Abstract
TRF1 is a component of the shelterin complex at mammalian telomeres; however, a role for TRF1 in telomere biology in the context of the organism is unclear. In this study, we generated mice with transgenic TRF1 expression targeted to epithelial tissues (K5TRF1 mice). K5TRF1 mice have shorter telomeres in the epidermis than wild-type controls do, and these are rescued in the absence of the XPF nuclease, indicating that TRF1 acts as a negative regulator of telomere length by controlling XPF activity at telomeres, similar to what was previously described for TRF2-overexpressing mice (K5TRF2 mice). K5TRF1 cells also show increased end-to-end chromosomal fusions, multitelomeric signals, and increased telomere recombination, indicating an impact of TRF1 on telomere integrity, again similar to the case in K5TRF2 cells. Intriguingly, K5TRF1 cells, but not K5TRF2 cells, show increased mitotic spindle aberrations. TRF1 colocalizes with the spindle assembly checkpoint proteins BubR1 and Mad2 at mouse telomeres, indicating a link between telomeres and the mitotic spindle. Together, these results demonstrate that TRF1, like TRF2, negatively regulates telomere length in vivo by controlling the action of the XPF nuclease at telomeres; in addition, TRF1 has a unique role in the mitotic spindle checkpoint.
Telomere repeat binding factor 1 (TRF1) is a component of the shelterin complex at mammalian telomeres (13, 18, 31). TRF1 is proposed to act as a negative regulator of telomere length by inhibiting telomerase activity in cis (1, 50, 52). In human cells, TRF1 overexpression leads to telomere shortening (1, 50), while displacement of TRF1 from telomeres using a TRF1 dominant-negative allele leads to telomere elongation (52). This role of TRF1 as a negative regulator of telomere length is proposed to be mediated by its interaction with Pot1, which together with TPP1 is proposed to regulate the access of telomerase to chromosome ends (32, 50, 56, 57). In human cells, TRF1 interacts with tankyrase 1 and 2, which poly-ADP-ribosylates TRF1, thereby controlling TRF1 binding to telomeres and regulating telomere length (12, 28, 49). TRF2, a homologue of TRF1, is also a negative regulator of telomere length in human cultured cells (1, 50). In addition, TRF2 is essential for telomere capping (11, 53). TRF2 overexpression results in telomere degradation, mediated by the TRF2-interacting XPF/ERCC1 nuclease, also involved in nucleotide excision repair (NER) (7, 17, 35, 54, 58). TRF2 overexpression in the skin of mice leads to defective NER, increased skin cancer, and premature aging (7, 35). TRF1 and TRF2 share the same architecture, characterized by a C-terminal Myb domain and a TRFH N-terminal domain (9, 14, 21). Interestingly, both proteins contain distinct binding sites for the shelterin protein Tin2, probably determining their different binding partners and functions at telomeres (14). When specifically targeted to telomeres, TRF1 overexpression releases the so-called telomere position effect, suggesting a role for TRF1 in controlling telomere silencing (30). Recently, an interaction between TRF1 and RNA polymerase II (Pol II) which seems to aid in telomere transcription was described (47). In addition, TRF1 regulates sister telomere cohesion at telomeres through posttranscriptional modification by tankyrase 1 (10, 49). TRF1 has also been shown to interact with components of the mitotic spindle, including the mitotic kinase NIMA and the spindle regulator Mad1 (40, 43). Furthermore, TRF1 has been shown to interact with microtubules and to control microtubule polymerization (40).
These different roles of TRF1 in telomere biology as well as in chromosome dynamics and genomic integrity suggest an impact of TRF1 on cancer and aging. Interestingly, TRF1 is upregulated in some human epithelial cancers (33, 41), suggesting that increased TRF1 expression may favor tumorigenesis. More recently, TRF1 was found to be altered in some cases of aplastic anemia (45), a disease characterized by premature loss of bone marrow regeneration and the presence of short telomeres. In addition, the TRF1-interacting protein Tin2 is found to be mutated in some cases of dyskeratosis congenita and Revesz syndrome, also characterized by defective bone marrow regeneration and the presence of short telomeres (46).
Interestingly, mice deleted for TRF1 show embryonic lethality due to unknown reasons but do not appear to have defects in telomere length maintenance or telomere capping (29), arguing that TRF1 is not involved in telomere length regulation and telomere capping, at least during the very early stages of mouse embryonic development. Similarly, mice deficient in the TRF1 regulator tankyrase also show normal telomere length and telomere capping (15, 27). The lack of viable TRF1 mouse models has prevented the study of the impact of altered TRF1 expression in normal development, cancer, and aging.
In this study, we set out to address the role of TRF1 in telomere biology by generating tissue-specific TRF1 transgenic mice. In particular, we targeted TRF1 overexpression to mouse stratified epithelia by using the keratin 5 (K5) promoter, generating K5TRF1 mice. K5TRF1 mice show a two- to threefold increase in the amount of TRF1 bound to telomeres, which is accompanied by a slight increase of tankyrase but not of TRF2. Augmented TRF1 abundance at telomeres results in telomere shortening in vivo, indicating that TRF1 is an essential telomere length regulator in the context of epithelial homeostasis. Furthermore, we show that similar to that previously described for K5TRF2 mice (35), telomere shortening in K5TRF1 mice is also mediated by the XPF nuclease, suggesting that TRF1 and TRF2 act in the same pathway of telomere length control in mammals. Finally, we show that TRF1 colocalizes with the spindle assembly checkpoint (SAC) proteins BubR1 and Mad2 at mouse telomeres and that TRF1 overexpression, but not TRF2 overexpression, results in aberrant mitotic spindles, demonstrating a specific role for TRF1 in mitosis.
MATERIALS AND METHODS
Generation and maintenance of mouse strains.
The full-length mouse TRF1 cDNA was cloned downstream of bovine K5 regulatory sequences (38). After digestion with NotI, DNA was microinjected at 2 μg/μl into the pronuclei of fertilized oocytes from C57BL/6 × CBA F1 mice. Five founder mice were identified by using the K5 bovine promoter as a probe. Four mice (K5TRF1-A to -D) transmitted the transgene to the offspring. Founders were backcrossed for three to five generations onto a pure C57BL/6 background to obtain the different K5TRF1 transgenic lines studied here. All wild-type and K5TRF1 mice used for the analyses were littermates. K5TRF2/K5TRF1 double mutant mice were obtained by crossing female K5TRF2 mice (35) with male K5TRF1 mice. K5TRF1/XPF−/− mice were generated by crossing K5TRF1 mice with XPF+/− mice, as previously described (35). XPF alleles were generated as described previously (35).
All mice were generated and maintained at the Spanish National Cancer Centre under specific-pathogen-free conditions in accordance with the recommendation of the Federation of European Laboratory Animal Science Associations.
Real-time quantitative RT-PCR.
Total RNA was extracted using Trizol (Invitrogen Life Technologies) from the tail and back skin of three or four age-matched (2 to 5 months old) K5TRF1-A, K5TRF1-B, K5TRF1-C, and K5TRF1-D transgenic mice as well as their corresponding wild-type controls. Reserve transcription (RT) was carried out following the manufacturer's instructions (Invitrogen Life Technologies). Real-time RT-PCR was performed as described previously (35). Primers TRF1FB (5′ TCT AAG GAT AGG CCA GAT GCC A 3′) and TRF1RB (5′ CTG AAA TCT GAT GGA GCA CGT C 3′) amplified a 186-bp fragment of mouse TRF1 cDNA. The ACTIN-F and ACTIN-R primers (35) were used in parallel RT-PCRs for actin detection. We determined the relative expression of TRF1 in each sample by calculating the ΔΔCT value, which expresses the difference between the cycle threshold with the TRF1 primer pair and that with the β-actin primer pair. Each RT-PCR was repeated twice.
Telomere length analyses of skin sections.
Quantitative telomere fluorescence in situ hybridization (Q-FISH) directly on skin sections was performed as previously described (35).
Isolation and culture of keratinocytes from newborn mouse skin.
Primary keratinocyte cultures from newborn mice (0 to 2 days old) were obtained as previously described (35).
Telomere length analyses of isolated skin keratinocytes. (i) Q-FISH analysis.
Primary keratinocytes were subjected to Q-FISH as previously described (35, 44). TFL-Telo software (59) was used to quantify the fluorescence intensity from at least 10 metaphases from each genotype.
For chromosomal aberration analysis, chromosomes were analyzed by superimposing the telomere image on the DAPI (4′,6-diamidino-2-phenylindole) image of each metaphase.
(ii) TRF analysis.
One million keratinocytes were included in agarose plugs, and TRF analysis was performed as described by Blasco et al. (8).
ChIP analysis.
For chromatin immunoprecipitation (ChIP) analysis, 1.5 × 106 keratinocytes were used per condition. ChIP analysis with the indicated antibodies was performed as described previously (6). In all cases, ChIP values were represented as percentages of the total input DNA, therefore correcting for differences in the numbers of telomere and centromere repeats.
Analysis of TelRNAs (or TERRAs).
For analysis of telomeric RNAs (TelRNAs or TERRAs), RNA dot blot analysis was performed using different amounts of RNA (Trizol; Invitrogen) as described previously (47).
Isolation of epidermal keratinocytes from adult mice.
Mice were sacrificed, and the skin from the tail and back was removed surgically by making a longitudinal incision. Adult skin keratinocytes were isolated basically as described previously (35).
Tumor induction experiments.
Wild-type and K5TRF1B littermate mice (8 to 12 weeks old) were shaved and treated with a single dose of 20 μg of DMBA [7,12-dimethylbenz(a)anthracene] (Sigma) in 200 μl of acetone. Mice were subsequently treated twice weekly with 12.5 μg of tetradecanoyl phorbol acetate (TPA; Sigma) in 200 μl of acetone each dose for 15 weeks. The number and size of papillomas were determined and registered for each mouse weekly.
Telomere length determination for skin tumor sections.
For Q-FISH, paraffin-embedded tumor (SCC) sections of the indicated genotypes obtained from the DMBA-TPA experiment were deparaffinized and hybridized with a peptide nucleic acid telomeric probe. Telomere fluorescence was determined as described previously (35). More than 420 telomere signals per squamous cell carcinoma (SCC) and at least three independent SCC per genotype were analyzed. The images were captured at a magnification of ×100, using a Leica CTR MIC microscope and a Cohu high-performance charge-coupled device camera. Telomere fluorescence was integrated using spot IOD analysis in the TFL-Telo program (kindly provided by P. Landsdorp, Terry Fox Laboratory, Vancouver, Canada) (59).
Histopathological analysis.
Skin samples and other mouse tissues were recovered after mouse sacrifice, fixed in 10% buffered formalin, dehydrated, and embedded in paraffin. For histopathological analysis, 4-mm sections were deparaffinized and stained with hematoxylin and eosin according to standard procedures. Images were captured with a DP-10 digital camera in an Olympus Vanox microscope at the indicated magnifications.
ESC clonogenic assays.
Five thousand keratinocytes isolated from wild-type and K5TRF1 newborn mice were seeded onto J2-3T3 fibroblasts (105 per well in six-well dishes) previously treated with mitomycin C (MMC) (10 mg/ml for 2 h). Keratinocytes were grown in Cnt-02 medium at 37°C and 5% CO2 for 2 weeks. Cells were then rinsed with phosphate-buffered saline (PBS), fixed with 10% formaldehyde, and stained with 1% rhodamine B (Sigma) to visualize colony formation. For each keratinocyte sample, epidermal stem cell (ESC) clonogenic assays were performed in triplicate. The number and size of colonies were measured for each genotype, and the average numbers of colonies obtained for different ranks of size were calculated.
Telomerase TRAP assay.
Telomerase activity was measured with a modified telomere repeat amplification protocol (TRAP) as described previously (8).
Telomere recombination measurements using CO-FISH.
Chromosome orientation FISH (CO-FISH) was performed as described previously (3, 7, 25), using first a (TTAGGG)7 probe labeled with Cy3 and then a (CCCTAA)7 probe labeled with rhodamine green (Applied Biosystems, Bedford, MA). Telomere fluorescence signals and DAPI-stained chromosome images of each metaphase were captured on a Leitz Leica DMRB fluorescence microscope.
B1-SINE Cobra analysis of global DNA methylation.
Global DNA methylation levels were determined using the B1-SINE Cobra method as previously described (6).
Analysis of genomic subtelomeric DNA methylation.
Methylation of subtelomeric repeats at chromosomes 1 and 2 was determined by bisulfite genomic sequencing as previously described (6). One to nine colonies were sequenced per culture.
Antimitotic drugs and quantification of mitotic index.
Primary keratinocytes were treated overnight with nocodazole (0.2 μg/ml; Sigma, St. Louis, MO) or paclitaxel (Taxol) (33 nmol; Sigma, St. Louis, MO) and fixed in PBS-buffered 4% paraformaldehyde at room temperature for 10 min, followed by permeabilization with PBS-0.1% Triton X-100 for 10 min. Cells were then blocked with 2% bovine serum albumin (Sigma, St. Louis, MO) in PBS for 1 h at room temperature and incubated for 1 h at room temperature with a rabbit anti-phospho-histone H3 (Ser10) antibody (Upstate; 1:500). After being labeled, cells were rinsed with PBS-0.1% Triton X-100 and incubated with anti-rabbit antibody-Alexa 488 (Molecular Probes), and DNA was counterstained with DAPI. The mitotic index was calculated by scoring the ratio of positive cells for phospho-histone H3.
Cytogenetic analysis.
Exponentially growing primary keratinocytes were incubated with 0.1 μg/ml colcemid (Gibco) for 5 to 6 h at 37°C and then fixed with methanol-acetic acid (3:1). For analysis of chromosomal number, at least 65 metaphases per genotype were analyzed by studying the DAPI image.
Immunofluorescence.
See the supplemental material for details of the different immunofluorescence experiments.
RESULTS
Transgenic mice with increased TRF1 expression in epithelial tissues.
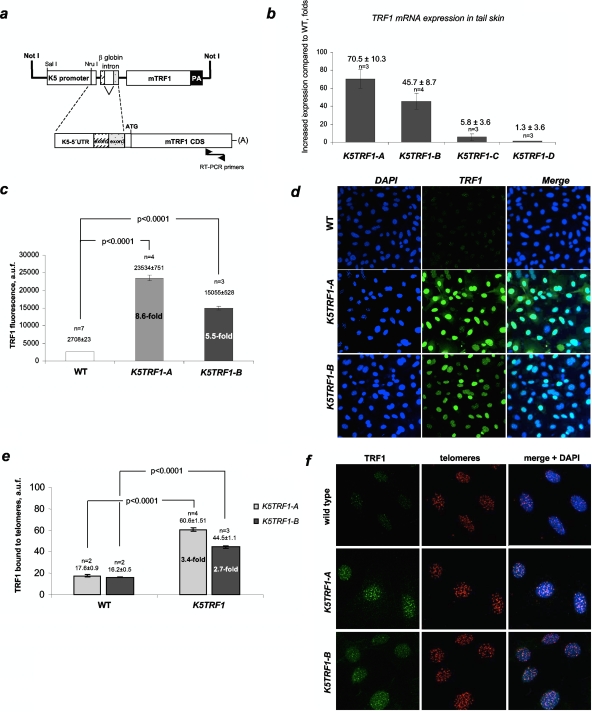
To address the role of TRF1 in telomere biology in the context of the adult organism, we targeted expression of mouse TRF1 to the basal layer and the stem cell compartments of various stratified epithelia, including the skin, by using the K5 promoter (K5TRF1 mice) (Fig. 1a) (38). We obtained four K5TRF1 founder mouse lines with different levels of TRF1 mRNA expression in the skin (Fig. 1b). Mice from the founder line K5TRF1-A showed the highest TRF1 mRNA overexpression (70-fold higher than that in littermate wild-type mice), followed by K5TRF1-B mice (45-fold higher than that in littermate wild-type mice) (Fig. 1b). Two additional mouse founder lines, K5TRF1-C and K5TRF1-D, showed only low levels of TRF1 mRNA overexpression and were not used thereafter (Fig. 1b). In agreement with increased TRF1 mRNA levels, primary keratinocytes derived from K5TRF1-A and K5TRF1-B newborn mice showed ninefold and sixfold increases in nuclear TRF1 protein, respectively, compared with wild-type controls (Fig. 1c and d) (see Materials and Methods). To quantify TRF1 abundance specifically at telomeric regions, chromatin-unbound TRF1 was extracted before performing TRF1 immunofluorescence (see Materials and Methods). We detected 3.5-fold and 2.7-fold increased TRF1 abundances at K5TRF1-A and K5TRF1-B telomeres compared to that in wild-type controls (Fig. 1e and f; see Fig. S1 in the supplemental material for representative images of colocalization of TRF1 with telomeres). A similar, twofold increase in TRF1 density at K5TRF1-A telomeres was detected by using telomere ChIP assays (see Fig. S2a in the supplemental material). Of interest, the densities of histone 3 lysine 9 trimethylation (H3K9-3m) and H3K9 acetylation (AcH3K9) marks at telomeres were not significantly altered in K5TRF1-A mice compared to wild-type controls after normalization by both H3 abundance and input of telomeric DNA (see Fig. S2a in the supplemental material). Both global DNA methylation, measured by DNA methylation of SINE repeats (see Fig. S2b in the supplemental material), and subtelomeric DNA methylation, as determined by bisulfite sequencing of subtelomeric regions in chromosomes 1 and 2 (see Fig. S2c in the supplemental material), were not altered by TRF1 overexpression. This indicates that increased TRF1 density at telomeres does not significantly alter the assembly of these heterochromatic marks.
FIG. 1.
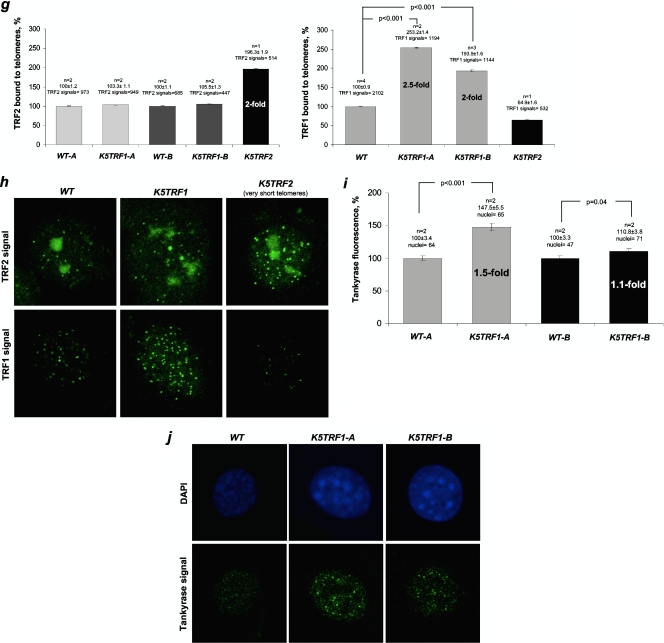
Increased TRF1 expression in K5TRF1 mice. (a) Scheme of the K5TRF1 construct. (b) Quantification of TRF1 mRNA levels in mice from the indicated founder lines. Values represent the increases in TRF1 expression in K5TRF1 mice compared to that in wild-type controls. The number of mice (n) and mean ± standard error (SE) are indicated above each bar. (c) Quantification of TRF1 protein levels in wild-type and K5TRF1 primary keratinocytes by immunofluorescence. The number of keratinocyte cultures (n) and the mean ± SE are indicated above each bar. (d) Representative images of nuclear TRF1 in K5TRF1 keratinocytes. The TRF1-specific signal is shown in green. Note the higher TRF1 immunofluorescence in K5TRF1-A cells than in K5TRF1-B cells. (e) Quantification of TRF1 protein bound to telomeres. Values are expressed as mean ± SE increases in TRF1 levels in transgenic keratinocytes relative to those in nontransgenic keratinocytes. n, number of keratinocyte cultures. (f) Representative images of telomere-bound TRF1 in the indicated genotypes. The TRF1-specific signal is shown in green, telomeric sequences detected by telomere Q-FISH are shown in red, and colocalization is shown in yellow. (g) Telomere-bound TRF2 protein is not increased in K5TRF1 cells. Values are expressed as means ± SE relative to the wild-type level (100%). n, number of keratinocyte cultures. At least 400 TRF2 and 500 TRF1 signals were analyzed per genotype. As a control, K5TRF2 cells showed increased TRF2 bound to telomeres compared to wild-type controls, while telomere-bound TRF1 was not increased in these cells. (h) Representative images of TRF2 and TRF1 protein levels in the indicated genotypes. (Top) TRF2 is shown in green. (Bottom) TRF1 is shown in green. (i) Increased telomere-bound tankyrase 1 protein levels in K5TRF1 cells. Values are expressed as means ± SE relative to the wild-type level (100%). n, number of keratinocyte cultures. At least 45 nuclei were analyzed per genotype. (j) Representative images of tankyrase 1 immunofluorescence in wild-type and K5TRF1 cells. Note the higher tankyrase 1 signal in K5TRF1 cells.
Importantly, the abundance of TRF2 at telomeres was not altered by TRF1 overexpression, as indicated both by ChIP analysis (see Fig. S2a in the supplemental material) and by TRF2 immunofluorescence (Fig. 1g and h). TRF1 immunofluorescence was increased two- to threefold in the same K5TRF1 cells tested for TRF2 expression. As expected, TRF2 immunofluorescence was increased (twofold compared to that in wild-type controls) in cells derived from K5TRF2 mice (Fig. 1g and h) (35). Of interest, telomere-bound TRF1 was decreased in K5TRF2 cells (Fig. 1g and h), maybe as a consequence of very short telomeres in these cells (35). Interestingly, we also detected a significant increase in telomere-bound tankyrase 1 in both K5TRF1-A and K5TRF1-B transgenic lines (Fig. 1i and j), indicating that TRF1 overexpression results in increased loading of tankyrase 1 at mouse telomeres (see Discussion) (49). Taken together, these results indicate a two- to threefold increase in TRF1 abundance at K5TRF1 telomeres, accompanied by an increase of telomere-bound tankyrase 1 but not of TRF2.
Telomere shortening in K5TRF1 epidermis.
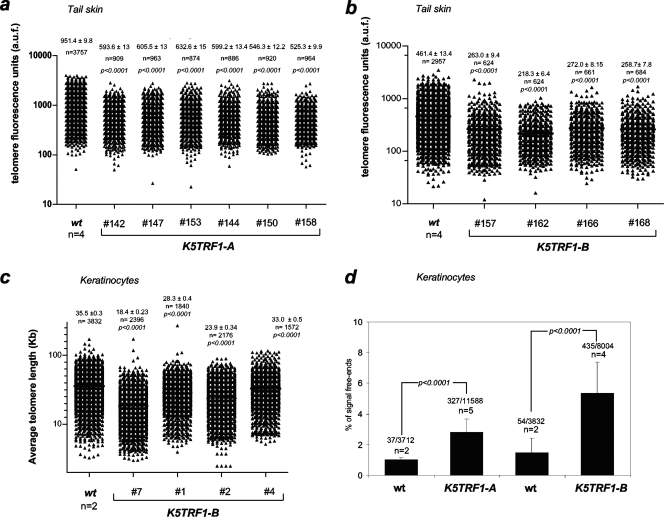
Next, we addressed whether a two- to threefold increase in TRF1 abundance at telomeres was sufficient to impact telomere length in the context of skin homeostasis. For this purpose, we measured telomere length directly in K5TRF1 tail skin sections by using Q-FISH (see Materials and Methods). Adult K5TRF1 mice (2 to 4 months old) from the K5TRF1-A and K5TRF1-B lines showed significantly shorter telomeres than the corresponding age-matched wild-type controls (P < 0.0001 for all comparisons) (Fig. 2a and b). These results were confirmed by conventional metaphase Q-FISH analysis of primary keratinocytes freshly isolated from newborn K5TRF1 and wild-type mice (Fig. 2c and d), as well as by telomere restriction fragment analysis, an independent technique based on Southern blotting (Fig. 2e). In agreement with shorter telomeres in K5TRF1 mice, we also detected a significant increase in signal-free ends (telomeres with undetectable fluorescence by Q-FISH) in both K5TRF1 mouse lines compared to their corresponding wild-type controls (Fig. 2d). Taken together, these results indicate that a two- to threefold increase in the amount of TRF1 bound to telomeres is sufficient to induce a significant telomere shortening in vivo, arguing for an important role for TRF1 in telomere length control in the context of epithelial homeostasis.
FIG. 2.
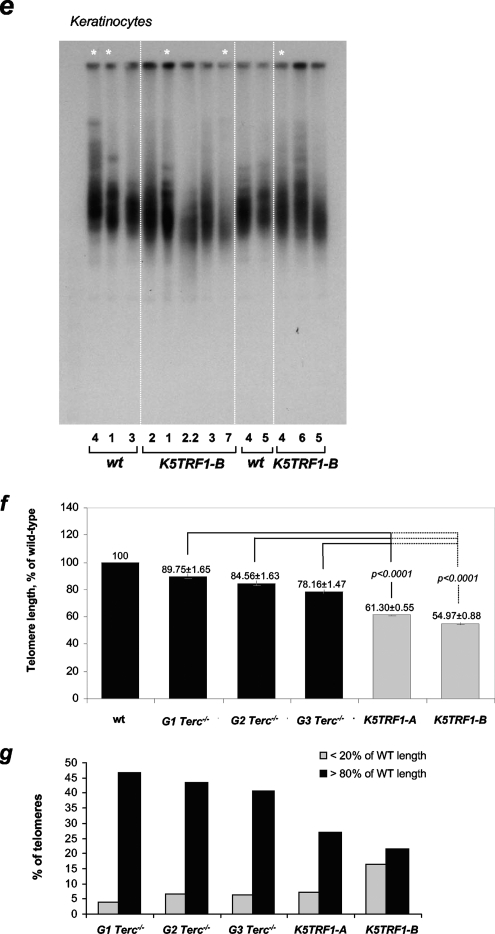
Telomere shortening in K5TRF1 mice. (a) Quantification of telomere fluorescence in tail skin sections from wild-type (wt) (n = 4) and K5TRF1-A (n = 6) mice. Mean ± SE telomere fluorescence (in arbitrary units) and the number of telomere dots analyzed (n) are indicated. (b) Quantification of telomere fluorescence in tail skin sections from wild-type (wt) (n = 4) and K5TRF1-B (n = 4) mice. Mean ± SE telomere fluorescence (in arbitrary units) and the number of telomere dots analyzed (n) are indicated. (c) Quantification of telomere length (kb) in metaphase spreads of primary keratinocytes isolated from wild-type (wt) (n = 2) and K5TRF1-B (n = 4) newborn mice. Mean ± SE telomere length (kb) and the number of telomeres analyzed (n) are shown. (d) The percentages of signal-free ends (telomeres with an undetectable fluorescence signal) in metaphase spreads of wild-type (wt) (n = 2), K5TRF1-A (n = 5), and K5TRF1-B (n = 4) primary keratinocytes are represented with bars. The number of signal-free ends of the total telomere signals analyzed per genotype is indicated above each bar. (e) Telomere length, determined by telomere restriction fragment analysis, in primary keratinocytes isolated from wild-type (wt) and K5TRF1-B newborn mice. Numbers at the bottom identify the cultures. Asterisks indicate keratinocyte cultures also analyzed by Q-FISH for panel c. (f) Comparison of telomere lengths determined by Q-FISH in skin sections from K5TRF1-A and K5TRF1-B mice, as well as from G1, G2, and G3 Terc−/− mice in a C57BL/6 genetic background. The average telomere fluorescence of each genotype is represented relative to that of the corresponding wild-type (wt) control. Telomere length values for K5TRF1-A and -B keratinocytes were obtained from panels a and b. Telomere length values for G1, G2, and G3 Terc−/− keratinocytes were previously described by Blanco et al. (7). Data indicating the mean ± SE and statistical significance are indicated above each bar. (g) Percentages of telomeres showing telomere fluorescence of <20% or >80% that of wild-type controls. Values were obtained as indicated for panel f.
For illustrative purposes, we compared telomere length in the skin of K5TRF1 mice from both founder lines to that of first- to third-generation (G1-G3) telomerase-deficient Terc−/− mice, both in a C57BL/6 genetic background (see Materials and Methods) (23, 26). To minimize possible differences in genetic background, all values are presented relative to telomere length in the corresponding wild-type controls (100%). As shown in Fig. 2f, K5TRF1 mice from the K5TRF1-A and K5TRF1-B lines presented shorter telomeres than G1-G3 Terc−/− mice. Furthermore, K5TRF1 mice showed an increase in the percentage of telomeres shorter than 20% of the wild-type telomere length compared to G1-G3 Terc−/− mice (Fig. 2g). These results indicate that a two- to threefold increase in TRF1 abundance at telomeres produces a more severe and rapid telomere shortening in vivo than that produced by complete telomerase ablation in the context of G1-G3 telomerase-deficient mice.
Normal telomerase activity in K5TRF1 mice.
TRF1 is proposed to act as a telomerase inhibitor in cis by limiting telomerase-mediated telomere elongation (50). To address this, we measured telomerase activity in keratinocytes from K5TRF1 and wild-type mice, using an in vitro TRAP assay (see Materials and Methods). As shown in Fig. S3 in the supplemental material, telomerase TRAP activities were similar in both genotypes, ruling out that increased TRF1 protein could be directly inhibiting the telomerase catalytic activity and in this way leading to telomere shortening. This does not exclude the possibility that TRF1 could inhibit the access of telomerase to the telomere. However, the fact that increased TRF1 abundance at K5TRF1 telomeres had a more severe effect on telomere length than that achieved by complete telomerase ablation during three generations of Terc-deficient mice (Fig. 2f and g) argues that TRF1 regulates telomere length, at least in part, by telomerase-independent mechanisms (see below). These findings indicate that short telomeres in K5TRF1 mice cannot be attributed solely to inhibition of telomerase-mediated telomere elongation by TRF1.
XPF nuclease mediates telomere shortening in K5TRF1 epidermis.
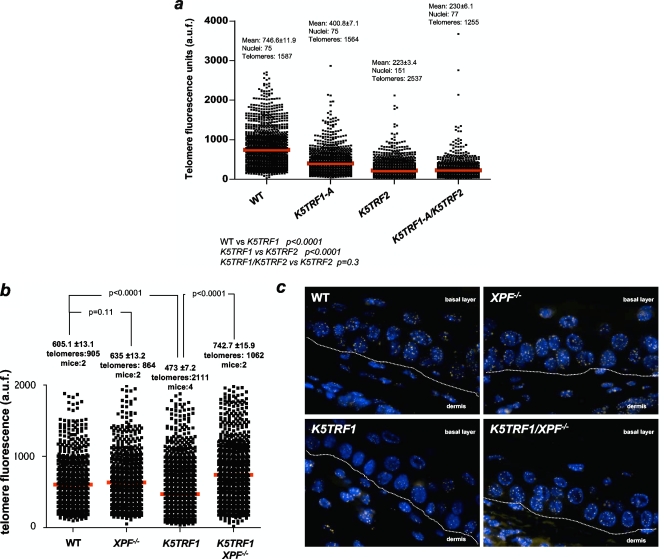
We recently described that a twofold increase in TRF2 abundance at telomeres in K5TRF2 mice results in severe telomere shortening mediated by the XPF/ERCC1 NER nuclease (7, 35) (Fig. 1g and h). Since TRF1 and TRF2 are part of the same protein complex at telomeres (18), we next addressed whether both proteins share the same molecular pathway of telomere length regulation. To this end, we generated double-transgenic K5TRF1/K5TRF2 mice. Interestingly, telomere length in double-transgenic mice was indistinguishable from that in K5TRF2 single transgenics (Fig. 3a), arguing that both proteins are likely to be in the same genetic pathway of telomere length regulation. These results also indicate that the K5TRF2 allele is epistatic over K5TRF1 in inducing telomere shortening, as both proteins were similarly increased (two- to threefold) at telomeres (Fig. 1g and h). Finally, it is relevant that both K5TRF1 and K5TRF2 alleles are epistatic over telomerase function in controlling telomere length, as indicated by the fact that K5TRF1 and K5TRF2 mice showed short telomeres in spite of having an intact telomerase pathway and normal levels of telomerase catalytic activity (Fig. 3a; also see Fig. S3 in the supplemental material).
FIG. 3.
XPF mediates telomere shortening in K5TRF1 epidermis. (a) Mean ± SE telomere fluorescence (in arbitrary units) and the numbers of telomeres and nuclei used are indicated. Mean telomere length is indicated with a red bar. Note that K5TRF1/K5TRF2 double-transgenic mice showed similarly short telomeres to those of K5TRF2 mice, suggesting that the TRF2 allele is dominant over TRF1, as both proteins were similarly overexpressed (twofold) in K5TRF1 and K5TRF2 cells compared to their respective wild-type controls (Fig. 1g). (b) Quantification of telomere fluorescence in tail skin sections from wild-type (WT), XPF−/−, K5TRF1, and double mutant K5TRF1/XPF−/− littermate mice (n = 2 to 4 mice per genotype). At least 864 telomere dots of each genotype were analyzed by Q-FISH. Average fluorescence (in arbitrary units) and SE are shown. Statistical significance is indicated in each comparison. (c) Representative images of telomere fluorescence in tail skin sections from wild-type (WT), XPF−/−, K5TRF1, and double mutant K5TRF1/XPF−/− littermate mice. The dermis and basal layer are indicated and separated by a white line.
Next, we investigated whether XPF deficiency was sufficient to rescue telomere shortening in K5TRF1 epidermis, similarly to that previously described for K5TRF2 mice (35). To this end, we generated K5TRF1/XPF−/− mice (see Materials and Methods). As shown in Fig. 3b and c, XPF deficiency rescued telomere length in K5TRF1/XPF−/− mice to the length in wild-type and XPF−/− controls. These findings indicate that increased TRF1 abundance at telomeres leads to XPF-dependent telomere degradation, further supporting the notion that TRF1 and TRF2 are in the same genetic pathway of telomere length control in mammals.
Moderately increased chromosomal aberrations in K5TRF1 mice.
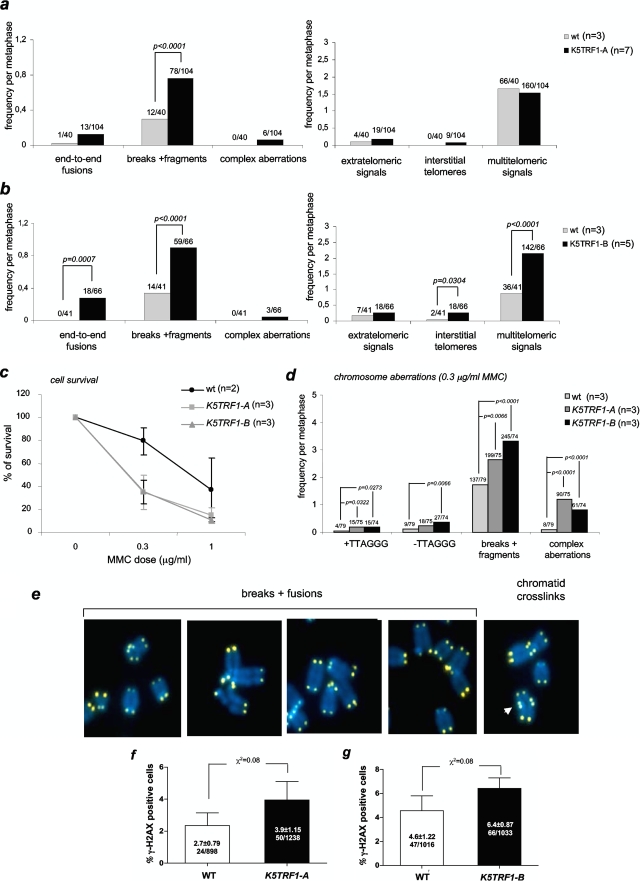
To address whether telomere shortening in K5TRF1 mice triggered chromosomal instability in these mice, we studied chromosomal aberrations involving telomeres by using conventional telomere Q-FISH on metaphase spreads (see Materials and Methods). Karyotypic analysis of primary keratinocytes from both K5TRF1 founder lines indicated a modest increase in the frequency of end-to-end fusions (Fig. 4a and b), suggesting that telomere shortening in TRF1 mice leads to dysfunctional telomeres (Fig. 4a and b). It is relevant to note that increased TRF1 expression also led to a significant increase in chromosome breaks and fragments, as well as other types of chromosomal aberrations previously associated with TRF2 overexpression (7, 35), such as complex chromosomal aberrations, interstitial telomeres, and multitelomeric signals. This further supports the notion that the K5TRF1 allele partially recapitulates K5TRF2 phenotypes. The fact that both K5TRF1 and K5TRF2 cells show complex chromosomal aberrations may be related to their interplay with XPF, known to be involved in NER and cross-link repair.
FIG. 4.
Increased chromosomal instability and MMC hypersensitivity in K5TRF1 cells. (a and b) Quantification of chromosomal aberrations per metaphase in primary keratinocytes isolated from K5TRF1-A (a) and K5TRF1-B (b) mice. Values above each bar indicate the total number of chromosomal aberrations out of the total number of metaphases analyzed. n, number of independent keratinocyte cultures analyzed per genotype. (c) K5TRF1 cells are hypersensitive to MMC treatment. Error bars represent SE. n, number of keratinocyte cultures used. (d) Quantification of chromosomal aberrations after MMC treatment for the indicated genotypes. The frequency of chromosomal aberrations out of the total number of metaphases analyzed is indicated above each bar. The chi-square test was used for statistical significance calculations. n, number of independent keratinocyte cultures examined per genotype. (e) Representative examples of the indicated chromosomal aberrations in K5TRF1 cells. (f and g) Increased γH2AX foci in K5TRF1 skin sections for both K5TRF1-A and K5TRF1-B founder lines. The percentage of γH2AX-positive cells per genotype is indicated. Values are expressed as means ± SE.
K5TRF1 cells are hypersensitive to the DNA cross-linking agent MMC.
The fact that K5TRF1 cells show a significant increase in chromosomal aberrations that are not directly associated with critically short telomeres suggests that these cells may have a global defect in DNA repair, most likely as a consequence of a genetic interaction with XPF at telomeres (Fig. 3b and c). In this regard, K5TRF2 mice have impaired NER, as indicated by an increased sensitivity to UV irradiation and to MMC (35, 58). To specifically address the putative impact of transgenic TRF1 on NER efficiency, we studied the sensitivity of K5TRF1-A and K5TRF1-B primary keratinocytes to treatment with MMC, a DNA cross-linking agent that specifically decreases the viability of ERCC1 or XPF mutant cells but not that of cells deficient in other NER components (42). K5TRF1-A and K5TRF1-B keratinocytes showed decreased viability after treatment with low doses of MMC compared to wild-type controls (Fig. 4c). Furthermore, MMC-treated K5TRF1 keratinocytes from both founder lines showed a higher frequency of chromosome breaks and fragments as well as other complex chromosomal aberrations than did similarly treated wild-type cells (Fig. 4d and e). These observations suggest that K5TRF1 cells have defective repair of MMC-induced DNA cross-links in the presence of normal TRF2 levels, similar to that previously described for both K5TRF2- and XPF/ERCC1-deficient cells (35). As previously hypothesized for K5TRF2 mice, increased TRF1 abundance at telomeres may overrecruit XPF to telomeres, resulting in XPF-mediated telomere shortening and suboptimal NER efficiency at nontelomeric sites (35). It is relevant that in spite of having a similar sensitivity to MMC to that of K5TRF2 mice, K5TRF1 mice do not show the dramatic degenerative skin phenotypes present in K5TRF2 mice, such as severe skin hyperpigmentation, hair loss, and spontaneous cancer in skin areas exposed to the light (35). This difference can be explained by longer telomeres in K5TRF1 mice than in K5TRF2 mice (Fig. 3a).
Increased DNA damage foci in the skin of K5TRF1 mice.
Next, we addressed whether short telomeres and increased chromosomal instability in K5TRF1 mice resulted in increased DNA damage in the skin of these mice. γH2AX foci have previously been shown to mark the presence of DNA double-strand breaks, including those associated with critically short/dysfunctional telomeres (16, 34, 51). We quantified the percentage of basal layer keratinocytes showing γH2AX foci in skin sections. We detected a modest increase in γH2AX-positive cells in the skin of K5TRF1 mice compared to wild-type skin (Fig. 4f and g), suggesting that TRF1 overexpression results in a slight increase in γH2AX-positive cells in the skin of these mice, similar to that previously shown for K5TRF2 epidermis (35).
K5TRF1 keratinocytes show increased telomere recombination.
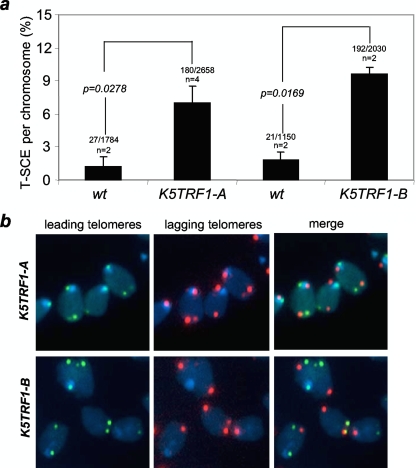
K5TRF2 mice show an increased frequency of sister chromatid exchange within telomeric DNA (T-SCE) (35), suggesting a role for TRF2 in regulating homologous recombination between sister telomeric sequences (20, 36). To address this possibility in TRF1-overexpressing mice, we determined the frequency of T-SCE events in primary keratinocytes from mice with different genotypes by two-color chromosome orientation CO-FISH (3, 5). The strand-specific nature of CO-FISH typically yields two telomeric signals of each color (red, lagging strand; green, leading strand) per chromosome in the absence of recombination events. T-SCE leads to a mixture of red and green fluorescence (7, 25), detected by the leading and lagging telomere probes (Fig. 5a and b). K5TRF1-A and K5TRF1-B cells showed significantly increased T-SCE frequencies compared to wild-type cells (P < 0.03 in both cases) (Fig. 5a), suggesting that increased TRF1 abundance at telomeres results in derepression of homologous recombination between sister telomeres. These results further support the notion that TRF1 and TRF2 are in a similar pathway for telomere length control. Notably, ablation of Pot1, a component of shelterin, also results in increased T-SCE frequencies (55), suggesting a more general role for shelterin in controlling DNA recombination at telomeres. We have previously shown that defects in both DNA methylation and histone trimethylation (H3K9 and H4K20 marks) are accompanied by increased T-SCE events at telomeres (6, 25), suggesting the intriguing possibility that increased TRF1 expression may indirectly affect telomere recombination by altering the status of these epigenetic marks. However, as previously discussed, we did not detect changes in global or subtelomeric DNA methylation in K5TRF1 cells (see Fig S2b and c in the supplemental material). Similarly, we did not detect significant changes in H3K93me and AcH3K9 marks at K5TRF1 telomeres, as determined by ChIP (see Fig. S2a in the supplemental material). Together, these results suggest that TRF1 affects telomere recombination independently of the status of these epigenetic marks.
FIG. 5.
Increased telomere recombination in K5TRF1 keratinocytes. (a) Quantification of frequency of T-SCE events in wild-type (wt), K5TRF1-A, and K5TRF1-B keratinocytes analyzed by CO-FISH. The number of T-SCE events and the total number of analyzed chromosome are indicated on top of each bar. n, number of independent keratinocyte cultures. Statistical significance, calculated by the chi-square test, is shown. (b) Representative CO-FISH images of metaphases of the indicated genotypes hybridized with specific probes against leading (green fluorescence) and lagging (red fluorescence) telomeres.
Decreased abundance of TelRNAs (or TERRAs) associated with shorter telomeres in K5TRF1 and K5TRF2 mice.
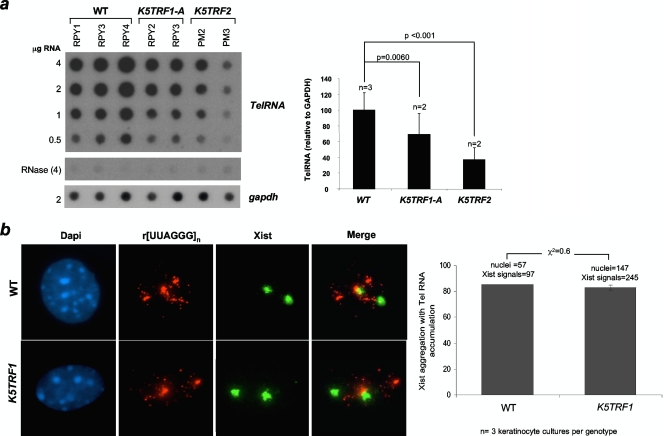
It was recently reported that telomeres are transcribed by the DNA-dependent RNA Pol II (47), generating long (UUAGGG)n-containing RNAs that remain associated with the telomeric chromatin (2, 47). TRF1 has been proposed to support telomere transcription through direct interaction with RNA Pol II (47), and the abundance of telomere transcripts has been proposed to be proportional to the length of telomeric repeats (47). In agreement with shorter telomeres in K5TRF1 mice, we observed decreased telomere transcription in skin keratinocytes derived from K5TRF1 transgenic mice compared to that in wild-type controls (Fig. 6a). Furthermore, telomere transcripts in these cells showed a normal dotted nuclear distribution, including their accumulation near the inactive X chromosome in female cells (Fig. 6b). Interestingly, K5TRF2 cells showed a further decrease in the abundance of telomere transcripts compared to K5TRF1 mice, in agreement with the fact that K5TRF2 cells had shorter telomeres (Fig. 6a). It is interesting that increased TRF1 occupancy of telomeres cannot rescue reduced transcription of short telomeres in K5TRF1 keratinocytes (47). This suggests that control of telomeric transcription by telomere length is dominant over the function of TRF1 in promoting the generation of TelRNAs (47). Together, these results suggest that shorter telomeres in K5TRF1 and K5TRF2 mice produce fewer telomeric transcripts than do wild-type telomeres, in accordance with a positive correlation between telomere length and telomere transcription (47).
FIG. 6.
Reduced telomere transcription in K5TRF1 and K5TRF2 cells. (a) Quantification of TelRNA levels. Average values and standard deviations were obtained for two or three independent keratinocyte cultures. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (b) Telomere RNAs accumulate near the Xist RNA marking the inactive X chromosome in both wild-type (WT) and K5TRF1 cells.
Slightly decreased clonogenic activity of K5TRF1 ESCs.
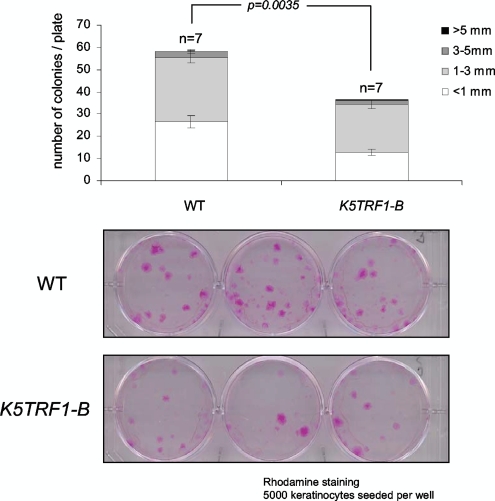
Short telomeres in telomerase-deficient mice lead to defective mobilization and clonogenic activity of ESCs, which are already noticeable at the first (G1) Terc−/− mouse generation (22, 48). Since K5TRF1 mice show an important reduction in average telomere length in the skin compared to their wild-type counterparts, we set out to study the impact of TRF1 overexpression on ESCs. To this end, we determined the clonogenic activities of primary keratinocytes derived from newborn wild-type and K5TRF1 mice (see Materials and Methods). The number and size of colonies in this assay have been proposed to reflect the ability of individual ESCs to proliferate in vitro (4). K5TRF1 mice showed a reduced clonogenic activity compared to wild-type cells (Fig. 7), suggesting a defective ability of K5TRF1 ESCs to proliferate and to form macroscopic colonies. This is in agreement with shorter telomeres and increased chromosomal instability in these cells.
FIG. 7.
Decreased clonogenic activity of K5TRF1 ESCs. The graph shows quantification of the size and number of macroscopic colonies obtained from keratinocytes isolated from wild-type (WT) and K5TRF1-B newborn mice. n, number of independent keratinocyte cultures. Representative examples of clones obtained from the indicated genotypes are shown in the bottom panel. Clones were visualized by staining with rhodamine.
This mild defect in ESC clonogenic activity, however, was not associated with severe skin abnormalities in K5TRF1 mice. These results are in contrast with a dramatic impact of TRF2 overexpression on both skin aging and skin cancer (7, 35). This difference may be due to longer telomeres in K5TRF1 cells than in K5TRF2 cells, thus highlighting the importance of short telomeres in the pathobiology of these diseases. Therefore, although both telomeric proteins are part of the same complex at telomeres, TRF2 is more potent than TRF1 in leading to dramatic telomere loss, which in turn severely impacts cancer and aging.
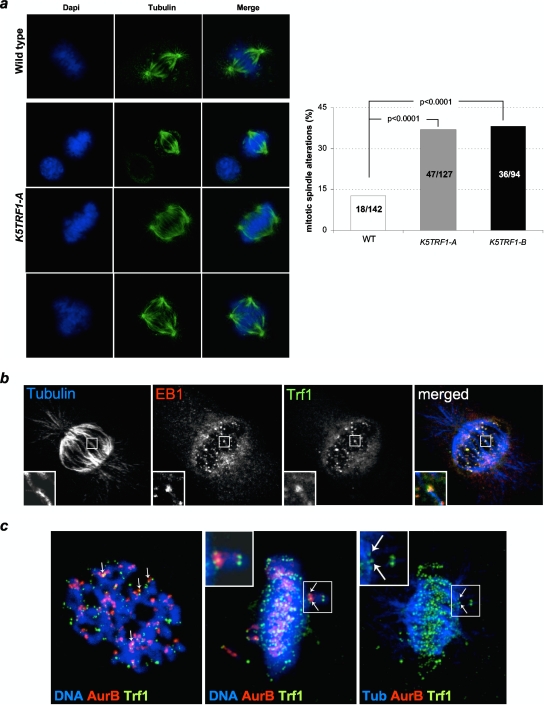
K5TRF1 but not K5TRF2 cells show aberrant mitotic spindles.
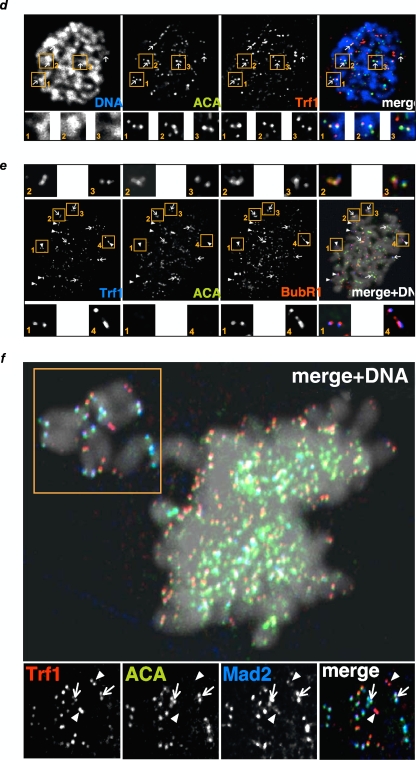
In addition to its telomeric roles, TRF1 has been suggested to be part of the mitotic spindle checkpoint (40, 43). To address whether K5TRF1 mice showed any defects in spindle formation during mitosis, we first studied the percentage of K5TRF1 keratinocytes showing mitotic spindle defects compared to wild-type controls. To this end, we performed confocal microscopy with TRF1 and alpha-tubulin antibodies to visualize the mitotic spindle (Fig. 8a; see Fig. S4a in the supplemental material). K5TRF1 cells showed significantly increased mitotic spindle defects, including multipolar spindles and misaligned chromosomes, compared to wild-type controls (Fig. 8a). It is important to note that we did not find colocalization of TRF1 with microtubules (alpha-tubulin) in any of the images analyzed for either wild-type or K5TRF1 cells (see Fig. S4a in the supplemental material). Instead, TRF1 was always located at chromosomes in all cases studied, even in cells with multipolar mitotic spindles (see Fig. S4a in the supplemental material). To analyze the specific localization of TRF1 at chromosomes, we costained TRF1 with different markers of centromeres (ACA or Aurora B), the plus end of microtubules (EB1), and the SAC proteins BubR1 and Mad2. As depicted in Fig. 8b to f, TRF1 partially colocalized with EB1, ACA, or Aurora B at the short p arms but not at the telomeric regions of the long q arms of chromosomes, probably as a consequence of the close proximity of centromeres and telomeres in murine chromosomal p arms. Similarly, the SAC regulator BubR1 colocalized with ACA and TRF1 at the p arms of the chromosomes (Fig. 8e). Interestingly, BubR1 also colocalized with TRF1 at the telomere region of the long q arms, where centromeric staining (ACA) was absent (Fig. 8e). In an analogous manner, Mad2, the critical switch of SAC activity, also colocalized with TRF1 (Fig. 8f), especially in nonaligned chromosomes, suggesting a possible link between TRF1 and deregulation of the SAC.
FIG. 8.
TRF1 colocalizes with SAC proteins in K5TRF1 cells. (a) Representative images of aberrant mitotic spindles in wild-type (WT) and K5TRF1 primary keratinocytes. α-Tubulin is shown in green. The graph shows the percentages of aberrant mitotic spindles in wild-type and K5TRF1 primary keratinocytes. The number of aberrant mitotic spindles and the total number of spindles analyzed are indicated in each bar. (b to d) TRF1 colocalizes with SAC proteins in K5TRF1 chromosomes. Colocalization was seen between TRF1 and EB1 (plus end of microtubules close to kinetochores) (b), TRF1 and Aurora B (kinetochores) (c), and TRF1 and the centromere antigen ACA (d). (e) The SAC protein BubR1 colocalizes with TRF1 at both centromeres and p-arm telomeres (panels 2 and 3), as well as at the q-arm telomeres (panels 1 and 4), where ACA is absent. (f) Colocalization of TRF1 and Mad2. Some misaligned chromosomes (square) in K5TRF1 keratinocytes maintain Mad2 staining that colocalizes with TRF1, not only at the p arms, where ACA is positive (arrows), but also at the q-arm telomeres, which are negative for ACA (arrowheads).
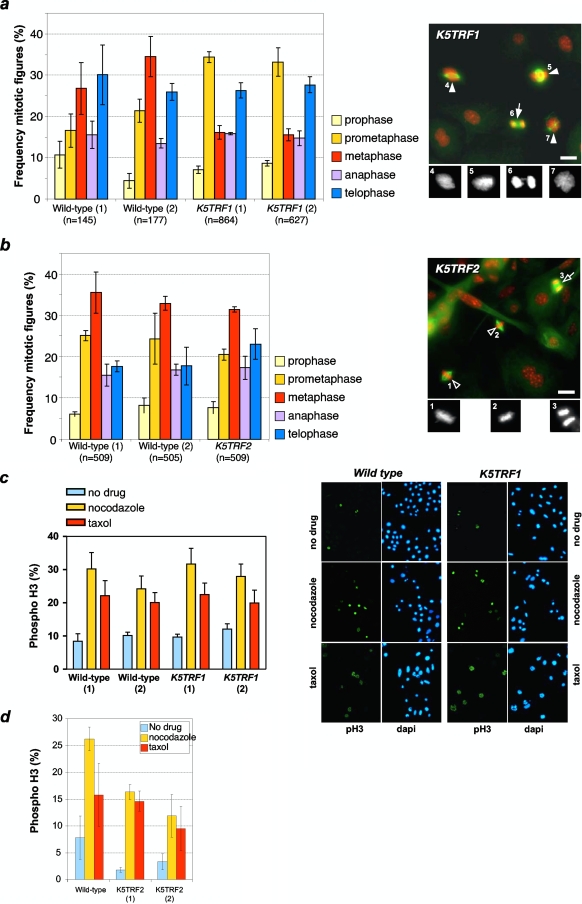
To specifically address this possibility, we determined the frequencies of different mitotic figures in wild-type and K5TRF1 cells. As shown in Fig. 9a, K5TRF1 cells displayed an abnormally high frequency of cells in prometaphase, accompanied by a decreased frequency of cells in metaphase. Furthermore, K5TRF1 cells showed misaligned chromosomes at the metaphasic plate, abnormal spindles, and the presence of anaphase bridges (see examples in the right panel of Fig. 9a), suggesting that TRF1 overexpression results in defective mitosis. Importantly, this was not observed in K5TRF2 cells (Fig. 9b), indicating that TRF1 overexpression, but not TRF2 overexpression, induces mitotic abnormalities and the subsequent activation of the SAC. Polyploidy was not significantly increased in cultured K5TRF1 keratinocytes compared to the wild-type controls (see Fig. S4b in the supplemental material), suggesting that the K5TRF1 mitotic defects cannot be attributed to increased polyploidy.
FIG. 9.
Transgenic TRF1 expression induces aberrant mitotic spindles. (a) (Left) Quantification of the five different mitotic phases (prophase, prometaphase, metaphase, anaphase, and telophase). Wild-type and K5TRF1 keratinocytes were stained for α-tubulin, phospho-histone H3, and DNA (DAPI), and mitotic cells were counted in three different experiments. K5TRF1 cells showed an increased number of prometaphases compared to wild-type cells. (Right) K5TRF1 cells showed dramatic mitotic aberrations, with aberrant metaphase plates (closed arrowheads) and chromosome bridges (closed arrow). Cells were stained for α-tubulin (green) and DNA (red). The bottom panels are a detailed representation of the DAPI staining. Bar, 20 μm. Numbers are for identification of individual mitoses. (b) (Left) Quantification of the five different mitotic phases (prophase, prometaphase, metaphase, anaphase, and telophase). Wild-type and K5TRF2 keratinocytes were stained for α-tubulin, phospho-histone H3, and DNA (DAPI), and mitotic cells were counted in three different experiments. Unlike K5TRF1 cells, K5TRF2 cells did not show any difference from the wild-type cells. (Right) In accordance with the previous observation, K5TRF2 metaphases were normal (open arrowheads) and anaphases did not show chromosome bridges (open arrow). Cells were stained for α-tubulin (green) and DNA (red). The bottom panels are a detailed representation of the DAPI staining. Bar, 20 μm. Numbers are for identification of individual mitoses. (c) K5TRF1 keratinocytes display an efficient mitotic assembly checkpoint in the presence of paclitaxel (Taxol) or nocodazole. (Left) Percentages of phospho-H3-positive cells after the indicated treatments. Numbers in parentheses refer to independent keratinocyte cultures. (Right) Representative images of wild-type and K5TRF1 phospho-H3-positive keratinocytes. (d) K5TRF2 cells have a robust spindle assembly checkpoint. When K5TRF2 cells were poisoned with either paclitaxel (Taxol) or nocodazole, they had a three- to fivefold increase in the mitotic index due to SAC activation.
Finally, it is important to note that these mitotic defects did not result in significant functional defects in the induction of the SAC in the presence of paclitaxel or nocodazole (Fig. 9c). Similarly, K5TRF2 cells also showed a normal sensitivity to the same mitotic poisons (Fig. 9d).
Moderately increased skin cancer and aging in K5TRF1 mice.
Since K5TRF1 mice partially phenocopy some of the defects previously described for K5TRF2 mice, such as XPF-mediated telomere shortening, increased end-to-end chromosome fusions, increased multitelomeric signals, increased complex chromosomal aberrations, and hypersensitivity to MMC, as well as increased telomere recombination, we next addressed whether K5TRF1 mice also showed increased cancer susceptibility upon skin carcinogenic treatments. To this end, we treated the skin with the carcinogen DMBA, followed by weakly treatments with the tumor promoter TPA, which results in the appearance of skin papillomas and, at later times, skin carcinomas (7). As shown in Fig. S5a to c in the supplemental material, K5TRF1 mice showed a slightly increased incidence of papilloma formation following the DMBA-plus-TPA treatment, as well as a slight increase in malignant skin tumors (SCC and BCC) (see Fig. S5c and f in the supplemental material). Interestingly, as shown in Fig. S5d in the supplemental material, we observed that TRF1 expression tended to be downregulated in DMBA-TPA-induced K5TRF1 tumors compared to normal tissue, suggesting a selection of cells with lower TRF1 expression during tumorigenesis, something that may favor telomere elongation within the tumor. To address this, we compared the telomere lengths of three independent wild-type and K5TRF1 SCC tumors. As shown in Fig. S5e in the supplemental material, wild-type SCC showed a quite homogeneous telomere length whereas K5TRF1 SCC were very heterogeneous in length, with some tumors showing very short telomeres and others showing longer telomeres than the wild-type counterparts. These results are in agreement with TRF1 overexpression leading to telomere shortening at the same time that it may favor activation of telomere recombination mechanisms. TRF1 downregulation in K5TRF1 tumors (see Fig. S5d in the supplemental material) may also favor telomere reelongation in some of these tumors. Finally, K5TRF1 mice showed a lifespan similar to that of wild-type mice (not shown) and a slight increase in degenerative skin lesions (atrophies, ulcers, and preneoplastic lesions) at the time of death compared to the wild-type controls (see Fig. S5g in the supplemental material), in agreement with shorter telomeres and increased DNA damage in the skin of K5TRF1 mice.
DISCUSSION
TRF1 and its distant homologue TRF2 are central components of the shelterin complex at mammalian telomeres. Both TRF1 and TRF2 bind directly to double-stranded DNA telomeric repeats and differentially recruit to telomeres a number of additional shelterin components, as well as other telomere-interacting proteins. Mounting evidence coming from overexpression of wild-type and dominant-negative TRF1 alleles in human cultured cells suggests that TRF1 is a key regulator of telomere length (18). However, the demonstration of a role of TRF1 or its interacting factor tankyrase (12, 19, 37, 49) in telomere maintenance in the context of mammalian organisms has been lacking to date. Mice genetically ablated for TRF1 are embryonically lethal but show a normal telomere length and normal telomere protection, arguing that TRF1 is not essential for telomere length regulation in early embryonic development. Similarly, mice that lack tankyrase also show a normal telomere length and telomere capping (15, 27).
Here we show that transgenic TRF1 expression induces a significant telomere shortening in the epidermis in two independent K5TRF1 transgenic mouse lines (K5TRF1-A and K5TRF1-B), demonstrating that a two- to threefold increase in the amount of TRF1 bound to telomeres is sufficient to provoke substantial telomere shortening in vivo. Strikingly, K5TRF1 transgenic expression had a more severe effect on telomere length than that achieved by complete ablation of telomerase activity during three mouse generations (G1-G3 Terc−/− mice), arguing that the effects of TRF1 on telomere length are at least partially independent of telomerase activity (50). In this regard, we show here that telomere shortening provoked by TRF1 overexpression is mediated by the XPF nuclease, previously shown to be involved in degrading telomeres as a consequence of increased TRF2 expression in mice (35). Indeed, telomere phenotypes in K5TRF1 mice are reminiscent of those previously described for K5TRF2 mice, including telomere shortening, increased chromosomal aberrations, increased sensitivity to MMC, and increased telomere recombination, suggesting that both shelterin components may share, at least partially, the same genetic pathway of telomere length regulation. Notably, telomere shortening was considerably more severe in K5TRF2 mice than in K5TRF1 mice, even though both proteins were overexpressed to similar levels, indicating that TRF2 is epistatic over TRF1 in negatively regulating telomere length. Consistent with this, while TRF2 overexpression resulted in very severe degenerative skin phenotypes (hair loss, skin dryness, and hyperpigmentation), as well as spontaneous skin cancer, TRF1 transgenic mice showed only modest increases in degenerative skin lesions and chemically induced skin tumorigenesis, highlighting the importance of telomere shortening in the pathogenesis of these diseases. Finally, the fact that K5TRF1 mice have a normal TRF2 abundance at telomeres suggests that the effects of TRF1 overexpression are not likely to be mediated indirectly by deregulated TRF2 expression in these mice. Interestingly, we observed a slight increase in tankyrase 1 binding to K5TRF1 telomeres in spite of the fact that the acidic domain of mouse TRF1 does not bind to mouse tankyrase 1 (28), suggesting that other domains in TRF1 may recruit tankyrase 1 to mouse telomeres. Taken together, the results shown here support a model in which increased TRF1 expression results in abnormal XPF activity at telomeres, thus leading to increased telomere shortening and suboptimal NER efficiency at nontelomeric sites. Intriguingly, a direct interaction between TRF1 and NER proteins has not been found to date (58).
We recently proposed that the abundance of TelRNAs (or TERRAs) is positively correlated with the length of telomere repeats, suggesting that they may be involved in telomere length regulation (47). In agreement with this notion, K5TRF1 mice showed a decreased abundance of telomere transcripts compared to wild-type controls, and this was further aggravated in K5TRF2 mice, which presented even shorter telomeres.
In addition to telomere length defects in K5TRF1 mice, we also found increased mitotic defects in cells derived from these mice. Furthermore, we found that TRF1 colocalizes with the SAC proteins BubR1 and Mad2 at mouse telomeres. In particular, K5TRF1 cells showed a clear colocalization of TRF1 and BubR1 at both the p-arm and q-arm telomeres. In addition, Mad2, a critical regulator of SAC inactivation, also colocalized with TRF1 at mouse telomeres. This is in agreement with previous findings showing a TRF1-Mad1 interaction in human cells (43). In contrast to K5TRF1 cells, K5TRF2 cells did not show mitotic defects, suggesting a putative role for TRF1 in spindle formation or chromosome alignment. Together, these findings suggest that an increase of TRF1 at mouse telomeres may directly or indirectly sustain the SAC response by interfering with BubR1 and Mad2. Similarly, we cannot rule out that increased tankyrase amounts at K5TRF1 telomeres could be interfering with SAC, as tankyrase has also been proposed to regulate spindle poles (28). These results are in line with recent findings showing that BubR1 localizes to unprotected telomeres in Drosophila (39). It is tempting to speculate that increased TRF1 recruitment at K5TRF1 telomeres may lead to a similar deregulation of SAC proteins and to mitotic spindle defects.
Supplementary Material
Acknowledgments
M. A. Blasco's laboratory is funded by the Spanish Ministry of Innovation and Science, the Regional Government of Madrid, the European Union, and the Spanish Association Against Cancer (AECC). M. Malumbres's laboratory is funded by the Spanish Ministry of Health, the Spanish Ministry of Innovation and Science, the Consolider-Ingenio 2010 Programme (to both M.M. and M.A.B.), and the Regional Government of Madrid.
Footnotes
Published ahead of print on 5 January 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 223474-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzalin, C. M., P. Reichenbach, L. Khoriauli, E. Giulotto, and J. Lingner. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318798-801. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, S. M., M. A. Brenneman, and E. H. Goodwin. 2004. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 323743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrandon, Y., and H. Green. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 842302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechter, O. E., Y. Zou, W. Walker, W. E. Wright, and J. W. Shay. 2004. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 643444-3451. [DOI] [PubMed] [Google Scholar]
- 6.Benetti, R., S. Gonzalo, I. Jaco, G. Schotta, P. Klatt, T. Jenuwein, and M. A. Blasco. 2007. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 178925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco, R., P. Munoz, J. M. Flores, P. Klatt, and M. A. Blasco. 2007. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 21206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 9125-34. [DOI] [PubMed] [Google Scholar]
- 9.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17231-235. [DOI] [PubMed] [Google Scholar]
- 10.Canudas, S., B. R. Houghtaling, J. Y. Kim, J. N. Dynek, W. G. Chang, and S. Smith. 2007. Protein requirements for sister telomere association in human cells. EMBO J. 264867-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celli, G. B., and T. de Lange. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7712-718. [DOI] [PubMed] [Google Scholar]
- 12.Cook, B. D., J. N. Dynek, W. Chang, G. Shostak, and S. Smith. 2002. Role for the related poly(ADP-ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, S. W., and E. H. Blackburn. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21553-563. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., Y. Yang, M. van Overbeek, J. R. Donigian, P. Baciu, T. de Lange, and M. Lei. 2008. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 3191092-1096. [DOI] [PubMed] [Google Scholar]
- 15.Chiang, Y. J., M. L. Nguyen, S. Gurunathan, P. Kaminker, L. Tessarollo, J. Campisi, and R. J. Hodes. 2006. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol. Cell. Biol. 262037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d'Adda di Fagagna, F., P. M. Reaper, L. Clay-Farrace, H. Fiegler, P. Carr, T. Von Zglinicki, G. Saretzki, N. P. Carter, and S. P. Jackson. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426194-198. [DOI] [PubMed] [Google Scholar]
- 17.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13768-785. [DOI] [PubMed] [Google Scholar]
- 18.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 19.Donigian, J. R., and T. de Lange. 2007. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J. Biol. Chem. 28222662-22667. [DOI] [PubMed] [Google Scholar]
- 20.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26447-450. [DOI] [PubMed] [Google Scholar]
- 21.Fairall, L., L. Chapman, H. Moss, T. de Lange, and D. Rhodes. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8351-361. [DOI] [PubMed] [Google Scholar]
- 22.Flores, I., M. L. Cayuela, and M. A. Blasco. 2005. Effects of telomerase and telomere length on epidermal stem cell behavior. Science 3091253-1256. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Cao, I., M. Garcia-Cao, A. Tomas-Loba, J. Martin-Caballero, J. M. Flores, P. Klatt, M. A. Blasco, and M. Serrano. 2006. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 7546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Gonzalo, S., I. Jaco, M. F. Fraga, T. Chen, E. Li, M. Esteller, and M. A. Blasco. 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8416-424. [DOI] [PubMed] [Google Scholar]
- 26.Herrera, E., E. Samper, J. Martin-Caballero, J. M. Flores, H. W. Lee, and M. A. Blasco. 1999. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 182950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao, S. J., M. F. Poitras, B. D. Cook, Y. Liu, and S. Smith. 2006. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol. Cell. Biol. 262044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao, S. J., and S. Smith. 2008. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 9083-92. [DOI] [PubMed] [Google Scholar]
- 29.Karlseder, J., L. Kachatrian, H. Takai, K. Mercer, S. Hingorani, T. Jacks, and T. de Lange. 2003. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 236533-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koering, C. E., A. Pollice, M. P. Zibella, S. Bauwens, A. Puisieux, M. Brunori, C. Brun, L. Martins, L. Sabatier, J. F. Pulitzer, and E. Gilson. 2002. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 31055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, D., M. S. O'Connor, J. Qin, and Z. Songyang. 2004. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 27951338-51342. [DOI] [PubMed] [Google Scholar]
- 32.Loayza, D., and T. De Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 4231013-1018. [DOI] [PubMed] [Google Scholar]
- 33.Matsutani, N., H. Yokozaki, E. Tahara, H. Tahara, H. Kuniyasu, K. Haruma, K. Chayama, and W. Yasui. 2001. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int. J. Oncol. 19507-512. [PubMed] [Google Scholar]
- 34.Modesti, M., and R. Kanaar. 2001. DNA repair: spot(light)s on chromatin. Curr. Biol. 11R229-R232. [DOI] [PubMed] [Google Scholar]
- 35.Munoz, P., R. Blanco, J. M. Flores, and M. A. Blasco. 2005. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 371063-1071. [DOI] [PubMed] [Google Scholar]
- 36.Muntoni, A., and R. R. Reddel. 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14(Spec. No. 2)R191-R196. [DOI] [PubMed] [Google Scholar]
- 37.Muramatsu, Y., T. Ohishi, M. Sakamoto, T. Tsuruo, and H. Seimiya. 2007. Cross-species difference in telomeric function of tankyrase 1. Cancer Sci. 98850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murillas, R., F. Larcher, C. J. Conti, M. Santos, A. Ullrich, and J. L. Jorcano. 1995. Expression of a dominant negative mutant of epidermal growth factor receptor in the epidermis of transgenic mice elicits striking alterations in hair follicle development and skin structure. EMBO J. 145216-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musaro, M., L. Ciapponi, B. Fasulo, M. Gatti, and G. Cenci. 2008. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat. Genet. 40362-366. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura, M., X. Z. Zhou, S. Kishi, I. Kosugi, Y. Tsutsui, and K. P. Lu. 2001. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr. Biol. 111512-1516. [DOI] [PubMed] [Google Scholar]
- 41.Oh, B. K., Y. J. Kim, C. Park, and Y. N. Park. 2005. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 16673-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petit, C., and A. Sancar. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 8115-25. [DOI] [PubMed] [Google Scholar]
- 43.Prime, G., and D. Markie. 2005. The telomere repeat binding protein Trf1 interacts with the spindle checkpoint protein Mad1 and Nek2 mitotic kinase. Cell Cycle 4121-124. [DOI] [PubMed] [Google Scholar]
- 44.Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul, and M. A. Blasco. 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savage, S. A., R. T. Calado, Z. T. Xin, H. Ly, N. S. Young, and S. J. Chanock. 2006. Genetic variation in telomeric repeat binding factors 1 and 2 in aplastic anemia. Exp. Hematol. 34664-671. [DOI] [PubMed] [Google Scholar]
- 46.Savage, S. A., N. Giri, G. M. Baerlocher, N. Orr, P. M. Lansdorp, and B. P. Alter. 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoeftner, S., and M. A. Blasco. 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10228-236. [DOI] [PubMed] [Google Scholar]
- 48.Siegl-Cachedenier, I., I. Flores, P. Klatt, and M. A. Blasco. 2007. Telomerase reverses epidermal hair follicle stem cell defects and loss of long-term survival associated with critically short telomeres. J. Cell Biol. 179277-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, S., I. Giriat, A. Schmitt, and T. de Lange. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 2821484-1487. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 201659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 131549-1556. [DOI] [PubMed] [Google Scholar]
- 52.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385740-743. [DOI] [PubMed] [Google Scholar]
- 53.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92401-413. [DOI] [PubMed] [Google Scholar]
- 54.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119355-368. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L., A. S. Multani, H. He, W. Cosme-Blanco, Y. Deng, J. M. Deng, O. Bachilo, S. Pathak, H. Tahara, S. M. Bailey, R. R. Behringer, and S. Chang. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 12649-62. [DOI] [PubMed] [Google Scholar]
- 56.Xin, H., D. Liu, M. Wan, A. Safari, H. Kim, W. Sun, M. S. O'Connor, and Z. Songyang. 2007. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445559-562. [DOI] [PubMed] [Google Scholar]
- 57.Ye, J. Z., D. Hockemeyer, A. N. Krutchinsky, D. Loayza, S. M. Hooper, B. T. Chait, and T. de Lange. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 181649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, X. D., L. Niedernhofer, B. Kuster, M. Mann, J. H. Hoeijmakers, and T. de Lange. 2003. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 121489-1498. [DOI] [PubMed] [Google Scholar]
- 59.Zijlmans, J. M., U. M. Martens, S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large interchromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 947423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.