Ser473 reflects a requirement for only mTORC2, which apparently binds PA more strongly, and therefore dissociations are rare—meaning that very high concentrations of rapamycin would be needed to bind the low concentrations of mTOR obtained when PA dissociates from mTORC2. Thus, the dissociation constant for PA and mTORC2 (Kd2) would be less than the dissociation constant for mTORC1 and PA (Kd1). This is depicted in a simplified model for the differential strength of association between mTORC1 and mTORC2 with PA in Fig. 8. We propose that rapamycin-FKBP12 binds the FRB domain of mTOR only when PA is dissociated from the FRB domain. However, since the association of PA with mTORC2 is stronger than the association with mTORC1, the dissociation of PA from mTORC2 is less frequent and higher concentrations of rapamycin are needed to interact with the rare mTOR released from mTORC2. The rate constants described in the proposed model are only for the interaction between mTOR and PA, and therefore the model represents an oversimplification, since the involvement of Rictor and Raptor, along with other components of the mTORC1 and mTORC2 complexes, has been neglected. However, the model does provide a first approximation of the differential stability of mTORC1 and mTORC2 complexes that is consistent with the observed differential sensitivities to rapamycin and PA.
FIG. 8.
Model of the differential effects of rapamycin (Rap) and 1-BtOH on mTORC1 and mTORC2. The constants of mTORC1 and mTORC2 dissociation (KDs) from PA represent the ratios of the rate constants of dissociation (kd) and formation (kf). The data provided here are consistent with a model whereby the rate constant of the dissociation of mTORC1 from PA and mTOR (kd1) is greater than the rate constant of the dissociation of mTORC2 from PA and mTOR (kd2). Thus, there are fewer dissociations of PA from mTORC2. The ability of rapamycin-FKBP12 to suppress mTORC1 and mTORC2 would be dependent on how frequently mTOR became available to bind rapamycin-FKBP12. Far more mTOR dissociated from mTORC1 than from mTORC2 would be generated, and therefore less rapamycin would be required to compete with PA for binding to the mTOR derived from mTORC1. In contrast, the rare dissociations of mTORC2 would require much more rapamycin-FKBP12 to compete with PA to capture the rare mTOR proteins derived from mTORC2. Reducing PA levels would shift the equilibrium in favor of dissociation of the mTOR complexes and therefore reduce the concentration of rapamycin-FKBP12 needed to bind to and suppress mTOR. PC, phosphatidylcholine.
We used two cell lines in this study, 786-O renal cancer cells and MDA-MB-231 breast cancer cells—both of which have high levels of PLD activity (32, 39). The PLD activity of MDA-MB-231 cells is highly elevated only in the absence of serum, whereas the elevated PLD activity in 786-O cells occurs both in the presence and in the absence of serum. While the significance of this difference is not clear, interestingly, Akt phosphorylation at Ser473 is high in 786-O cells and low in MDA-MB-231 cells. The low level of Akt phosphorylation in MDA-MB-231 cells in the absence of serum, where PLD activity is high (39), clearly reveals that PLD activity and PA are not sufficient by themselves to activate mTORC2 and cause the phosphorylation of Akt at Ser473. In contrast, the introduction of an exogenous PLD gene did stimulate the phosphorylation of S6 kinase (1), suggesting that mTORC1 can be activated by elevated levels of PA. Exogenously provided PA has been reported to stimulate S6 kinase, but only in the presence of amino acids (4) or suppressed TSC2 (31). Thus, PA stimulates mTORC1 in conjunction with other input. It will be of interest to determine what signals, in addition to those that activate PLD, are necessary for the activation of mTORC2. The data provided here showing that insulin can stimulate Akt phosphorylation in a PLD-dependent manner in MDA-MB-231 cells may provide a lead as to the additional signals needed to activate mTORC2. While insulin increases PLD activity (8), the already elevated PLD activity in MDA-MB-231 cells makes it likely that another aspect of insulin signaling is required for the activation of mTORC2. Recently, Rosen and colleagues demonstrated that suppression of mTORC1 led to an increase in Akt phosphorylation at Ser473 that was dependent on IGF-1 (20). A similar observation made by Wan et al. (36) demonstrated that suppression of S6 kinase increased the phosphorylation of Akt at Ser473. Thus, components of IGF-1 and insulin signaling are likely to be an important component, in addition to PA, for mTORC2 activation.
There are two major classes of PLD isoforms—PLD1 and PLD2. We have found that dominant negative mutant forms of both PLD1 and PLD2 suppress S6 kinase and Akt phosphorylation and that using both mutant forms together is more effective than using either one by itself. This implies that both PLD1 and PLD2 are involved. We have noticed the same phenomenon for receptor endocytosis (26) and HIFα expression (32). Thus, we believe that both PLD isoforms are involved. In this regard, there are two recent reports that support this hypothesis—at least for mTORC1. Sung Ho Ryu and colleagues reported that PLD2 associates functionally with Raptor (15), implicating PLD2 in the activation of mTORC1, and very recently Chen and colleagues reported that PLD1 is a direct target of Rheb (28), which functions upstream from mTORC1—implicating PLD1 in the activation of mTORC1. While this study used short hairpin RNA for PLD2 to rule out a role for PLD2 in Rheb-induced S6 kinase phosphorylation, it is possible that this procedure did not reduce PLD2 protein levels sufficiently to completely suppress its effect. We have found that PLD2 is especially resistant to downregulation when using siRNA approaches (32; our unpublished observations). Thus, at present, there is evidence supporting a role for both PLD1 and PLD2 in the activation of mTORC1. The data presented here indicate that both PLD1 and PLD2 are also involved in the activation of mTORC2, although the mechanism remains to be worked out.
Rapamycin and rapamycin derivatives have been widely employed in clinical trials, with mostly disappointing results (25). A recent clinical study focused on glioblastoma, where there are commonly defects in PTEN (3). This study indicated that there was cell cycle arrest in response to rapamycin and effects on S6 kinase phosphorylation—implicating mTORC1. As indicated here, mTORC1 is much more sensitive to rapamycin than is mTORC2. However, Akt phosphorylation at Ser473 is dependent on mTORC2, indicating that mTORC2 may more critical in cancer—in that Akt phosphorylates many key substrates critical for cancer cell survival (19). Thus, targeting mTOR effectively may require strategies that suppress mTORC2. Consistent with this hypothesis, we recently found that mTORC2 is required for the expression of HIF2α in kidney cancer cells (33).
Kaelin and colleagues have reported that HIF2α is required for tumor formation by kidney cancer cells (18). These two studies reinforce the concept that mTORC2 could be an important target of anticancer therapies. As indicated in this study, suppressing PLD activity makes rapamycin effective in suppressing mTORC2 in 786-O cells, which have high levels of PLD activity (32). It is therefore possible that combining strategies that suppress PLD activity with rapamycin could improve the efficacy of rapamycin. While there are no drugs currently being used to target PLD directly, targeting the intracellular signals that increase PLD activity remains a possibility. We just recently reported that a natural product from Magnolia grandiflora known as honokiol suppresses PLD activity (12) and might therefore be used in combination with rapamycin to suppress mTORC2. It will therefore be of interest to determine whether honokiol can improve the efficacy of rapamycin in cancer cells with elevated PLD activity. This study provides a rationale for targeting the signals that regulate PLD activity to increase the efficacy of rapamycin, which has produced mixed results in clinical trials.
Acknowledgments
We thank Michael Ohh (University of Toronto) and Paige Yellen for comments on the manuscript. We thank Michael Frohman (State University of New York, Stony Brook) for the PLD genes used in this study.
This work was supported by grants from the National Cancer Institute (CA46677) and a SCORE grant from the National Institutes of Health (GM60654). Research Centers in Minority Institutions (RCMI) award RR-03037 from the National Center for Research Resources of the National Institutes of Health, which supports infrastructure and instrumentation in the Biological Sciences Department at Hunter College, is also acknowledged. A.G. was supported by a Gene Center fellowship from the RCMI.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Chen, Y., V. Rodrik, and D. A. Foster. 2005. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene 24672-679. [DOI] [PubMed] [Google Scholar]
- 2.Chen, Y., Y. Zheng, and D. A. Foster. 2003. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene 223937-3942. [DOI] [PubMed] [Google Scholar]
- 3.Cloughesy, T. F., K. Yoshimoto, P. Nghiemphu, K. Brown, J. Dang, S. Zhu, T. Hsueh, Y. Chen, W. Wang, D. Youngkin, L. Liau, N. Martin, D. Becker, M. Bergsneider, A. Lai, R. Green, T. Oglesby, M. Koleto, J. Trent, S. Horvath, P. S. Mischel, I. K. Mellinghoff, and C. L. Sawyers. 2008. Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 5139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang, Y., M. Vilella-Bach, R. Bachmann, A. Flanigan, and J. Chen. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 2941942-1945. [DOI] [PubMed] [Google Scholar]
- 5.Foster, D. A. 2004. Targeting mTOR-mediated survival signals in anticancer therapeutic strategies. Exp. Rev. Anticancer Ther. 4691-701. [DOI] [PubMed] [Google Scholar]
- 6.Foster, D. A. 2006. Phospholipase D survival signals as a therapeutic target in cancer. Curr. Signal Transduction Ther. 1295-303. [Google Scholar]
- 7.Foster, D. A. 2007. Regulation of mTOR by phosphatidic acid? Cancer Res. 671-4. [DOI] [PubMed] [Google Scholar]
- 8.Foster, D. A., and L. Xu. 2003. Phospholipase D in cell proliferation and cancer. Mol. Cancer Res. 1789-800. [PubMed] [Google Scholar]
- 9.Frias, M. A., C. C. Thoreen, J. D. Jaffe, W. Schroder, T. Sculley, S. A. Carr, and D. M. Sabatini. 2006. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 161865-1870. [DOI] [PubMed] [Google Scholar]
- 10.Gadir, N., D. Jackson, E. Lee, and D. A. Foster. 2008. Defective TGF-β signaling sensitizes human cancer cells to rapamycin. Oncogene 271055-1062. [DOI] [PubMed] [Google Scholar]
- 11.Gadir, N., E. Lee, A. Garcia, A. Toschi, and D. A. Foster. 2007. Suppression of TGF-β signaling by phospholipase D. Cell Cycle 62840-2845. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, A., Y. Zheng, C. Zhao, A. Toschi, J. Fan, N. Schreibman, H. A. Brown, D. Bar-Sagi, D. A. Foster, and J. Arbiser. 2008. Honokiol suppresses survival signals mediated by Ras-dependent phospholipase D activity in human cancer cells. Clin. Cancer Res. 144267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guertin, D. A., and D. M. Sabatini. 2007. Defining the role of mTOR in cancer. Cancer Cell 129-22. [DOI] [PubMed] [Google Scholar]
- 14.Guertin, D. A., and D. M. Sabatini. 2005. An expanding role for mTOR in cancer. Trends Mol. Med. 11353-361. [DOI] [PubMed] [Google Scholar]
- 15.Ha, S. H., D. H. Kim, I. S. Kim, J. H. Kim, M. N. Lee, H. J. Lee, J. H. Kim, S. K. Jang, P. J. Suh, and S. H. Ryu. 2006. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 182283-2291. [DOI] [PubMed] [Google Scholar]
- 16.Hresko, R. C., and M. Mueckler. 2005. mTOR/RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 28040406-40416. [DOI] [PubMed] [Google Scholar]
- 17.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110163-175. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning, B. D., and L. C. Cantley. 2007. AKT/PKB signaling: navigating downstream. Cell 1291261-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Reilly, K. E., F. Rojo, Q. B. She, D. Solit, G. B. Mills, D. Smith, H. Lane, F. Hofmann, D. J. Hicklin, D. L. Ludwig, J. Baselga, and N. Rosen. 2006. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 661500-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabatini, D. M. 2006. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer 6729-734. [DOI] [PubMed] [Google Scholar]
- 22.Sancak, Y., C. C. Thoreen, T. R. Peterson, R. A. Lindquist, S. A. Kang, E. Spooner, S. A. Carr, and D. M. Sabatini. 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25903-915. [DOI] [PubMed] [Google Scholar]
- 23.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 3071098-1101. [DOI] [PubMed] [Google Scholar]
- 24.Sarbassov, D. D., S. M. Ali, S. Sengupta, J. H. Sheen, P. P. Hsu, A. F. Bagley, A. L. Markhard, and D. M. Sabatini. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22159-168. [DOI] [PubMed] [Google Scholar]
- 25.Sawyers, C. L. 2003. Will mTOR inhibitors make it as cancer drugs? Cancer Cell 4343-348. [DOI] [PubMed] [Google Scholar]
- 26.Shen, Y., L. Xu, and D. A. Foster. 2001. Phospholipase D requirement for receptor-mediated endocytosis. Mol. Cell. Biol. 21595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, M., Y. Zheng, A. Garcia, and D. A. Foster. 2007. Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 258268-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, Y., Y. Fang, M. S. Yoon, C. Zhang, M. Roccio, F. J. Zwartkruis, M. Armstrong, H. A. Brown, and J. Chen. 2008. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc. Natl. Acad. Sci. USA 1058286-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung, T. C., R. L. Roper, Y. Zhang, S. A. Rudge, R. Temel, S. M. Hammond, A. J. Morris, B. Moss, J. Engebrecht, and M. A. Frohman. 1997. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 164519-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung, T. C., Y. M. Altshuller, A. J. Morris, and M. A. Frohman. 1999. Molecular analysis of mammalian phospholipase D2. J. Biol. Chem. 274494-502. [DOI] [PubMed] [Google Scholar]
- 31.Tee, A. R., R. Anjum, and J. Blenis. 2003. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 27837288-37296. [DOI] [PubMed] [Google Scholar]
- 32.Toschi, A., J. Edelstein, P. Rockwell, M. Ohh, and D. A. Foster. 2008. HIF2α expression in VHL-deficient renal cancer cells is dependent on phospholipase D. Oncogene 272746-2753. [DOI] [PubMed] [Google Scholar]
- 33.Toschi, A., E. Lee, N. Gadir, M. Ohh, and D. A. Foster. 2008. Differential dependence of HIF1α and HIF2α on mTORC1 and mTORC2. J. Biol. Chem. 28334495-34499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vander Haar, E., S. I. Lee, S. Bandhakavi, T. J. Griffin, and D. H. Kim. 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9316-323. [DOI] [PubMed] [Google Scholar]
- 35.Veverka, V., T. Crabbe, I. Bird, G. Lennie, F. W. Muskett, R. J. Taylor, and M. D. Carr. 2008. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene 27585-595. [DOI] [PubMed] [Google Scholar]
- 36.Wan, X., B. Harkavy, N. Shen, P. Grohar, and L. J. Helman. 2007. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 261932-1940. [DOI] [PubMed] [Google Scholar]
- 37.Wang, L., T. E. Harris, R. A. Roth, and J. C. Lawrence, Jr. 2007. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 28220036-20044. [DOI] [PubMed] [Google Scholar]
- 38.Zeng, Z., D. D. Sarbassov, I. J. Samudio, K. W. Yee, M. F. Munsell, C. Ellen Jackson, F. J. Giles, D. M. Sabatini, M. Andreeff, and M. Konopleva. 2007. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood 1093509-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, Y., V. Rodrik, A. Toschi, M. Shi, L. Hui, Y. Shen, and D. A. Foster. 2006. Phospholipase D couples survival and migration signals in response to stress in human breast cancer cells. J. Biol. Chem. 28115862-15868. [DOI] [PubMed] [Google Scholar]