Abstract
Initiation of protein synthesis in eukaryotes requires recruitment of the ribosome to the mRNA and its translocation to the start codon. There are at least two distinct mechanisms by which this process can be achieved; the ribosome can be recruited either to the cap structure at the 5′ end of the message or to an internal ribosome entry segment (IRES), a complex RNA structural element located in the 5′ untranslated region (5′-UTR) of the mRNA. However, it is not well understood how cellular IRESs function to recruit the ribosome or how the 40S ribosomal subunits translocate from the initial recruitment site on the mRNA to the AUG initiation codon. We have investigated the canonical factors that are required by the IRESs found in the 5′-UTRs of c-, L-, and N-myc, using specific inhibitors and a tissue culture-based assay system, and have shown that they differ considerably in their requirements. The L-myc IRES requires the eIF4F complex and the association of PABP and eIF3 with eIF4G for activity. The minimum requirements of the N- and c-myc IRESs are the C-terminal domain of eIF4G to which eIF4A is bound and eIF3, although interestingly this protein does not appear to be recruited to the IRES RNA via eIF4G. Finally, our data show that all three IRESs require a ternary complex, although in contrast to c- and L-myc IRESs, the N-myc IRES has a lesser requirement for a ternary complex.
The proteins encoded by the Myc family of genes are basic helix-loop-helix leucine zipper proteins that function as transcription factors and interact with another basic helix-loop-helix leucine zipper protein known as Max (3). Myc-Max heterodimers bind to DNA sequences called E boxes (CANNTG) and thereby facilitate activation of gene expression (3). All members of this family of genes contain three exons and two introns, and the main initiation codon is located toward the 5′ end of exon 2 such that exon 1 forms the majority of the 5′ untranslated region. We and others have shown previously that members of the Myc gene family contain a complex RNA structural element within exon 1 termed an internal ribosome entry segment (IRES), which allows the mRNAs to be translated by cap-independent internal ribosome entry (17, 18, 31, 43). IRESs were first identified in picornaviruses (2), but it has been shown that many cellular mRNAs also use these elements to initiate translation (39). Indeed, microarray studies have suggested that up to 10% of all cellular mRNAs contain IRESs (6, 15, 35, 39). In general, cellular IRESs are used during situations when the cap-dependent scanning mechanism of translation initiation is compromised (39).
The majority of viral IRESs require both canonical initiation factors (eukaryotic initiation factors [eIFs]) (eIF4F is comprised of eIF4G [the scaffold protein], eIF4E [the cap binding protein], and eIF4A [a DEAD box helicase]; eIF3, which also binds to eIF4G, is required for ribosome recruitment) and specific trans-acting factors for function, although each IRES has distinct requirements. For example, hepatitis A virus IRES function requires the association of eIF4G with eIF4E (1). In contrast, the cricket paralysis virus IRES can directly assemble 80S ribosomes in the absence of canonical initiation factors and initiator tRNA by directly recruiting the 40S ribosomal subunit (38).
The precise canonical and trans-acting factor requirements for the majority of cellular IRESs are unknown, although we have identified a set of proteins required specifically by the Myc family of IRESs, including YB-1, PSF, p54nrb, PTB-1, and GRSF (7). However, the canonical eIFs that are required by cellular IRESs have yet to be defined, although previous studies suggest that c-myc IRES function does not need full-length eIF4GI or poly(A) binding protein (PABP) (42, 45, 46). Here we show that the IRESs found in c-, L-, and N-myc mRNAs differ in their canonical eIF requirements. The L-myc IRES requires the eIF4F complex, and the interaction of both PABP and eIF3 with eIF4G is necessary for IRES activity. In contrast, the c- and N-myc IRESs can function with only the C-terminal domain of eIF4GI plus eIF4A and eIF3, although eIF3 does not interact with the IRESs via eIF4G. Interestingly, the data suggest that the N-myc IRES is less dependent than the c- or L-myc IRES on the level of ternary complex that is required to bring the initiator methionine into the P site of the ribosome.
MATERIALS AND METHODS
Plasmid constructs.
Plasmids used for this study were described previously (8).
The hairpin-containing vectors used for eIF3 knockdown were Mission short hairpin RNAs (Sigma-Aldrich). The control was a nontarget Mission control plasmid (Sigma-Aldrich).
Chemical structure probing.
The chemical structure probing protocol used was adapted from a previously described method (41), modified as described previously (23) and described in detail in a previous study (29).
Primer extension.
The procedure for primer extension was adapted from a previously described method (41) and was described in detail by Mitchell et al. (29).
Secondary structure prediction.
Secondary structure predictions were generated using the online implementation of the Mfold algorithm (48), incorporating version 3.0 of the Turner rules (26).
Cell culture.
HeLa cells were cultured under the conditions described by the ATCC (www.atcc.org). All cell transfections were performed in triplicate. HeLa cells were transfected using FuGene 6 (Roche) following the supplier's instructions.
Cells were transfected with the knockdown plasmid pSilencer31 (si31) or the control plasmid pSilencer 31 M, with or without the 4GIf expression plasmid, and with reporter plasmids for 24 h before harvest of the cells. Cells were cotransfected with pcDNA EGFP-NSP3 or pcDNA EGFP-NSPdelta4G (34) and the reporter plasmids for 24 h. The activities of firefly and Renilla luciferases in lysates prepared from transfected cells were measured using a dual-luciferase reporter assay system (Promega), and light emission was measured for 10 s in an Optocomp I luminometer. Cotransfection with the RNA interference (RNAi)-based plasmids to reduce eIF4GI levels reduced the levels of both firefly luciferase and Renilla luciferase. Therefore, it was not possible to normalize firefly luciferase expression to Renilla luciferase expression, since both of these proteins have similar half-lives of around 4 h. However, the transfection control β-galactosidase, expressed from pcDNA3.1His/BLacZ, has a much longer half-life in HeLa cells (around 40 h), and we found that its levels were relatively unaffected by the knockdown. The levels of firefly luciferase were therefore expressed relative to this transfection control. All assays were performed in triplicate on at least three independent occasions. Errors were calculated as standard deviations for the three calculated IRES activities and expressed as percentages of the average activity relative to the level in the control, which was set at 100%. In addition, firefly and Renilla luciferase activities were also expressed relative to the levels of RNA. These were determined by performing real-time PCR using a Stratagene MX3005P QPCR system. RNA was isolated from each transfection mix, and 5% of the RNA was reverse transcribed using C.therm polymerase (Roche) and analyzed using a QuantiTech SYBR green PCR kit following the manufacturer's instructions (Qiagen). Renilla luciferase cDNA was amplified using the primers 5′-GGAATTATAATGCTTATCTACGTGC-3′ and 5′-CTTGCGAAAAATGAAGACCTTTTAC-3′. Firefly luciferase cDNA was amplified using the primers 5′-CTCACTGAGACTACATCAGC-3′ and 5′-TCCAGATCCACAACCTTCGC-3′. Actin cDNA was amplified using the primers 5′-GGCATGGGTCAGAAGGATT-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′.
Real-time data were analyzed by relative quantification using the 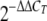 method as described by Livak and Schmittgen (24a).
method as described by Livak and Schmittgen (24a).
To examine the requirement for the ternary complex, cells were treated with 75 μM Salubrinal for 24 h. Alternatively, cells were cotransfected with plasmid construct pcDNA3.1eIF2αS51D or -A.
Hippuristanol was prepared as described by Shen et al. (36) and dissolved in dimethyl sulfoxide (DMSO) as a 10 mM stock. Cells were treated with hippuristanol for 6 h posttransfection and then harvested.
SDS-polyacrylamide gel electrophoresis and Western blotting.
Cell extracts were prepared as described previously (30) and subjected to electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and the proteins were then transferred to a polyvinylidene difluoride membrane (Millipore). Proteins were detected with the relevant antisera, using chemiluminescent reagents (47). Total cell extracts were prepared as described previously (47), and proteins were detected using antisera to eIF2α, phospho-eIF2α, actin, c-Myc, and eIF4GI.
RESULTS
The Myc family of IRESs requires eIF4G and PABP for activity in vivo.
Previous studies have shown that c-myc IRES-mediated translation in vitro and in vivo is dependent upon the C-terminal domain of eIF4G for activity (42, 45, 46) and that in vitro IRES activity is independent of PABP binding (45, 46). A plasmid-based RNAi approach (si31) was used to reduce eIF4GI levels in conjunction with a plasmid that generates an mRNA encoding eIF4GI that is resistant to the effects of RNAi as a consequence of a number of silent point mutations (4GIf) or with variants of this plasmid in which the binding sites for PABP, eIF4E, eIF3, and eIF4A were mutated (9, 12) (Fig. 1). This system was tested using two viral IRESs for which the canonical trans-acting factor requirements were described previously (20, 22, 25). Cells were transfected with the plasmids shown and then transfected with pREMCVF or pRHCVF, and luciferase activity was determined. As expected, the encephalomyocarditis virus (EMCV) IRES was affected by a reduction in eIF4GI and by replacement with the plasmid which had mutations in the eIF4A binding site yet was unaffected and indeed slightly stimulated by replacement plasmids with a mutated eIF4E or PABP binding site (Fig. 1A, panel i). In contrast, the hepatitis C virus (HCV) IRES was unaffected by the eIF4G knockdown (Fig. 1A, panel ii). These plasmids were then used to test the roles of eIF4G and PABP in N- and L-myc IRES-mediated translation in vivo (Fig. 2). Transfection of HeLa cells with si31 reduced eIF4GI levels to approximately 15% of the control level, while cotransfection with the RNAi-resistant vector 4GIf or 4GIf-PABP increased the level of eIF4GI to 120% of the control value (Fig. 2A, panel i). HeLa cells were cotransfected with si31 and the dicistronic reporter vector pRLsF, pRMF, or pRNF, cells were harvested and lysed, and luciferase activity was determined (Fig. 2A, panel ii, and B, panels i to iii; see Fig. S1 in the supplemental material). There was a decrease in IRES-dependent initiation in cells with reduced levels of eIF4GI that was comparable to the reduction in overall cellular translation rates that was previously observed in HeLa cells (9, 12). The IRES-dependent translation was restored by cotransfection with wild-type eIF4G, suggesting that this family of IRESs are highly dependent on eIF4G for activity in vivo. One of the strategies used by certain viruses to inhibit the majority of host-mediated protein synthesis is to cleave the scaffold protein eIF4GI such that the eIF4E binding site is removed (14). Previous data suggested that the c-myc IRES is active following viral infection (16) and functions with only the C-terminal region of eIF4G (42, 45). To test whether this was also true for the L- and N-myc IRESs, the level of eIF4GI was reduced by RNAi and the cells were then transfected with an RNAi-resistant plasmid encoding the C-terminal domain of eIF4GI (CT), which arises following cleavage with the poliovirus 2A protease (32), in conjunction with either pRLsF, pRMF, or pRNF (Fig. 2A and B, panels i to iii). The data show that the IRES activity of c- and N-myc but not of L-myc was restored by CT (Fig. 2B, panels i to iii).
FIG. 1.
(i) Schematic representation of constructs generated as described previously (9) to replace wild-type eIF4G with either a form that was resistant to knockdown by siRNA (eIf4GIf) or versions of eIF4G where the PABP (eIF4GIf-PABP), eIF4E (eIF4GIf-4E), eIF4A (eIF4GIf-4A), or eIF3 (eIF4GIf-eIF3) binding site was mutated (12). Ct, the fragment of eIF4GI generated in cells infected with poliovirus. (ii) HeLa cells were cotransfected with si31 (eIF4G knockdown plasmid), either the resistant plasmid 4GIf or a resistant plasmid which harbored mutations in the PABP, eIF4A, eIF3, or eIF4E binding site, and a dicistronic plasmid harboring either the EMCV or HCV IRES. Firefly and Renilla luciferase assays were performed, and the data are expressed relative to those for RNAs generated from the relevant vectors (see Materials and Methods). All assays were performed in triplicate on at least three independent occasions. Relative luciferase activity is expressed relative to those in control cells, which are set at 100%. The numbers of light units generated by these assays were between 1 × 105 and 2 × 106.
FIG. 2.
(A) (i) HeLa cells were transfected with si31, which reduces eIF4G levels, and optionally with an eIF4G-containing plasmid which is resistant to si31 (4GIf) or with 4GIf containing a mutated PABP binding site (Fig. 1). Western blot analysis showed that eIF4G levels decreased upon si31 transfection and increased with si31 and 4GIf transfection. Cells were also transfected with si31 and a plasmid which encodes the C-terminal domain of eIF4G (CT), and Western blot analysis was used to demonstrate expression of this protein. (ii) Schematic analysis of the bicistronic construct used (pRF). The L-, N-, and c-myc IRESs were inserted between the Renilla and firefly luciferase cistrons to create pRLsF, pRNF, and pRMF, respectively. (B) HeLa cells were cotransfected with si31 (eIF4G knockdown plasmid), either the resistant plasmid (4GIf), the C-terminal fragment of eIF4G (CT), or the resistant plasmid harboring a mutation in the PABP binding site (Fig. 1), and either the L-myc (i), N-myc (ii), or c-myc (iii) IRES. Firefly and Renilla luciferase assays were performed, and the data are expressed relative to those for RNAs generated from the relevant vectors (see Materials and Methods). All assays were performed in triplicate on at least three independent occasions. Firefly luciferase activity (gray bars) and Renilla luciferase activity (black bars) were expressed relative to those in the control cells, which were set to 100%. The numbers of light units generated by these assays were between 1 × 105 and 2 × 106. These data show that all three IRESs are dependent on eIF4GI for function, but the N- and c-myc IRESs are able to operate with the C-terminal domain of eIF4GI. All three IRESs appear to require PABP for maximal activity, although there is partial restoration of N- and c-myc IRES activity with plasmid 4GIf containing a mutated PABP binding site. (iv to vi) Cells were additionally transfected with monocistronic hairpin vectors (8) which contained the three IRESs. These vectors were transfected into HeLa cells in conjunction with si31 (eIF4G knockdown plasmid) and either the resistant plasmid (4GIf) or the resistant plasmid harboring a mutation in the PABP binding site. Luciferase assays were performed, and the data show that all three IRESs require PABP for activity. (vii) HeLa cells were cotransfected with the three IRES-containing plasmids and either a plasmid which expresses the NSP3 protein or a mutant version where the eIF4G binding site is deleted (34). Luciferase activity was determined and normalized to the level of the RNA control. The data show that all three IRESs are inhibited by the presence of the wild-type NSP3 protein but not by the mutated version. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity. (C) (i) To determine the effect of transfection with the si31 eIF4G knockdown and mutated recovery plasmids described for panel A on the expression of endogenous c-Myc, transfected cell samples were lysed and subjected to SDS-polyacrylamide gel electrophoresis, and Western blot analysis was performed. The data show that c-Myc levels were reduced to approximately 10% of the control values in the presence of si31 and were restored to slightly above wild-type levels following cotransfection with 4GIf. In comparison, c-Myc levels were slightly reduced following transfection with the plasmid containing the mutated version of PABP. The c-Myc protein levels were restored, but not to fully wild-type levels, in the presence of the C-terminal fragment of eIF4G (CT) (ii) (32).
The c-myc IRES does not require PABP for function in vitro (45, 46). However, our data show that full IRES activity was not restored to the L-, N-, or c-myc IRES when the eIF4GI mutant plasmid that lacks the PABP binding site was introduced following eIF4GI depletion (Fig. 2B, panels i to iii). These data were also confirmed using monocistronic vectors (Fig. 2B, panels iv to vi). To investigate these data further, HeLa cells were cotransfected with a plasmid that encodes the rotavirus nonstructural protein 3 (NSP3) (a rotaviral functional homolog of PABP; as a control, a mutated version of this plasmid, NSP3del, was used), which has previously been shown to inhibit cap-dependent translation by competing with PABP for eIF4G binding (34), and the IRES-containing vectors. The data show that in the presence of functional NSP3 there is a large reduction in the activity of all three IRESs, providing further evidence that PABP is required for function (Fig. 2B, panel vii).
The effect of reduction of the eIF4GI level on the endogenous c-Myc protein was investigated (Fig. 2C). The data show that c-Myc expression is reduced in cells which lack eIF4GI and that the levels are not fully restored by cotransfection with the eIF4G plasmid in which the PABP binding site has been mutated (Fig. 2C, panel i), thereby confirming the data obtained using the reporter vectors. Cotransfection with the CT fragment of eIF4GI increased the c-Myc levels, but these were again lower than those in the control cells (Fig. 2C, panel ii), probably reflecting the fact that c-Myc protein can be synthesized by both cap-dependent scanning, which requires full-length eIF4G, and internal ribosome entry (44).
The c- and N-myc IRESs are dependent on eIF4A but not eIF4E for activity.
Studies of IRESs derived from picornaviruses show that the mechanisms that are used for internal ribosome entry vary between different groups of viruses. For example, certain viral IRESs have been defined as “land and start” IRESs, as the ribosome appears to land at or near the start codon, while others have been proposed to use a “land and scan” mechanism (2). In this case, the data strongly suggest that ribosomes land at an upstream AUG codon and then scan to the next AUG downstream, where translation is initiated (13). This difference in mechanism is likely to be reflected in the dependence of IRESs on certain canonical initiation factors, and one would predict that “land and scan” IRESs have a greater requirement for eIF4A. Little is known about how the ribosome interacts with cellular IRESs, although the data suggest that these can also be classified partially as “land and scan” and “land and start” IRESs, with the c-myc, L-myc, and Apaf-1 IRESs being in the former category (17, 23, 27) and the c-myb and Mnt IRESs (28) complying with the latter description. It was important to define the structure of the N-myc IRES (see Fig. S2 in the supplemental material) and to determine whether this IRES also falls mechanistically within the “land and scan” group of IRESs (Fig. 3). This was carried out by introducing AUG initiation codons at various positions throughout the sequence, using site-directed mutagenesis (Fig. 3A). A naturally occurring AUG codon is present in the N-myc IRES (position −24), but this does not appear to be recognized, since mutation of this sequence had no net effect (Fig. 3B). If the ribosome entry window lies upstream of the inserted AUG, polypeptide synthesis will initiate at the upstream open reading frame, resulting in a concomitant decrease in firefly luciferase expression, but if the point of ribosome entry lies between the upstream AUG and the reporter gene initiation codon, there will be no change in firefly luciferase synthesis. HeLa cells were transfected with these constructs, and luciferase activity was determined (Fig. 3B). The data show that the N-myc IRES uses a “land and scan” mechanism to initiate translation, although it was difficult to define the ribosome entry window precisely. However, it can be stated with a degree of certainty that it is between positions −232 and −173 within a pyrimidine-rich region (Fig. 3C). Control UUG mutations introduced into the sequence at the −16 and −56 positions did not reduce the synthesis of firefly luciferase, suggesting that the inhibition observed with the AUG mutations was not due to a change in RNA structure.
FIG. 3.
(A) To determine whether the N-myc IRES was similar to the L- and c-myc IRESs and used a “land and scan” mechanism, site-directed mutagenesis was used to insert out-of-frame AUG codons into the IRESs within the vector pRNF. The position of each mutated sequence is numbered from the A of ATGG or the U of UUGG. (B) The resulting constructs were transfected into HeLa cells in conjunction with pcDNA3.1/HisB/lacZ. The data are presented as percentages of firefly luciferase activity compared to the activity from the wild-type IRES. (C) The location of the ribosome entry site is shown on a secondary structure model of the N-myc IRES (see Fig. S2 in the supplemental material). It was difficult to define the ribosome entry window precisely, although it can be stated with a degree of certainty that it lies between positions −232 and −173 within a pyrimidine-rich region.
Tests were performed to examine whether ribosomes scanning on the Myc family of IRESs require eIF4A. Cells were transfected with the plasmid constructs pRLsF, pRMF, and pRNF and were treated with hippuristanol, a specific and potent inhibitor of eIF4A (4). In cells treated with 10 μM hippuristanol for 6 h, overall protein synthesis was inhibited approximately 60% (Fig. 4A, panel i). The c-, L-, and N-myc IRES-driven firefly luciferase activity was also inhibited in all cases (Fig. 4A, panel ii), and these IRESs are similar in this regard to certain viral IRESs (4). In the case of the c-myc IRES, these data are in agreement with previous studies (45, 46). The effect of eIF4A inhibition on endogenous c-Myc levels was also tested (Fig. 4A, panel iii).
FIG. 4.
(A) (i) To test the effect of hippuristanol on cellular translation, cells were incubated with 1 mM hippuristanol for 6 h and then pulse labeled with [35S]methionine. The data show a 60% reduction in total protein synthesis after this period. (ii) The IRES-containing constructs were transfected into HeLa cells, and after 24 h, either hippuristanol or DMSO was added to the medium. After 6 h, luciferase activity was determined, and the values are expressed relative to those for the DMSO-treated control. In all cases, IRES activity was reduced in the presence of hippuristanol. (iii) To determine the effect of hippuristanol on the expression of endogenous c-Myc, Western blot analysis was performed. The level of c-Myc protein was reduced to approximately 20% following exposure of cells to hippuristanol. (B) HeLa cells were transfected with si31 (−eIF4G) and with either the eIF4GI-containing plasmid which is resistant to si31 (4GIf) or 4GIf containing a mutated eIF4A binding site. Western blot analysis shows that eIF4GI levels decreased upon si31 transfection and increased with 4GIf transfection. Cell lysates were then immunoblotted to determine the effect of mutant eIF4G(−4A) expression on the levels of endogenous c-Myc protein. The data show that these levels were reduced. (C) HeLa cells were cotransfected with si31 (−eIF4G) and either the resistant plasmid (4GIf) or the resistant plasmid harboring a mutation in the eIF4A or eIF4E binding site (see Fig. S1 in the supplemental material) and then transfected with either the L-myc (i), N-myc (ii), or c-myc (iii) IRES. Luciferase assays were performed, and the data are expressed relative to those for RNAs generated from these vectors. Luciferase activity in the control cells was set to 100%, and all numbers are expressed relative to the control cells. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity. All assays were performed in triplicate on at least three independent occasions. The data show that all three IRESs require eIF4A bound to eIF4G for activity and that the L-myc IRES, but not the N- and c-myc IRESs, requires eIF4E. (D) (i) HeLa cells were transfected with a plasmid which contained a short hairpin sequence (si-eIF4E) against eIF4E or a control sequence. Western blot analysis shows that it was possible to reduce the levels of eIF4E to approximately 15% of the value in the control cells. (ii) HeLa cells were transfected with the si-eIF4E plasmid and then with the IRES-containing vectors shown. Luciferase activity was determined, and values are expressed relative to those for RNAs derived from the vectors. The value in the control cells was set at 100%, and changes in activity are expressed relative to the controls. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity.
It has been shown that the EMCV IRES requires the C-terminal domain of eIF4GI to which eIF4A is bound for activity (25, 33) and that eIF4GI recruits eIF4A to a defined location on the IRES (20). To determine whether the eIF4A-eIF4G interaction was also required for Myc family IRES activity, cells were depleted of eIF4GI as described above and transfected with a recovery plasmid in which the eIF4A binding site on eIF4G was mutated and with the dicistronic IRES-containing constructs (4GIf-4A) (Fig. 1 and 4B). These data show that the mutant protein was unable to restore activity to any of the IRESs, demonstrating that the eIF4GI-eIF4A interaction is required for IRES function (Fig. 4C, panels i to iii). In addition, the level of c-Myc protein was reduced following transfection of cells with these constructs (Fig. 4B). The majority of viral IRESs do not require eIF4E for function, and it has generally been assumed that this would also be true for cellular IRESs. Therefore, the levels of endogenous eIF4GI were reduced as described before and then replaced with a mutant version of eIF4GI that lacks the eIF4E binding site (4GIf-4E). The mutant version of the plasmid was able to fully restore IRES function for the c- and N-myc IRESs (Fig. 4C, panels ii and iii) but, interestingly, not for the L-myc IRES (Fig. 4C, panel i). To confirm these data, the levels of eIF4E were reduced by small interfering RNA (siRNA) (Fig. 4D, panel i), and cells were then transfected with the IRES-containing plasmids (Fig. 4D, panel ii). The data again showed that only the L-myc IRES was affected by the reduction in the level of eIF4E (Fig. 4D, panel ii). Taken together, these data suggest that the L-myc IRES requires the eIF4F complex for activity, whereas the N- and c-myc IRESs do not, although the interaction of eIF4G with eIF4A is necessary for their activity.
The c- and N-myc IRESs are able to function in the absence of an eIF3-eIF4F interaction.
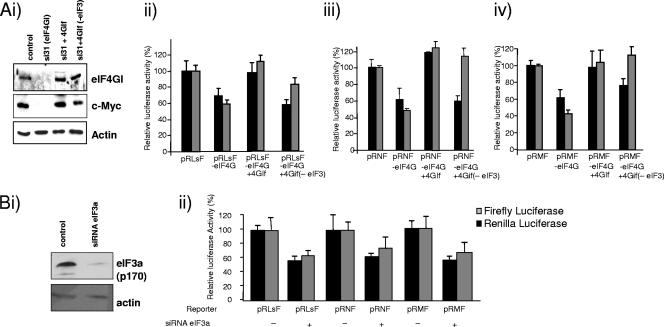
The majority of viral IRESs require eIF3 for activity, and in some cases, such as the HCV and classical swine fever virus IRESs, this protein interacts directly with the RNA (37). Therefore, the eIF4GI replacement system was used to test the requirement of the Myc family of IRESs for eIF3. HeLa cells were transfected with the eIF4GI knockdown plasmid and then cotransfected with 4GIf-eIF3, in which the eIF3 binding site was mutated (Fig. 5A, panel i), and with the reporter constructs that harbor the three IRESs (Fig. 5A, panels ii to iv). Rather surprisingly, the data show that N- and c-myc IRES function was restored by plasmids that express eIF4G but lack the eIF3 binding site (Fig. 5A, panels iii and iv). In agreement with these data, the endogenous levels of c-Myc were restored to around 50% with this plasmid, again explained by the fact that c-Myc can be translated by two distinct mechanisms (44). One explanation for these data could be that these cellular IRESs require eIF3 for function but that this protein is not recruited via the eIF4F complex. To investigate this hypothesis, cells were transfected with a short hairpin RNA-expressing plasmid that reduces the expression of the large subunit of eIF3 (termed eIF3a or p170) (Fig. 5B, panel i) and then transfected with the IRES-containing constructs to assess the effect on internal ribosome entry (Fig. 5B, panel ii). In all cases, IRES activity was reduced, showing that eIF3 is required for IRES function (Fig. 5B, panel ii).
FIG. 5.
(A) HeLa cells were cotransfected with si31 (−eIF4G) and either the resistant plasmid (4GIf) or the resistant plasmid harboring a mutation in the eIF3 binding site. (i) Cell lysates were then immunoblotted to determine the effects on the expression of endogenous c-Myc protein. As before, a reduction in eIF4G levels resulted in a very large reduction in c-Myc expression, which was partially relieved by transfection with the recovery plasmid which contained the mutated version of eIF3. (ii to iv) Cells were transfected with si31, 4GIf harboring a mutation in the eIF3 binding site, and the IRES-containing vectors. Luciferase activity was determined and expressed relative to the level of RNA generated from the vectors. The value obtained in the control cells was set to 100%, and all values are expressed relative to the appropriate control, e.g., pRLsF-eIF4G activity is expressed relative to that of pRLsF. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity. The L-myc IRES activity was not restored with the plasmid which harbors the eIF3 mutant, in contrast to the N- and c-myc IRESs. (B) (i) HeLa cells were transfected with a plasmid which contained a short hairpin sequence (sheIF3) against the p170 subunit of eIF3 (eIF3a) or a control sequence. Western blot analysis shows that it was possible to reduce the levels of eIF3a to approximately 20% of the value in the control cells. (ii) HeLa cells were cotransfected with sheIF3 plasmids and the bicistronic IRES-containing vectors. Luciferase activity was determined, and values are expressed relative to those for RNAs generated from the reporter vectors. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity. The values obtained in the control cells were set to 100%, and all values are expressed relative to those for the appropriate controls. The data show that IRES activity was reduced in all cases.
The Myc family of IRESs show differences in their dependence on the ternary complex.
The ternary complex (comprised of eIF2, GTP, and Met-tRNAi) delivers Met-tRNAi to the 40S ribosome and is essential for cap-dependent translation. However, it is not required by the cricket paralysis virus IRES (38), and under some conditions the HCV IRES is also able to function in the absence of the ternary complex (22). We therefore tested the role of this complex in Myc family IRES activity. Cells were transfected with the plasmids which harbored the three IRESs and then treated with Salubrinal, which is a selective inhibitor of complexes that dephosphorylate eIF2α (Fig. 6A, panel i) (5), or cotransfected with a dominant-negative mutant of eIF2α to provide two different mechanisms of eIF2α inhibition (Fig. 6B, panel i, S51D mutant) (40). A reduction in the activity of all three IRESs was observed in the presence of both Salubrinal and the dominant-negative version of eIF2α (S51D), although the N-myc IRES was less affected than the L- and c-myc IRESs (Fig. 6A, panel ii, and B, panel ii).
FIG. 6.
(A) HeLa cells were transfected with the IRES-containing plasmids and then, after 24 h, exposed to 75 μM Salubrinal. (i) Western blot analysis was performed to confirm that exposure of cells to this compound causes a net increase in phosphorylation of the alpha subunit of eIF2. (ii) Cells were lysed, and luciferase activity was determined and shown as the percentage of luciferase produced from treated cells compared to the amount produced from the control cells, which was set at 100%. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity. Assays were performed in triplicate on at least three independent occasions. The data show that the N-myc IRES is less sensitive to the effects of Salubrinal than the L- and c-myc IRESs. (B) The IRES-containing plasmids pRLsF, pRNF, and pRMF were cotransfected into HeLa cells with pcDNA3.1eIF2αS51D or -A (which harbors a constitutively active or dominant-negative version of eIF2α) or pcDNA3.1. (i) Western blot analysis was performed to show that these proteins were expressed. (ii) The data are presented as percentages of luciferase produced from control cells (pcDNA3.1) relative to the amount produced from the IRESs in cells transfected with the mutant plasmids (S51D and S51A). Assays were performed in triplicate on at least three independent occasions. Black bars denote Renilla luciferase activity, and gray bars denote firefly luciferase activity.
DISCUSSION
The scanning ribosome model of translation initiation proposes that the primed 40S subunit with associated initiation factors and Met-tRNAi first binds to the mRNA close to the 5′ cap structure and then migrates in a 5′-to-3′ direction (21). In contrast, for internal ribosome entry, the data suggest that a complex RNA structural element is used to recruit the ribosome in the presence of certain eIFs and ITAFs. We recently identified a group of ITAFs that are required by the Myc family of IRESs, including PSF, p54nrb, YB1, and GRSF1 (7), and it was therefore timely to carry out a systematic analysis of this group of IRESs to identify canonical eIFs that are also required for their activity. Interestingly, our data show that there are some similarities between the members of this group of IRESs and viral IRESs in their requirement for canonical initiation factors. There are two major isoforms of eIF4G in mammalian cells, namely, eIF4GI and the homologue eIF4GII, which shares 46% identity, and a functionally related protein termed death-associated protein 5 (DAP5) which is able to replace eIF4G under certain cellular conditions (e.g., endoplasmic reticulum stress) (24). However, the data shown suggest that all three IRESs are dependent upon eIF4GI for activity and that a reduction in eIF4GI levels correlates with a decrease in the amount of firefly luciferase that is produced by the IRES, particularly compared to the smaller decrease in Renilla luciferase activity (Fig. 2 to 4). However, the IRESs differ since only the L-myc IRES requires full-length eIF4GI and eIF4E (Fig. 2 and 4) for function and is similar in this regard to the IRES found in hepatitis A virus (1). In contrast, the c- and N-myc IRESs are similar to the EMCV IRES, as they require the C-terminal domain of eIFGI to which eIF4A is bound (Fig. 2 and 4) (25, 33). In the case of the EMCV IRES, both eIF4GI and eIF4A are required to bring about conformational changes in the RNA near the ribosome binding site (20), and it is possible that these proteins function in a similar manner on the c- and N-myc IRESs.
The results suggest that all three IRESs require eIF3 for activity (Fig. 5), in agreement with recent data which identified subunits of eIF3 in purified 48S complexes assembled on c-myc IRES RNA (46). However, since no effect was observed on the activities of the c- and N-myc IRESs with the constructs that contained the mutated eIF3 binding site on eIF4G, yet their activity was decreased following transfection of cells with siRNA specific to a subunit of eIF3, our data could suggest that the c- and N-myc IRESs are similar to certain viral IRESs, such as that of HCV, in that eIF3 is recruited directly to the RNA (10, 19). However, further work is required to investigate this interaction.
In vitro data have shown that the c-myc IRES requires a poly(A) tail for activity, but not PABP (45, 46), which would appear to differ from the in vivo data shown herein. However, these different findings are not mutually exclusive considered in conjunction with data which show that the c-and N-myc IRESs are fully active with the C-terminal domain of eIF4GI which lacks the PABP binding site yet are inhibited by NSP3 (Fig. 2B, panel vii). Moreover, it has been suggested that major conformational changes occur when eIF4G is part of the eIF4F complex, since this protein is a relatively poor substrate for either the L protease from foot-and-mouth disease virus (32) or the rhinovirus 2A protease (11) yet it is readily cleaved when present as part of the eIF4F complex. Therefore, we suggest a model where eIF4GI is in a conformationally inactive closed state when not bound to PABP (Fig. 7A, panel i) and where binding of NSP3 to the N-terminal domain of eIF4G locks the protein in this inactive state (Fig. 7A, panel ii). The L-myc IRES, which requires the eIF4F complex for function, is active only with full-length eIF4G to which PABP is bound (Fig. 7B, panel i), but c- and N-myc IRESs are able to function with a complex where PABP has altered the conformation of eIF4G to an open state (Fig. 7B, panel ii) or with the C-terminal half of eIF4G, which by its nature is already in the open state (Fig. 7B, panel iii).
FIG. 7.
Model for the interaction of canonical initiation factors with the three Myc family IRESs. (A) In the absence of PABP (i) or in the presence of NSP3 (ii), eIF4G is unable to form the correct conformation to allow interaction with the IRESs. (B) (i) The L-myc IRES requires the scaffold protein eIF4G, the interaction of eIF4E, eIF4A, PABP, and eIF3 with this scaffold, and trans-acting factors (X) for function. The c- and N-myc IRESs are able to function with complexes that are comprised of trans-acting factors (X) and eIF4G (ii) or the C-terminal fragment of eIF4G (CT) (iii) in the presence of PABP and eIF4A. eIF3 may be recruited directly to the IRES RNA.
The data show that all three IRESs, like the majority of viral IRESs, require the ternary complex for activity, although the N-myc IRES appeared to be less affected than the L- and c-myc IRESs by a reduction in the amount of ternary complex (Fig. 6). This could be related to the apparent difference in the mechanism of ribosome recruitment to the N-myc IRES (Fig. 3; see Fig. S2 in the supplemental material).
Overall, we show that different cellular IRESs, even those from closely related genes, have different requirements for components of the canonical translation initiation machinery. It will be interesting to extend these studies further to other cellular IRESs.
Supplementary Material
Acknowledgments
This work was funded by grants from the BBSRC (L.C.C.) and CRUK (L.A.W.). Catherine Jopling and Martin Bushell are David Phillips Fellows, and Anne Willis holds a BBSRC Professorial Fellowship.
Footnotes
Published ahead of print on 5 January 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus internal ribosome entry site requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 757854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Blackwood, E. M., B. Luscher, and R. N. Eisenman. 1992. Myc and Max associate in vivo. Genes Dev. 671-80. [DOI] [PubMed] [Google Scholar]
- 4.Bordeleau, M. E., A. Mori, M. Oberer, L. Lindqvist, L. S. Chard, T. Higa, G. J. Belsham, G. Wagner, J. Tanaka, and J. Pelletier. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2213-220. [DOI] [PubMed] [Google Scholar]
- 5.Boyce, M., K. F. Bryant, C. Jousee, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron, and J. Yuan. 2005. A selective inhibitor of eIF2a dephosphorylation protects cells from ER stress. Science 307935-939. [DOI] [PubMed] [Google Scholar]
- 6.Bushell, M., M. Stoneley, Y. W. Kong, T. L. Hamilton, K. A. Spriggs, H. C. Dobbyn, X. L. Qin, P. Sarnow, and A. E. Willis. 2006. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell 23401-412. [DOI] [PubMed] [Google Scholar]
- 7.Cobbold, L. C., K. A. Spriggs, S. J. Haines, H. C. Dobbyn, C. Hayes, C. H. de Moor, K. S. Lilley, M. Bushell, and A. E. Willis. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell. Biol. 2840-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coldwell, M. J., M. L. deSchoolmeester, C. A. Fraser, B. M. Pickering, G. Packham, and A. E. Willis. 2001. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 204095-4100. [DOI] [PubMed] [Google Scholar]
- 9.Coldwell, M. J., and S. J. Morley. 2006. Specific isoforms of translation initiation factor 4GI show differences in translational activity. Mol. Cell. Biol. 268448-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier, A. J., J. Gallego, R. Klinck, P. T. Cole, S. J. Harris, G. P. Harrison, F. Aboul-ela, G. Varani, and S. Walker. 2002. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 9375-380. [DOI] [PubMed] [Google Scholar]
- 11.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 708444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinton, T. M., M. J. Coldwell, G. A. Carpenter, S. J. Morley, and V. M. Pain. 2007. Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 2821695-1708. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, R. J. 2000. A comparative view of initiation site selection mechanisms, p. 127-183. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1985-1000. [PMC free article] [PubMed] [Google Scholar]
- 15.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap-binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 9613118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, Bip, and eIF4G conferred by internal ribosome entry sites. RNA 41500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jopling, C. L., K. A. Spriggs, S. A. Mitchell, M. Stoneley, and A. E. Willis. 2004. L-Myc protein synthesis is initiated by internal ribosome entry. RNA 10287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jopling, C. L., and A. E. Willis. 2001. N-myc translation is initiated via an internal ribosome entry segment that displays enhanced activity in neuronal cells. Oncogene 202664-2670. [DOI] [PubMed] [Google Scholar]
- 19.Kieft, J. S., K. H. Zhou, A. Grech, R. Jubin, and J. A. Doudna. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9370-374. [DOI] [PubMed] [Google Scholar]
- 20.Kolupaeva, V. G., I. B. Lomakin, T. V. Pestova, and C. U. T. Hellen. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol. Cell. Biol. 23687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Genes Dev. 234187-208. [DOI] [PubMed] [Google Scholar]
- 22.Lancaster, A. M., E. Jan, and P. Sarnow. 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12894-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Quesne, J. P. C., M. Stoneley, G. A. Fraser, and A. E. Willis. 2001. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 310111-126. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, S. M., S. Cerquozzi, T. E. Garber, N. H. Ungureanu, M. Andrews, and M. Holcik. 2007. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res. 36168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Livak, K. J., and T. D. Schmittgen. 2002. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 25.Lomakin, I. B., C. U. T. Hellen, and T. V. Pestova. 2000. Physical association of eIF4G with eIF4A strongly enhances binding of eIF4G to EMCV and is required for internal initiation of translation. Mol. Cell. Biol. 206019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews, D., J. Sabina, M. Zuker, and D. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, S. A., E. C. Brown, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2001. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 213364-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell, S. A., K. A. Spriggs, M. Bushell, J. R. Evans, M. Stoneley, J. P. C. le Quesne, R. V. Spriggs, and A. E. Willis. 2005. Identification of a motif that mediates polypyrimidine tract binding protein dependent internal ribosome entry. Genes Dev. 191556-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell, S. A., K. A. Spriggs, M. J. Coldwell, R. J. Jackson, and A. E. Willis. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell 11757-771. [DOI] [PubMed] [Google Scholar]
- 30.Morley, S. J., L. McKendrick, and M. Bushell. 1998. Cleavage of translation initiation factor 4G during anti-Fas IgM induced apoptosis does not require signalling through the p38 mitogen-activated protein kinase. FEBS Lett. 43841-48. [DOI] [PubMed] [Google Scholar]
- 31.Nanbru, C., I. Lafon, S. Audiger, G. Gensac, S. Vagner, G. Huez, and A.-C. Prats. 1997. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 27232061-32066. [DOI] [PubMed] [Google Scholar]
- 32.Ohlmann, T., M. Rau, V. M. Pain, and S. J. Morley. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF-4E. EMBO J. 151371-1382. [PMC free article] [PubMed] [Google Scholar]
- 33.Pestova, T., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43s preinitiation complex. Mol. Cell. Biol. 166870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 175811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin, X. L., and P. Sarnow. 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 27913721-13728. [DOI] [PubMed] [Google Scholar]
- 36.Shen, Y.-C., C. V. S. Prakash, Y.-T. Chang, M.-C. Hung, S.-J. Chen, H.-J. Chen, and M.-C. Hsu. 2000. Bioactive steroids from the Formosan gorgonian Isis hippuris. Chin. Pharm. J. (Taipei) 52341-351. [Google Scholar]
- 37.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. T. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 724775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spahn, C., E. Jan, A. Mulder, R. Grassucci, P. Sarnow, and J. Frank. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118465-475. [DOI] [PubMed] [Google Scholar]
- 39.Spriggs, K. A., M. Stoneley, M. Bushell, and A. E. Willis. 2008. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 10027-38. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava, S. P., K. U. Kumar, and R. J. Kaufmann. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 2732416-2423. [DOI] [PubMed] [Google Scholar]
- 41.Stern, S., D. Moazed, and H. Noller. 1998. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164481-489. [DOI] [PubMed] [Google Scholar]
- 42.Stoneley, M., S. Chappell, C. Jopling, M. Dickens, M. MacFarlane, and A. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 201162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoneley, M., F. E. M. Paulin, J. P. C. Le Quesne, S. A. Chappell, and A. E. Willis. 1998. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16423-428. [DOI] [PubMed] [Google Scholar]
- 44.Stoneley, M., T. Subkhankulova, J. P. C. Le Quesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thoma, C., G. Bergamini, B. Galy, P. Hundsdoerfer, and M. W. Hentze. 2004. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell 15925-935. [DOI] [PubMed] [Google Scholar]
- 46.Thoma, C., S. Fraterman, M. Gentzel, M. Wilm, and M. W. Hentze. 2008. Translational initiation by the c-myc mRNA internal ribosome entry sequence and the poly(A) tail. RNA 141579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West, M. J., M. Stoneley, and A. E. Willis. 1998. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17769-780. [DOI] [PubMed] [Google Scholar]
- 48.Zucker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 24448-52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.