Abstract
Eukaryotic mRNAs possess a 5′-terminal cap structure (cap), m7GpppN, which facilitates ribosome binding. The cap is bound by eukaryotic translation initiation factor 4F (eIF4F), which is composed of eIF4E, eIF4G, and eIF4A. eIF4E is the cap-binding subunit, eIF4A is an RNA helicase, and eIF4G is a scaffolding protein that bridges between the mRNA and ribosome. eIF4G contains an RNA-binding domain, which was suggested to stimulate eIF4E interaction with the cap in mammals. In Saccharomyces cerevisiae, however, such an effect was not observed. Here, we used recombinant proteins to reconstitute the cap binding of the mammalian eIF4E-eIF4GI complex to investigate the importance of the RNA-binding region of eIF4GI for cap interaction with eIF4E. We demonstrate that chemical cross-linking of eIF4E to the cap structure is dramatically enhanced by eIF4GI fragments possessing RNA-binding activity. Furthermore, the fusion of RNA recognition motif 1 (RRM1) of the La autoantigen to the N terminus of eIF4GI confers enhanced association between the cap structure and eIF4E. These results demonstrate that eIF4GI serves to anchor eIF4E to the mRNA and enhance its interaction with the cap structure.
The cap structure, m7GpppN, is present at the 5′ terminus of all nuclear transcribed eukaryotic mRNAs. Cap-dependent binding of the ribosome to mRNA is mediated by the cap-binding protein eukaryotic translation initiation factor 4E (eIF4E), which forms a complex termed eIF4F together with eIF4G and eIF4A. Mammalian eIF4G, which has two isoforms, eIF4GI and eIF4GII, is a modular, multifunctional protein that binds to poly(A)-binding protein (PABP) (14) and eIF4E (18, 20) via the N-terminal third region. Mammalian eIF4G binds to eIF4A and eIF3 (15) via the middle third region and to eIF4A and Mnk protein kinase at the C-terminal region. eIF4GI also possesses an RNA-binding sequence (2, 9, 33) in the middle region. There are two RNA-binding sites on eIF4GI; one is located amino terminal to the first HEAT domain, and the other is located within the first HEAT domain (23). Mammalian and Saccharomyces cerevisiae eIF4E are similar in size (24 kDa), but mammalian eIF4GI (220 kDa) is larger than its yeast counterpart (150 kDa), as the latter lacks a C-terminal domain corresponding to mammalian eIF4GI (38).
The affinity of eIF4E for the cap structure has been a matter of dispute for some time. The earlier works of Carberry et al. (4) and Ueda et al. (39) estimated the equilibrium dissociation constant (Kd) of the eIF4E-cap complex by fluorescence titration to be 2 × 10−6 to 5 × 10−6 M depending on the nature of the cap analog. Later on, development of a new methodology for the fluorescence titration experiments yielded Kd values of 10−7 to 10−8 (29, 41). The source of the difference with the previous reports was thoroughly analyzed (29, 30). The interaction between the cap structure and eIF4E is dramatically enhanced by eIF4GI. This was first reported by showing that cross-linking of mammalian eIF4E to the cap structure is more efficient when it is a subunit of the eIF4F complex (19) or when it is complexed to eIF4GI (11). A similar enhancement of the binding of eIF4E to the cap structure was observed in yeast (40). However, two very different mechanisms were proposed to explain these observations. For the mammalian system, it was postulated that the middle segment of eIF4GI, which binds RNA, stabilizes the eIF4E interaction with the cap structure (11). This model was based primarily on the finding that in poliovirus-infected cells, eIF4GI is cleaved between its N-terminal third and the middle third, and consequently, eIF4E remains attached to the N-terminal eIF4GI fragment lacking the RNA-binding region. Under these conditions, cross-linking of eIF4E to the cap structure was poor (19, 31). In contrast, in yeast, a strong interaction between the cap structure and eIF4E was achieved using an eIF4G fragment containing the eIF4E-binding site that lacks the RNA-binding region (34, 40). Also, the yeast eIF4G fragment from amino acids 393 to 490 (fragment 393-490), which does not contain the RNA-binding site, forms a right-handed helical ring that wraps around the N terminus of eIF4E. This conformational change was suggested in turn to engender an allosteric enhancement of the association of eIF4E with the cap structure (10). Such an interaction between mammalian eIF4GI and eIF4E has not been reported.
To understand the mechanism by which eIF4GI stimulates the interaction of eIF4E with the cap structure in mammals, we reconstituted the eIF4E-cap recognition activity in vitro with purified eIF4E and eIF4GI recombinant proteins. Using a chemical cross-linking assay, we demonstrate that only mammalian eIF4GI fragments possessing RNA-binding activity enhance the cross-linking of eIF4E to the cap structure. Our data provide new insight into the mechanism of cap recognition by the eIF4E-eIF4GI complex.
MATERIALS AND METHODS
Expression and purification of eIF4GI deletion mutants.
The full-length human eIF4GI cDNA clone DKFZp762O191Q3 (GenBank accession no. AL120751) (pSP4GI) was a kind gift from Richard E. Lloyd (Baylor College of Medicine) (3). The hemagglutinin (HA)-tagged full-length eIF4GI fragment was amplified from pSP4GI by PCR and inserted into the pcDNA3 vector (pcDNA3-HA-eIF4GI[1-1599]). Open reading frames encoding eIF4GI fragments 84-1599, 84-1129, 84-674, 197-1599, 197-1129, and 197-674 were fused with a six-His sequence at the N terminus and a FLAG sequence at the C terminus and were subcloned into pUC-T7-EMCV IRES (26). To express the eIF4GI fragments, BHK-21 cells (30 100-mm dishes) were infected with vTF7-3 (8) and then transfected with a pUC-T7-EMCV IRES vector that encoded the eIF4GI fragments using a polyethylenimine-based method (35). After 20 to 24 h, cells were harvested and frozen at −80°C. Cells were thawed and disrupted by homogenization in 30 ml of buffer A (20 mM HEPES-KOH [pH 7.5], 10% glycerol, 5 mM 2-mercaptoethanol) containing 0.1 M KCl, 0.1% Triton X-100, and Complete EDTA-free protease inhibitor cocktail (Roche), and the resulting solution was incubated on ice for 20 min. After centrifugation at 9,000 rpm for 10 min, the supernatant was applied to SP-Sepharose (5 ml) (GE Healthcare). After the column was washed with buffer A containing 0.1 M KCl but no protease inhibitors (100 ml), proteins were eluted with 20 ml of buffer A containing 1 M KCl and 0.1% Triton X-100. The eluate was supplemented with imidazole (20 mM) and applied to Ni-nitrilotriacetic acid agarose resin (Qiagen; 0.6 ml). After the column was washed with buffer A containing 1 M KCl and 20 mM imidazole, proteins were eluted with buffer containing 250 mM imidazole, 0.15 M KCl, 20 mM HEPES-KOH (pH 7.5), 10% glycerol, and 5 mM 2-mercaptoethanol (2 ml). To dissociate eIF4E from eIF4GI, the eluate was incubated with 4E-BP1 (30 μg/ml) (26) at 25°C for 30 min and then applied to an anti-FLAG agarose column (Sigma; 0.2 ml). After the column was washed with buffer containing 0.1 M KCl, 20 mM HEPES-KOH (pH 7.5), 10% glycerol, and 5 mM 2-mercaptoethanol (20 ml), proteins were eluted with the same buffer containing 0.1 mM EDTA (600 μl) and FLAG peptide (Sigma; 100 μg/ml) and dialyzed against the same buffer using a Slide-A-Lyzer mini-dialysis unit (molecular weight cutoff of 10,000) (Pierce). To express eIF4GI fragment 84-674 [eIF4GI(84-674)] and eIF4GI(197-674), a HeLa cell-based cell-free protein expression system (26, 27) was employed. Expressed proteins were purified as described above.
Expression and purification of eIF4E.
A cDNA containing the entire open reading frame and a partial 3′ noncoding region of the mouse eIF4E was inserted between the NdeI and BamHI restriction sites of pET-3b (Novagen) to generate pET-3b-eIF4E. Escherichia coli BL21(DE3) was transformed with pET-3b-eIF4E and grown in Luria broth (LB) at 25°C. eIF4E expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. Cells were suspended in sonication buffer (20 mM HEPES-KOH [pH 7.5], 0.5 mM EDTA, 100 mM KCl, 7 mM 2-mercaptoethanol, and 100 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with NP-40 (0.5%) and lysed by sonication. Cellular debris was removed by centrifugation, and the supernatant was incubated with cap analog affinity matrix. Recombinant eIF4E was purified as described previously (7).
Expression and purification of La RRM1 fusion with eIF4GI fragments.
pET-La cDNA (a gift from J. Keene, Duke University) (5) and pcDNA3-HA-eIF4GI(1-1599) were used for the generation of La RNA recognition motif 1 (RRM1)-fused eIF4GI. To construct pGex-eIF4GI(197-674)-La-His and pGEX-eIF4GI(602-674)-La-His, the La fragment containing a His tag at the C terminus, eIF4GI(197-674) and eIF4GI(602-674) fragments were generated using PCR. The N-terminal eIF4GI fragments were cloned between the EcoRI and NotI restriction sites of pGEX-6P-1 (GE Healthcare) with the His-tagged N-terminal La fragment. GST-eIF4GI(197-674)-La-His and GST-eIF4GI(602-674)-La-His were expressed in Escherichia coli BL21(DE3) and purified by Ni-nitrilotriacetic acid agarose (Qiagen) and glutathione Sepharose 4B (GE Healthcare) chromatography as described previously (14, 24). After dialysis in cleavage buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.01% Triton X-100), protein solutions were supplemented with PreScission protease (GE Healthcare) and incubated at 4°C for 4 h. Protein solutions were incubated with glutathione Sepharose 4B to remove glutathione S-transferase (GST). Full-length human La protein was expressed in Escherichia coli BL21(DE3) using pET-La and purified as described previously (37).
In vitro transcription.
The plasmid encoding luciferase T3luc(A)+ was described previously (13). T3luc(A)+ was linearized with BamHI and transcribed with T3 RNA polymerase. Synthesis of capped RNA transcripts was performed with the MAXIscript T3 kit (Ambion). For electrophoretic mobility shift assay (EMSA), the plasmid encoding rabbit β-globin (pBS-β-globin; a gift from C. U. Hellen, State University of New York Downstate Medical Center) (12) was linearized with NcoI and transcribed in vitro in the presence of [α-32P]UTP with T7 RNA polymerase using MAXIscript T7 kit (Ambion). The RNA transcript (163 nucleotides) was purified using denaturing 5% acrylamide-8 M urea polyacrylamide gel electrophoresis (PAGE) in Tris-boric acid-EDTA (TBE) buffer. RNA was eluted from the gel with RNase-free water.
In vitro translation.
Rabbit reticulocyte lysate (Promega) was treated with rhinovirus 2Apro (25 μg/ml) for 30 min at 30°C as described previously (28), followed by incubation for 10 min on ice with 0.8 mM elastatinal (Sigma). The lysate was supplemented with recombinant eIF4GI fragments (200 nM) and incubated with capped luciferase RNA (1 ng/μl) for 60 min at 30°C. Aliquots of the translation mixture (1/100th) were assayed for luciferase activity using the luciferase assay system (Promega) in a Lumat LB 9507 bioluminometer (EG&G Bertold).
EMSA.
[32P]UTP-labeled β-globin mRNA (0.2 nM) and recombinant eIF4GI fragments were incubated for 15 min at 30°C in binding buffer (100 mM KCl, 20 mM HEPES-KOH [pH 7.5], 10% glycerol, 3 mM MgCl2, and 0.1% Triton X-100). The reaction mixtures were supplemented with sample buffer and analyzed on a nondenaturing polyacrylamide gel (4% acrylamide and 0.1% bisacrylamide) prepared with running buffer (50 mM Tris [pH 8.5], 380 mM glycine, and 5% glycerol). The gel was prerun for 2 h at 50 V at 4°C prior to sample loading. Electrophoretic separation was performed at 50 V at 4°C overnight.
Chemical cross-linking assay.
Chemical cross-linking was performed as previously described (25, 36). Uncapped firefly luciferase (Luc) RNA poly(A)+ was radioactively labeled at the cap using vaccinia virus guanylyltransferase (Ambion) with [α-32P]GTP (Perkin Elmer Life Science) and S-adenosylmethionine according to the manufacturer's instructions. Cap-labeled mRNA was oxidized using 0.2 mM NaIO4 for 2 h at 4°C. Oxidized mRNA (105 cpm) in a total reaction mixture volume of 20 μl was incubated with purified recombinant proteins in the cross-linking buffer (8.75 mM HEPES-KOH [pH 7.3], 17.5 mM KCl, 26.25 mM potassium acetate, and 0.7 mM MgCl2) at 30°C for 10 min. Reaction mixtures were incubated at 4°C overnight in the presence of 0.2 mM NaCNBH3. After the addition of an RNase mixture (2 μg/μl RNase A, 2 units/μl RNase T1, and 0.01 units/μl RNase V1) and incubation at 37°C for 30 min to digest the RNA, samples were subjected to sodium dodecyl sulfate (SDS)-10% PAGE and autoradiography.
RESULTS
Purification of recombinant eIF4GI fragments.
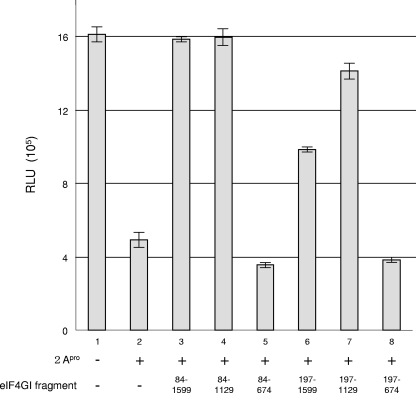
To investigate the effect of eIF4GI on the interaction between eIF4E and the cap structure, purified recombinant murine eIF4E and human eIF4GI fragments (depicted in Fig. 1A) were generated. The purified proteins were intact and migrated as expected according to their molecular masses upon an SDS-polyacrylamide gel (Fig. 1B, lanes 1 to 7). To assess the activity of the eIF4GI fragments, they were examined in an in vitro translation experiment in a rabbit reticulocyte lysate using a capped poly(A)+ luciferase RNA. The lysate was treated with rhinovirus 2A protease (2Apro) to cleave endogenous eIF4GI, which results in inhibition of cap-dependent translation. Rhinovirus 2Apro treatment reduced cap-dependent translation (to 30% of untreated control; Fig. 2, compare bar 1 to bar 2). The addition of eIF4GI fragments containing the full-length protein, except for the first 83 amino acids, restored cap-dependent translation (Fig. 2, compare bar 1 to bar 3). (This fragment will be referred to as full length throughout the manuscript. We used this fragment because the full-length protein is insoluble.) Importantly, the addition of a fragment containing just the N-terminal third and the middle third (fragment 84-1129) of eIF4GI also restored translation in agreement with a previous report (28). However, eIF4GI fragments that lacked the middle segment failed to restore cap-dependent translation (Fig. 2, compare bar 1 to bars 5 and 8). It is noteworthy that the eIF4GI fragment lacking the PABP-binding region (fragments 197-1599 and 197-1129) restored translation less effectively (62.5% to 88.1%) than eIF4GI fragments containing the PABP-binding site (Fig. 2, compare bars 3 and 4 to bars 6 and 7), consistent with the important role of PABP in facilitating translation. These results demonstrate that all the eIF4GI fragments except for those lacking the middle third are functional in translation.
FIG. 1.
Recombinant human eIF4GI proteins. (A) Diagram of eIF4GI constructs. PABP-, eIF4E-, eIF4A-, and eIF3-binding sites and the RNA-binding region are indicated. (B) Purified murine eIF4E and human eIF4GI fragments (2 μg) were analyzed by SDS-12% PAGE followed by Coomassie blue staining. The amino acid numbers of eIF4GI fragments are indicated above the lanes. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
FIG. 2.
Functional analysis of recombinant eIF4GI fragments. Rabbit reticulocyte lysates treated with rhinovirus 2Apro (+) or control buffer (−) were supplemented with recombinant eIF4GI fragments (200 nM) as indicated. Capped poly(A)+ luciferase mRNA (1 μg/ml) was added to the extract, and in vitro translation and luciferase assays were performed as described in Materials and Methods. The amino acid numbers of eIF4GI fragments are indicated below the graph. RLU, relative light units.
Binding of eIF4GI fragments to β-globin RNA.
Because the central third of eIF4GI binds to RNA (32), we wished to document the RNA-binding activity of the different eIF4GI fragments. This was performed by analyzing the interaction between an uncapped [32P]UTP-labeled fragment of β-globin RNA and eIF4GI by an EMSA (Fig. 3). The formation of the RNA-protein complex was analyzed with increasing concentrations of recombinant eIF4GI fragments. eIF4GI fragments containing the middle third formed an RNA-protein complex in a dose-dependent manner (Fig. 3, lanes 2 to 4 and 5 to 7), while N-terminal eIF4GI fragments failed to do so (lanes 8 to 10). This result demonstrates that eIF4GI binds to RNA through its middle segment.
FIG. 3.
The interaction of recombinant eIF4GI fragments with β-globin RNA as assayed by an EMSA. [32P]UTP-labeled β-globin RNA was incubated with recombinant eIF4GI fragments. In lane 1, no protein was added. eIF4GI fragments were added at the indicated concentrations. The positions of free RNA and the RNA-eIF4GI complex are shown to the right of the gel.
The interaction between the cap structure and eIF4E is enhanced by eIF4GI fragments containing the RNA-binding region.
To study the effects of different eIF4GI fragments on the binding of eIF4E to the cap structure, eIF4E was cross-linked to cap-labeled RNA by oxidation of the 2′, 3′ cis-diol bond of the m7G-ribose, thus converting it to a dialdehyde, followed by formation of a Schiff base between the aldehyde group and ɛ-amino groups in eIF4E. The Schiff base was reduced to form a covalent bond between the cap structure and eIF4E, and the RNA was degraded using a mixture of RNases (36). We chose to use the chemical cross-linking method rather than UV-induced cross-linking because the former is more efficient in cross-linking eIF4E to the cap structure (31). The cross-linking between the cap structure and eIF4E was enhanced by increasing the amount of eIF4E in a dose-dependent manner (Fig. 4A and B). To investigate the effect of eIF4GI on the cross-linking of eIF4E to the cap structure, 10 ng of eIF4E (20 nM) was used, because this amount of eIF4E was far below saturating (Fig. 4A, lane 3).
FIG. 4.
Recombinant eIF4GI fragments containing the RNA-binding region enhance the association between eIF4E and the cap structure. (A) Dose-dependent interaction of eIF4E with the cap structure. Cap-labeled luciferase poly(A)+ RNA was incubated with the amounts (nanomolar concentrations) of recombinant eIF4E indicated in the figure. eIF4E was cross-linked to the RNA as described in Materials and Methods. m7GDP was added (+) or not added (−) as indicated. (B) Quantitative analysis of the eIF4E bands in panel A. The intensities of the bands were measured using NIH ImageJ. The value obtained for the band in lane 1 was set at 1 (bar 1). (C) Recombinant full-length eIF4GI fragment enhances the interaction between the cap structure and eIF4E. Cap-labeled RNA was incubated with recombinant eIF4E and eIF4GI(84-1599). Increasing amounts of eIF4E were incubated with eIF4GI (20 nM) at the molar ratios indicated in the figure. (D) Quantitative analysis of the enhancement of eIF4E cross-linking to the cap in panel C. The band intensities in lanes 1, 5, and 9 were set at 1 (gray bars). (E) Recombinant eIF4GI fragments containing the RNA-binding region enhance the interaction between the cap structure and eIF4E. Cap-labeled RNA was incubated with recombinant eIF4E and eIF4GI fragments at a molar ratio of 1:1. The eIF4GI fragments are indicated above the figure. (F) Quantitative analysis of eIF4E bands in panel E. The band intensity in lane 1 was set at 1. The data shown in panels A, C, and E are representative of three experiments. Quantitative data with means plus standard deviations (error bars) are shown in panels B, D, and F.
We next examined the potentiation of eIF4E cross-linking by eIF4GI(84-1599) using different molar ratios of the proteins. A molar ratio of 1:1 of full-length eIF4GI to eIF4E resulted in optimal enhancement of eIF4E cross-linking to the cap structure (Fig. 4C and D). Therefore, this molar ratio was used for further experiments. Recombinant eIF4GI fragments containing an RNA-binding region (fragments 84-1599, 84-1129, 197-1599, and 197-1129) enhanced eIF4E cross-linking to the cap structure (Fig. 4E and F, lanes 3, 5, 9, and 11), while N-terminal eIF4GI fragments lacking the RNA-binding region (fragments 84-674 and 197-674) failed to enhance eIF4E cross-linking to the cap structure (lanes 7 and 13). These results demonstrate that the RNA-binding region of eIF4GI is essential for the enhancement of the interaction between the cap structure and eIF4E.
La RRM1 confers on the N-terminal region of eIF4GI the ability to stimulate the interaction between the cap structure and eIF4E.
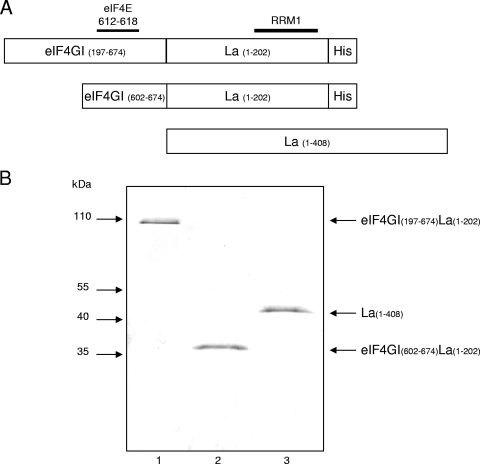
The middle third of eIF4GI is multifunctional, as it exhibits eIF3-, eIF4A-, and RNA-binding activity. If the RNA-binding activity per se were to be required for the enhancement of eIF4E binding to the cap by anchoring eIF4E to the mRNA, then any sequence nonspecific RNA-binding region would be expected to substitute for the middle segment of eIF4GI. To test this hypothesis, the RRM1 of the autoantigen La (amino acids 1 to 202) (6, 17) was fused to the N terminus of eIF4GI (Fig. 5A). Since the PABP-binding site of eIF4GI is dispensable for eIF4E binding to the cap (Fig. 4E, compare lanes 3 and 5 to lanes 9 and 11), an N-terminal fragment of eIF4GI lacking the PABP-binding site was chosen for this study. The full-length La protein was used as a negative control. SDS-PAGE followed by Coomassie blue staining showed that the recombinant proteins were of high purity (Fig. 5B).
FIG. 5.
Analysis of recombinant La RRM1-fused eIF4GI fragments. (A) Schematic representations of La RRM1-fused eIF4GI fragments. The positions of the eIF4E-binding site on eIF4GI and the RRM1 of the La autoantigen are indicated above the fragments. (B) Recombinant La RRM1-fused eIF4GI fragments and the full-length La(1-408) were analyzed by SDS-12% PAGE and Coomassie blue staining. Lane 1, eIF4GI(197-674)La(1-202); lane 2, eIF4GI(602-674)La(1-202); lane 3, La(1-408). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
Recombinant eIF4E and cap-labeled RNA were incubated with recombinant La-eIF4GI fusion fragments or the full-length La protein in a chemical cross-linking assay. The La-eIF4GI fragments, eIF4GI(197-674)La and eIF4GI(602-674)La, enhanced eIF4E binding to the cap structure 15- and 10-fold, respectively (Fig. 6A and B, lanes 7 and 9), while the full-length La protein failed to enhance cross-linking (lane 11). Strikingly, the eIF4GI(197-674)La fragment enhanced the interaction between the cap structure and eIF4E to the same extent as the eIF4GI(197-1129) fragment did (compare lane 7 to lane 3). The eIF4GI(602-674)La fragment also enhanced eIF4E cross-linking to the cap structure, albeit less efficiently (65%) than the eIF4GI(197-674)La fragment and the eIF4GI(197-1129) fragment (Fig. 6A and B, compare lanes 3 and 7 to lane 9). This result clearly demonstrates that the RNA-binding ability of eIF4GI is required and sufficient for efficient interaction between the cap structure and eIF4E.
FIG. 6.
La RRM1 confers on the N-terminal region of eIF4GI the ability to stimulate the interaction between the cap structure and eIF4E. (A) Cap-labeled RNA and eIF4E were incubated with recombinant eIF4GI fragments as indicated in the figure at a molar ratio of eIF4E to eIF4GI of 1:1. Proteins were cross-linked and analyzed as described in Materials and Methods. m7GDP was added (+) or not added (−) as indicated. (B) Quantitative analysis of eIF4E bands in panel A. The band intensity in lane 1 was set at 1. (C) Dose-dependent interaction between the cap structure and eIF4E by the addition of increasing concentrations of eIF4GI and La RRM1-fused N-terminal eIF4GI fragments. Cap-labeled RNA and eIF4E were incubated with recombinant eIF4GI fragment and La RRM1-fused N-terminal eIF4GI fragments, respectively, at the molar ratios of eIF4E to eIF4GI indicated in the figure. (D) Quantitative analysis of eIF4E bands in panel C. The band intensity of lane 1 in panel C was set at 1. The data shown in panels A and C are representative of three experiments. Quantitative data with means and standard deviations (error bars) for these experiments are shown in panels B and D. (E) RNA-binding ability and the interaction between the cap structure and eIF4E of La RRM1-fused N-terminal eIF4GI fragments. The RNA-binding ability of the three fragments and the ability of the three fragments to enhance the cap-eIF4E interaction are shown (+++, strong; ++, moderate, −, none). (F) Model of the stimulatory effect of La RRM1 on the interaction between the cap structure and eIF4E.
We next investigated the eIF4GI fragment dose-dependent enhancement of eIF4E cross-linking to the cap structure (Fig. 6C). Cap-labeled RNA and eIF4E were incubated in the presence of the increasing concentrations of the recombinant eIF4GI(197-1129) fragment or La RRM1-fused N-terminal eIF4GI fragments (fragment 197-674 or 602-674), respectively, at the following molar ratio of eIF4E to eIF4GI: 1:0.25 (Fig. 6C, lanes 3, 7, and 11), 1:0.5 (lanes 4, 8, and 12), 1:1 (lanes 5, 6, 9, 10, 13, and 14). The interaction between the cap structure and eIF4E was increased with increasing amounts of eIF4GI and La-eIF4GI fragments in a dose-dependent manner. The eIF4GI(602-674)La fragment enhanced eIF4E cross-linking to the cap structure less efficiently (62%) than the eIF4GI(197-1129) fragment and the eIF4GI(197-674)La fragment did (Fig. 6C and D, compare lanes 11 to 13 to lanes 3 to 5 and 7 to 9). Taken together, these results demonstrate that the RNA-binding region of eIF4GI is necessary to enhance eIF4E binding to the cap structure (see summary of results [Fig. 6E and F]).
DISCUSSION
In this paper, we demonstrate that the RNA-binding region of eIF4GI dramatically increases the interaction between the cap structure and mammalian eIF4E. These findings appear to be at variance with those reported for yeast, in which it was concluded that a yeast eIF4G fragment lacking the RNA-binding region is capable of enhancing the cap-binding affinity of eIF4E (40). The explanations for differences in the mechanism of stimulation of eIF4E-cap interaction by eIF4G between these systems might be manifold. First, while the readout for chemical cross-linking is the interaction of eIF4E with the cap structure, the magnitude of the cross-linking is strongly dependent on the interaction of eIF4G with the RNA, so that the affinity of eIF4E for the cap is only a minor factor. The interaction between eIF4E and the cap structure is a function of equilibrium conditions, but when eIF4E is kept close to the cap, the effective concentration of eIF4E in the vicinity of the cap is greatly increased, which shifts the equilibrium toward the formation of the cap-eIF4E complex, as the dissociation of the cap from eIF4E is prevented. In other words, the RNA-binding segment of eIF4GI dramatically enhances the stability of the eIF4E-eIF4G complex with the capped mRNA, but not via the interaction of eIF4E with the cap itself.
Although different methods were used in the mammalian versus yeast studies, this is unlikely to be the source of difference inasmuch as the cross-linking assay has been proven a reliable measure of cap-binding affinity in yeast (1). Another possible explanation for the disparate results is the possibility of a difference between the yeast and mammalian mechanism of eIF4E-eIF4G interactions. There are several lines of evidence that are consistent with this idea. In mammals, a short polypeptide (16-residue polypeptide from eIF4GII) binds to the dorsal surface of eIF4E with a strong affinity (26 ± 6 nM). Importantly, the conformation of eIF4E is not changed when it is bound to this peptide (21). In contrast, in yeast, the corresponding eIF4G peptide binds to eIF4E with low affinity (2 μM), but a larger fragment of eIF4G (393-490) binds to eIF4E with high affinity (3 nM) (10). The binding of eIF4G to eIF4E causes a mutual folding of both proteins that leads to a stable complex. In this complex, eIF4G wraps around the N terminus of eIF4E to form a molecular bracelet (10). Residues 23 to 32 of eIF4E become folded as a result of the interaction with eIF4G. Significantly, the formation of the stable eIF4E-eIF4G complex in yeast enhances the interaction between eIF4E and the cap (10).
Alignment of the sequence spanning amino acids 25 to 35 of mammalian and yeast eIF4E shows weak homology (see Fig. S1 in the supplemental material). This is consistent with the observation that mammalian eIF4G binding to eIF4E does not cause a coupled change in the conformations of both proteins (G. Wagner, personal communication) and also with our finding that binding of the N-terminal sequence of eIF4GI to eIF4E does not increase the cross-linking of the latter to the cap structure. Similarly, the on rate and off rate for binding of m7GpppG to human eIFE, and therefore the equilibrium binding constant, are the same in the presence and absence of human eIF4G(557-646) (35a).
The species-specific differences in the eIF4E-eIF4G interaction and their influence on cap recognition are also consistent with the findings that deletion of the first 32 amino acids from the N terminus of eIF4E had no effect on translation of a reporter mRNA in reticulocyte lysate (22), while deletion of the first 30 amino acids in yeast eIF4E had a dramatic deleterious effect on translation and growth (10). However, it is not immediately clear what might be the biological significance of the biochemical differences between yeast and mammals in the manner by which eIF4G enhances cap recognition by eIF4E.
In summary, mammalian eIF4G dramatically enhances the interaction of eIF4E with the cap by anchoring eIF4E to the mRNA via eIF4G. It is striking that eIF4G stimulates translation by interacting with two regions of the mRNA to stimulate eIF4E activity by circularizing the mRNA. One region is RNA sequence nonspecific as discussed in this paper, while the other is the poly(A) tail, through which eIF4G interacts with the PABP, resulting in enhanced binding of eIF4E to the cap structure (Fig. 7 and our unpublished data) (16).
FIG. 7.
Model of the mechanism by which eIF4GI stimulates the interaction between the cap structure and eIF4E. Binding of eIF4GI to the mRNA body brings eIF4E to the vicinity of the cap structure and consequently stabilizes the interaction between the cap and eIF4E. The thickness of the double-headed arrow represents the strength of the interaction. eIF4GI further enhances this interaction by interacting with PABP.
Supplementary Material
Acknowledgments
We thank Colin Lister for technical assistance.
A.Y. is the recipient of Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. This work was supported by grants from the National Institutes of Health (GM66157) and the Canadian Institute of Health Research to N.S.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altmann, M., I. Edery, H. Trachsel, and N. Sonenberg. 1988. Site-directed mutagenesis of the tryptophan residues in yeast eukaryotic initiation factor 4E. Effects on cap binding activity. J. Biol. Chem. 26317229-17232. [PubMed] [Google Scholar]
- 2.Berset, C., A. Zurbriggen, S. Djafarzadeh, M. Altmann, and H. Trachsel. 2003. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 224499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carberry, S. E., R. E. Rhoads, and D. J. Goss. 1989. A spectroscopic study of the binding of m7GTP and m7GpppG to human protein synthesis initiation factor 4E. Biochemistry 288078-8083. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. N., D. J. Kenan, J. D. Keene, A. Gatignol, and K. T. Jeang. 1994. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 687008-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig, A. W., Y. V. Svitkin, H. S. Lee, G. J. Belsham, and N. Sonenberg. 1997. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol. Cell. Biol. 17163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edery, I., M. Altmann, and N. Sonenberg. 1988. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74517-525. [DOI] [PubMed] [Google Scholar]
- 8.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyer, C., M. Altmann, H. S. Lee, A. Blanc, M. Deshmukh, J. L. Woolford, Jr., H. Trachsel, and N. Sonenberg. 1993. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 134860-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, J. D., N. J. Moerke, T. von der Haar, A. A. Lugovskoy, A. B. Sachs, J. E. McCarthy, and G. Wagner. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115739-750. [DOI] [PubMed] [Google Scholar]
- 11.Haghighat, A., and N. Sonenberg. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 27221677-21680. [DOI] [PubMed] [Google Scholar]
- 12.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 907642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 147322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 176940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M. N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenan, D. J., and J. D. Keene. 2004. La gets its wings. Nat. Struct. Mol. Biol. 11303-305. [DOI] [PubMed] [Google Scholar]
- 18.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 27021975-21983. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. A., I. Edery, and N. Sonenberg. 1985. Isolation and structural characterization of cap-binding proteins from poliovirus-infected HeLa cells. J. Virol. 54515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 154990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3707-716. [DOI] [PubMed] [Google Scholar]
- 22.Marcotrigiano, J., A. C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89951-961. [DOI] [PubMed] [Google Scholar]
- 23.Marcotrigiano, J., I. B. Lomakin, N. Sonenberg, T. V. Pestova, C. U. Hellen, and S. K. Burley. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7193-203. [DOI] [PubMed] [Google Scholar]
- 24.Masutani, M., N. Sonenberg, S. Yokoyama, and H. Imataka. 2007. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 263373-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrick, W. C., and N. Sonenberg. 1997. Assays for eukaryotic translation factors that bind mRNA. Methods 11333-342. [DOI] [PubMed] [Google Scholar]
- 26.Mikami, S., T. Kobayashi, S. Yokoyama, and H. Imataka. 2006. A hybridoma-based in vitro translation system that efficiently synthesizes glycoproteins. J. Biotechnol. 12765-78. [DOI] [PubMed] [Google Scholar]
- 27.Mikami, S., M. Masutani, N. Sonenberg, S. Yokoyama, and H. Imataka. 2006. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr. Purif. 46348-357. [DOI] [PubMed] [Google Scholar]
- 28.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niedzwiecka, A., J. Marcotrigiano, J. Stepinski, M. Jankowska-Anyszka, A. Wyslouch-Cieszynska, M. Dadlez, A. C. Gingras, P. Mak, E. Darzynkiewicz, N. Sonenberg, S. K. Burley, and R. Stolarski. 2002. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319615-635. [DOI] [PubMed] [Google Scholar]
- 30.Niedzwiecka, A., J. Stepinski, J. M. Antosiewicz, E. Darzynkiewicz, and R. Stolarski. 2007. Biophysical approach to studies of cap-eIF4E interaction by synthetic cap analogs. Methods Enzymol. 430209-245. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier, J., and N. Sonenberg. 1985. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5′ secondary structure. Mol. Cell. Biol. 53222-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., and C. U. Hellen. 2003. Coupled folding during translation initiation. Cell 115650-652. [DOI] [PubMed] [Google Scholar]
- 33.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 166870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ptushkina, M., T. von der Haar, S. Vasilescu, R. Frank, R. Birkenhager, and J. E. McCarthy. 1998. Cooperative modulation by eIF4G of eIF4E-binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J. 174798-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed, S. E., E. M. Staley, J. P. Mayginnes, D. J. Pintel, and G. E. Tullis. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 13885-98. [DOI] [PubMed] [Google Scholar]
- 35a.Slepenkov, S. V., N. L. Korneeva, and R. E. Rhoads. 2008. Kinetic mechanism for assembly of the m7GpppG · eIF4E · eIF4G complex. J. Biol. Chem. 28325227-25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonenberg, N., M. A. Morgan, W. C. Merrick, and A. J. Shatkin. 1978. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. USA 754843-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svitkin, Y. V., K. Meerovitch, H. S. Lee, J. N. Dholakia, D. J. Kenan, V. I. Agol, and N. Sonenberg. 1994. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 681544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarun, S. Z., Jr., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 157168-7177. [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda, H., H. Iyo, M. Doi, M. Inoue, and T. Ishida. 1991. Cooperative stacking and hydrogen bond pairing interactions of fragment peptide in cap binding protein with mRNA cap structure. Biochim. Biophys. Acta 1075181-186. [DOI] [PubMed] [Google Scholar]
- 40.von Der Haar, T., P. D. Ball, and J. E. McCarthy. 2000. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem. 27530551-30555. [DOI] [PubMed] [Google Scholar]
- 41.Zuberek, J., J. Jemielity, A. Jablonowska, J. Stepinski, M. Dadlez, R. Stolarski, and E. Darzynkiewicz. 2004. Influence of electric charge variation at residues 209 and 159 on the interaction of eIF4E with the mRNA 5′ terminus. Biochemistry 435370-5379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.