Abstract
The receptor for activated C-kinase (RACK1), a conserved protein implicated in numerous signaling pathways, is a stoichiometric component of eukaryotic ribosomes located on the head of the 40S ribosomal subunit. To test the hypothesis that ribosome association is central to the function of RACK1 in vivo, we determined the 2.1-Å crystal structure of RACK1 from Saccharomyces cerevisiae (Asc1p) and used it to design eight mutant versions of RACK1 to assess roles in ribosome binding and in vivo function. Conserved charged amino acids on one side of the β-propeller structure were found to confer most of the 40S subunit binding affinity, whereas an adjacent conserved and structured loop had little effect on RACK1-ribosome association. Yeast mutations that confer moderate to strong defects in ribosome binding mimic some phenotypes of a RACK1 deletion strain, including increased sensitivity to drugs affecting cell wall biosynthesis and translation elongation. Furthermore, disruption of RACK1's position at the 40S ribosomal subunit results in the failure of the mRNA binding protein Scp160 to associate with actively translating ribosomes. These results provide the first direct evidence that RACK1 functions from the ribosome, implying a physical link between the eukaryotic ribosome and cell signaling pathways in vivo.
Cells alter protein synthesis in response to stimuli whose effects are transmitted through established cell signaling pathways. Although the mechanisms of signal transduction to ribosomes remain unclear, the receptor for activated C-kinase (RACK1) has emerged as a possible molecular link that connects the signaling and translation machinery. RACK1, a highly conserved homologue of the β-subunit of heterotrimeric G proteins, was first identified over a decade ago as an anchoring protein for protein kinase C (33). Implicated as a scaffold in PDE4D5- and Src kinase-based signaling pathways (28), it functions in diverse developmental processes, such as sexual differentiation in Schizosaccharomyces pombe (29) and the control of cell proliferation in Drosophila melanogaster (26). The more recent discovery that RACK1 is a core component of the eukaryotic 40S ribosomal subunit (20, 24, 32) suggested that its signaling functions might directly influence the efficiency and specificity of translation.
In support of this possibility, cryo-electron microscopy (cryo-EM) studies showed that RACK1 binds the 40S subunit near the mRNA exit tunnel in a location that is conserved from yeast to humans (35). The cryo-EM data verified RACK1's architecture as a seven-bladed β-propeller and positioned the protein on the ribosome in such a way that much of its surface is exposed and available for interaction with other proteins and ligands. These structural data are consistent with the hypothesis that RACK1 might assemble signaling or other regulatory complexes directly on the ribosome (31). Indeed, various functions for RACK1 at the ribosome have been proposed, including roles in 40S and 60S subunit joining (8), the regulated translation of specific mRNAs (6, 36), and the localization of ribosomes for translation at specific sites within the cell (9, 10). Despite this abundance of hypothetical roles, the functional significance of RACK1 localization on the ribosome remains speculative.
Here, we provide the first experimental evidence that RACK1's position at the ribosome has biological importance in vivo. We determined the crystal structure of the full-length Saccharomyces cerevisiae RACK1 ortholog, Asc1p (henceforth RACK1), at 2.1-Å resolution. Using this structure and the cryo-EM model of the protein on the 40S ribosomal subunit, we analyzed the putative RACK1-40S subunit interface and generated eight RACK1 variants that have differing effects on ribosome binding in vivo. We show that yeast strains harboring even the most severely binding-defective RACK1 mutant fail to exhibit all of the phenotypes associated with RACK1 deletion. However, the efficiency of RACK1 binding to ribosomes correlates with a subset of growth behaviors observed for RACK1 deletion strains. These results indicate that although not required for all RACK1 activities, localization at ribosomes is integral to some aspects of RACK1 function.
MATERIALS AND METHODS
Plasmid construction.
To generate pRS316-RACK1, we cloned the yeast RACK1 (Asc1p; Stanford Yeast Genome Database sequence for YMR116C), including roughly 500 nucleotides upstream and downstream of the genomic sequence, into pRS316 (a yeast self-replicating plasmid [37]) as a PCR-generated SacII/XhoI fragment. PCR-mediated site-directed mutagenesis on this vector was used to make individual point mutants or to replace loops or insertions with a glycine linker. Integrating plasmid versions of these constructs were made by subcloning the SacII/XhoI fragments from the pRS316 vector into the integrating vector pRS306 (37). An intronless RACK1 construct derived from pRS316-RACK1 was used as a template to clone RACK1 as an SfoI/XhoI PCR fragment into the His6-maltose-binding protein (MBP) fusion expression vector pSV272 (a gift of Ian MacRae) for protein expression.
RACK1 expression and purification.
A selenomethionyl derivative of the full-length yeast orthologue of RACK1, Asc1p, was expressed in BL21(DE3) Escherichia coli cells as a His6-MBP fusion protein using the pSV272 vector (a gift from Ian MacRae) and standard techniques. Briefly, cells were grown in M9 minimal medium supplemented with kanamycin to an optical density (OD) of 0.5, at which point amino acids were added to inhibit methionine biogenesis and allow for selenomethionine incorporation (Leu, Ile, and Val at 50 mg/liter; Phe, Lys, and Thr at 100 mg/liter; selenomethionine at 75 mg/liter). After 20 min, expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a concentration of 1 mM and cells were left shaking at 37°C for 5 h.
The His6-MBP-RACK1 protein was initially purified using Ni-nitrilotriacetic acid affinity chromatography followed by cleavage of the His6-MBP tag with tobacco etch virus protease. The resulting mixture was applied to a Ni-nitrilotriacetic acid chromatography column once again; the flowthrough contained large amounts of very pure, untagged RACK1. Depending on the purity at this step, the protein was either used directly for crystallization or was further purified by additional MonoQ and size exclusion column chromatography.
Crystallization and structure determination.
Purified RACK1 was dialyzed into buffer containing 10 mM Tris, pH 7.4, and 1 mm dithiothreitol and concentrated to 8 mg/ml. The protein was crystallized using the hanging drop vapor diffusion method, combining 0.55 μl of recombinant protein with 0.55 μl of a reservoir solution containing 13.3% polyethylene glycol 2000-monomethyl ether, 22 mM MnO-acetate (Ac), and 100 mM NaOAc. Clusters of large needles grew within 1 to 5 days. These clusters were transferred from the hanging drop to a 2-μl drop of the mother liquor and agitated gently to obtain single crystals. The crystals were flash-cooled in cryo-protectant solution consisting of 4 parts mother liquor and 1 part 100% ethylene glycol and mounted for subsequent analysis.
We had previously attempted unsuccessfully to solve the structure of the yeast RACK1 by molecular replacement using a 3.2-Å diffraction data set obtained from native crystals. These crystals appeared to belong to the I41 space group and were predicted to contain two copies of RACK1 in the asymmetric unit. Although molecular replacement solutions using several different β-propeller structures were obtained, none could be refined. We originally attributed this to difficulties in correctly placing the molecules, given the low resolution of the data set and the sevenfold pseudosymmetry of the search model. Attempts to resolve the phase problem using a highly redundant data set measured from a selenomethionyl derivative for single-wavelength anomalous dispersion analysis revealed that the crystals were not I41 but monoclinic (C2) with pseudotetragonal symmetry. With the space group correctly assigned, a complete 2.1-Å data set was measured from a well-diffracting selenomethionyl-derived crystal and processed and scaled with XDS and XSCALE (21). Molecular replacement with Molrep (42) using a polyalanine derivative of the yeast Gβ protein (PDB entry 1GOT) as a search model successfully identified four copies of the protein in the asymmetric unit and produced high-quality electron density maps. The original search model was then discarded and the Phenix AutoBuild package (2) was used to build as much of the molecular model as possible. The remaining parts of the model were hand-built using Coot (15). The structure was refined using Phenix to R and Rfree values of 19.4% and 23.9%, respectively; the final model consisted of four chains containing residues 4 to 159 and166 to 318, 1 to 159 and 166 to 319, 5 to 159 and 166 to 318, and 2 to 159 and 166 to 319.
Construction of RACK1 yeast mutants.
RACK1 mutants were derived from a Σ1278b yeast strain (MATa ura3 leu2 trp1 his3). A RACK1 knockout was made using PCR-based gene manipulation methods (25). Transformation of Σ1278b-RACK1Δ with the pRS316 replicating plasmid containing either wild-type or mutant versions of RACK1 yielded strains harboring mutant forms of the protein. As invasive growth assays are not robust on synthetic media, we also generated RACK1 mutants by replacing the endogenous RACK1 locus with the mutant one. This was done by incorporating the mutant at the endogenous RACK1 locus of Σ1278b using the integrating plasmid pRS306 containing the desired mutant and selecting on synthetic defined medium lacking uracil (SCD-Ura) followed by counterselection on 5-fluoroorotic acid as described elsewhere (4). The integrity of the mutant locus was verified by sequencing.
Preparation and fractionation of polysomes.
Polysomes were prepared as described previously (7). Briefly, a 500-ml culture of cells was grown to an OD600 of 1.0 and transferred into prechilled centrifuge bottles containing 2 ml of 10-mg/ml freshly made cycloheximide. Cells were harvested and washed twice in lysis buffer (20 mM HEPES-KOH, pH 7.4, 2 mM MgOAc, 100 mM KOAc, 0.1 mg/ml cycloheximide, 3 mM dithiothreitol) and transferred to 50-ml Falcon tubes. Pellets were resuspended in 1.5 ml lysis buffer plus 0.2 U/μl RNasin per gram of cell pellet, and then 5 g of class beads per gram of cell pellet was added. Cells were lysed by vortexing for 2 min. The crude extracts were spun at 10,000 rpm in a microcentrifuge at 4°C for 20 min, and the resulting supernatant was flash-frozen and stored at −80°C for subsequent analysis.
To fractionate, 10 OD260 units of polysomal extract was applied to an 11-ml 10 to 50% sucrose (in lysis buffer) gradient. The gradients were spun in a Beckman ultracentrifuge using the SW41 rotor at a speed of 40,000 rpm for 1 h 45 min. The resulting samples were fractionated using Fluorinert and visualized using a UA6 UV/Vis detector. Fraction (750 μl) were collected and used either directly for analysis (for RACK1 Western blot assays) or concentrated (Scp160 Western blot assays) by trichloroacetic acid and acetone precipitation followed by resuspension in 75 μl of 1× sodium dodecyl sulfate (SDS) loading buffer.
Western blot assays.
Samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane, blocked in 5% milk in Tris-buffered saline with Tween (TBS-T; 20 mM Tris-HCl, pH 7.4, 500 mM NaCl, 0.05% Tween 20) for 30 min, and then incubated with primary antibody at 4°C overnight. A 1/10,000 dilution of rabbit anti-RACK1 (prepared by our lab) or a 1/3,000 dilution of rabbit anti-Scp160p (gift of Matthias Seedorf) in TBS-T was used for the primary hybridization. Membranes were subsequently incubated with horseradish peroxidase-conjugated secondary anti-rabbit antibody (Sigma) at a 1/10,000 dilution for 1 hour and detected with Amersham chemiluminescent reagents. Western blots were quantified using ImageJ (1), and in each case a cross-reacting, unrelated band was used for normalization between lanes when necessary.
Yeast phenotyping.
With the exception of determining growth rates in rich media and invasive growth, all yeast phenotypes were determined using “frogging assays” conducted by plating serial dilutions of RACK1Δ yeast strains carrying a wild-type or mutant RACK1 on the pRS316 replicating plasmid onto SCD-Ura solid medium supplemented with various chemical compounds. The concentrations used were 2.5 mM potassium disulfite, 9 nM rapamycin, and 25 mg/ml calcofluor white. Plates were inspected periodically over the course of a week to determine the phenotypes. For invasive growth assays, strains in which the endogenous copy of RACK1 were replaced with a mutant one were used. Cultures were spotted onto yeast extract-peptone-dextrose (YPD) plates and allowed to grow for 1 to 5 days before assaying using a standard plate washing technique (34).
RESULTS
Crystal structure of RACK1.
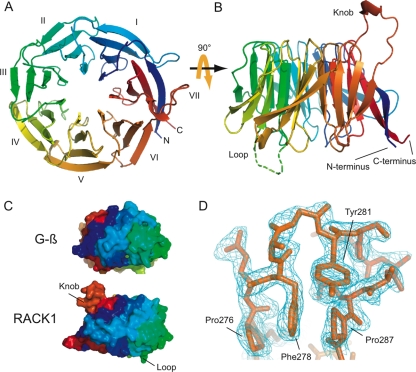
We determined the crystal structure of the full-length yeast orthologue of RACK1 (Asc1p) at 2.1-Å resolution by molecular replacement using a polyalanine derivative of the yeast Gβ protein (22) (PDB entry 1GOT) as a search model. The asymmetric unit contained four copies of RACK1, and excellent electron density was observed for the entire main chain in each copy except in a small disordered loop region spanning residues 160 to 166. The yeast RACK1 (PDB entry 3FRx), like the recently solved Arabidopsis thaliana RACK1-MBP fusion structure (41), is a markedly asymmetric seven-bladed β-propeller (Fig. 1A) with two notable large loop insertions, one occurring between blades III and IV and the other between blades VI and VII (Fig. 1B).
FIG. 1.
Crystal structure of RACK1. (A) Cartoon representation of the RACK1 crystal structure viewed from the top, the ribosome-binding face, to show the overall seven-bladed β-propeller architecture. The individual blades have been labeled I to VII and are colored using the CHAINBOWs scheme of MacPymol. (B) Cartoon representation of the RACK1 crystal structure viewed from the side. From this view, the two dramatic insertions are visible, labeled knob and loop, respectively. The twisting, extended interaction between the N (blue) and C (red) termini is also visible from this view. (C) Space-filling views of the seven-bladed β-propeller architecture of the yeast Gβ and our RACK1 crystal structure, showing the unusual asymmetry of the RACK1 structure. (D) Stick diagram of the structured knob region of the RACK1 structure with the corresponding 2Fobs-Fcalc electron density map shown contoured at 1.0 sigma. All ray-traced images were generated using MacPymol (13). Surface electrostatics were also calculated by using MacPymol. The conservation heat plot of the RACK1 surface was generated by using ConSurf (3) using a multiple sequence alignment we generated (unpublished data) using MUSCLE (14).
While the majority of our structure agrees with previous models for yeast RACK1 obtained either computationally (39) or by cryo-EM (35), the data reveal several unique structural features that had not been predicted. Although the overlap between the main chain of the crystal structure and that of the cryo-EM homology model (PDB entry 1TRJ) is good (C-α root mean square deviation over residues 1 to 159, 167 to 275, and 287 to 319 of 1.29 Å) throughout most of the seven blades, we observed a striking reorganization of the N and C termini in our structure from what was expected based on modeling. The homology model derived from Gβ predicted that the closure of the propeller would be mediated by the N terminus, which in Gβ forms a β-sheet with a portion of the C terminus in blade VII before extending toward blade VI, where it forms a β-sheet between blades I and VI. Our structure shows that the closure of the propeller does not involve the sixth blade, and instead the N terminus engages the C terminus exclusively in a β-sheet (Fig. 1A). Consequently, this portion of the protein does not wrap around the perimeter of the propeller neatly as in Gβ but instead bulges dramatically out and away from the rest of the propeller to create a protruding surface unique to RACK1 (Fig. 1C).
We also found that the large insertion between blades VI and VII—previously modeled as an unstructured, extended loop—actually folds into a well-ordered knob-like structure. Density for the main chain and many of the side chains was easily visible in all four copies in the asymmetric unit, and the resulting loop model could be refined well with reasonable temperature factors (average B-factor including side chains, 37.8 Å2). Furthermore, this structured knob did not participate in extensive crystal contacts. Inspection of the knob showed that the structure is stabilized through the sandwiching of Phe278 between two conserved prolines (Pro276 and Pro287) that flank the insertion as well as an edge-face π-π interaction between Tyr281 and Phe278 (Fig. 1D). Although Phe278 is only somewhat conserved between yeast and other eukaryotes and Tyr281 appears exclusive to fungi (unpublished data), an insertion of some sort at this position is universal among RACK1 sequences. This observation suggests that this loop may be an evolutionarily conserved feature of RACK1 with potential functional significance.
Phylogenetically conserved charged amino acids confer high-affinity binding of RACK1 to 40S subunits in vivo.
To test the biological role of RACK1's localization at the ribosome, we first sought to generate mutant forms of RACK1 that would fail to associate with ribosomes in vivo. We designed a suite of RACK1 mutants that were predicted to disrupt ribosome binding based on both the RACK1 crystal structure and a previous cryo-EM model of RACK1 bound to the 40S ribosomal subunit. The orientation of the RACK1 homology model in this cryo-EM structure had originally been determined by placing the region of the protein predicted to break the seven-fold symmetry of the β-propeller (the loop insertion in blade III) into an asymmetric region of the cryo-EM density (35). Given the additional asymmetries we observed in the RACK1 crystal structure, we examined other possible orientations of RACK1 in this density but found that the original orientation produced the most plausible interface between RACK1 and the ribosome.
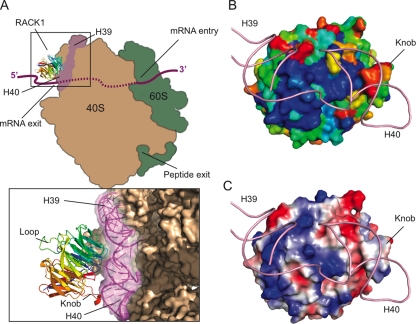
Superimposition of our crystal structure onto the 11.7-Å cryo-EM model places RACK1 at the head of the 40S ribosome near the mRNA exit tunnel, as originally proposed (Fig. 2A). The only portion of the 40S subunit that is close enough to RACK1 to anchor it to the ribosome are helices 39 and 40 of the 18S rRNA. The residues along the interface between these rRNA helices and RACK1 are well-conserved (Fig. 2B), and inspection of the surface electrostatic potential of RACK1 along this interface reveals several patches of positive charge generated by basic residues (Fig. 2C). Previous work showed that at least one pair of these basic residues (Arg38 and Lys40) contributes to RACK1's affinity for ribosomes in vitro (35), suggesting that these basic residues could mediate ribosome binding by interacting with the negatively charged backbone of the rRNA.
FIG. 2.
The RACK1-40S interface. (A) Cartoon schematic showing the positioning of RACK1 on the 40S ribosomal subunit as aligned using a least-squares superimposition function in Coot (15). The model positions RACK1 near the mRNA exit tunnel in close proximity to helices 39 and 40 of the 18S rRNA. A close-up view of this interaction with cryo-EM density for the 40S ribosome modeled shows that the top portion of RACK1, including the structured knob, faces the ribosome, while the bottom and sides of the protein are solvent facing and generally accessible. (B) Space-filling view of RACK1, colored according to electrostatic surface potential, with the positions of helices 39 and 40 modeled. Blue, positive charge; red, negative charge. (C) Space-filling view of RACK1, colored according to sequence conservation with the positions of helices 39 and 40 modeled. A gradient of blue to red indicates the degree of phylogenetic conservation, with dark blue indicating high conservation and red indicating low conservation.
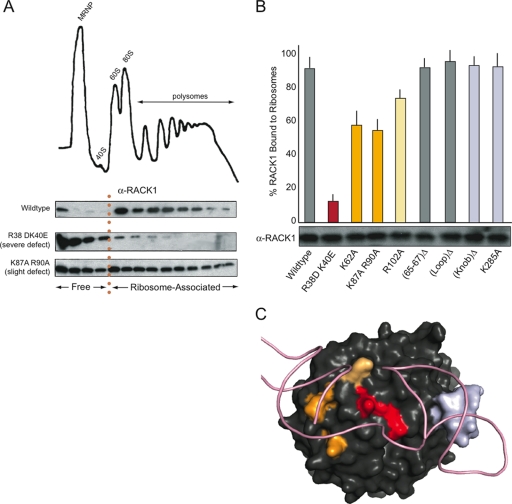
To examine this possibility more thoroughly and in an in vivo context, eight RACK1 mutants, consisting of point mutations or small deletions, were generated in a yeast self-replicating vector under the control of the native RACK1 promoter. Transformation of these constructs into RACK1Δ yeast strains generated strains harboring mutant forms of the protein. The relative amount of free RACK1 versus RACK1 bound to ribosomes could then be measured by determining the relative distribution of the protein by Western blotting of polyribosomes isolated from log-phase cell cultures (Fig. 3A). In wild-type cells, the majority of RACK1 is bound to ribosomes and appears in fractions as heavy or heavier than 40S subunits; only a small fraction appears in the lighter, mRNP-containing fractions. In contrast, RACK1 mutants that have binding defects shift out of ribosomes and into the lighter fractions.
FIG. 3.
Effects of RACK1 mutations on ribosome binding in vivo. (A) Representative data from polysome Western blot assays performed to determine the extent of ribosome association in vivo. A model polysome trace is shown, with the mRNP, 40S, 60S, 80S, and polysome fractions indicated. Western blots against RACK1 for fractions collected across the gradient from the wild-type, R38D K40E, and K87A R90A mutants are shown; fractions corresponding to free and ribosome-associated RACK1 are indicated. (B) Bar graph displaying the extent of ribosome association in each mutant tested, with error bars extending ± 2 standard deviations. Bars were colored to highlight the extent of the binding defect and for reference in the color-coding of the RACK1 surface in panel C. A Western blot showing the total RACK1 protein levels for each mutant is shown beneath each corresponding bar. (C) Space-filling view of RACK1 colored according to severity of binding defect in accordance with the findings shown in panel B, with the positions of helices 39 and 40 modeled. Residues corresponding to the (65-67)Δ and (Loop)Δ mutants are not colored.
The RACK1 mutations we tested were expressed at nearly the same level as the wild-type protein, as determined by Western blotting of total cell extracts with a polyclonal antibody, but they had varied effects on polyribosome association (Fig. 3B). Mutations in basic residues lying along the interface between RACK1 and the rRNA (Arg38, Lys40, Lys62, Lys87, Arg90, and Arg102) disrupted the protein's ability to associate with ribosomes in vivo, consistent with the previous report of a RACK1 mutant affecting ribosome binding in vitro (35). In contrast, neither a mutation on the opposite side of the protein (LoopΔ) nor mutation of uncharged residues along the rRNA interface (65-67Δ) produced a binding defect. This suggests that the predicted location of the ribosome binding interface is correct and implicates positively charged residues as critical mediators of the interaction. Although the surprisingly structured knob (residues 277 to 286) lies along this binding interface, neither deletion of the entire knob (KnobΔ) nor mutation of the highly conserved basic residue (K285A) at the tip of the knob had consequences for binding.
The most substantial binding defects were observed for mutations lying at the center of the of the platform generated by the junction between helices 39 and 40 (Arg38 and Lys40) (Fig. 3C). Mutations slightly further away from this central location (Lys62, Lys87, Arg90, and Lys102) but still lying on the interface produced less severe binding defects, and the mutations along the interface most distal to this central location (KnobΔ and Lys282) did not show a detectable binding defect. Together, these data clarify the RACK1-40S binding interface, confirming that RACK1 is anchored to the ribosome at the center of the rRNA platform by the universally conserved residues Arg38 and Lys40 and implicating other nearby basic residues in assisting in the interaction.
RACK1 has both ribosome localization-dependent and -independent functions in vivo.
Although RACK1 is involved in a number of cellular processes, the biological function of RACK1's position at the ribosome has not, until now, been explored. Our study of the RACK1-40S binding interface generated several RACK1 mutants whose ability to associate with ribosomes was compromised in vivo, providing us with the tools to address this issue directly.
We investigated the possibility of biological functions for RACK1's localization at the ribosome broadly by using a top-down approach to test our entire library of mutants for a number of phenotypes associated with a RACK1Δ strain. Some of the phenotypes we chose to test were related to specific proposed aspects of RACK1 function in yeast, such as cell wall maintenance (sensitivity to calcofluor white) (5), glucose sensing (invasive growth) (43, 44), and amino acid starvation and translation (sensitivity to rapamycin) (16, 17), while other phenotypes were more general.
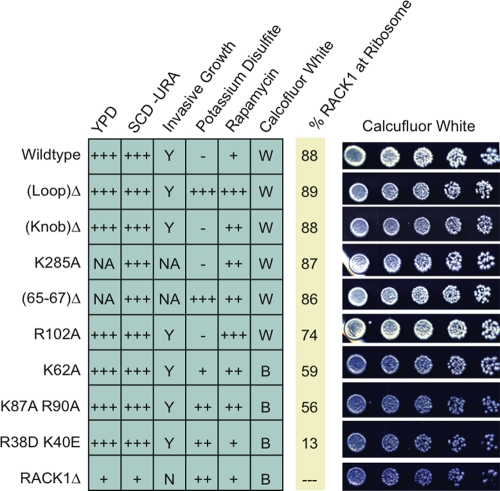
We observed three categories of behavior of our RACK1 mutant strains (Fig. 4). Regarding growth on rich medium (YPD) or synthetic medium (SCD-Ura) or the capacity to undergo haploid invasive growth, all of our RACK1 mutant strains behaved like wild-type cells. Because the RACK1Δ strain grows slowly on rich or minimal medium and is incapable of invasive growth, these observations suggest that the presence of RACK1 but not its localization is important for these behaviors. A second category of behavior resulted from exposure to potassium disulfite or rapamycin, where phenotypes were observed but did not correlate with RACK1 localization. In these cases, specific sites of RACK1 mutation rather than its association with the ribosome are important. A third class of behavior was observed upon exposure to calcofluor white, in which a clear correlation with the RACK1-ribosome association was apparent. This phenotype indicates a ribosome localization-dependent function for RACK1.
FIG. 4.
RACK1 mutant phenotyping. Summary of phenotypes observed for the RACK1 mutant, RACK1Δ, and wild-type yeast strains under a variety of environmental conditions. For YPD, SCD-Ura, potassium disulfite, and rapamycin, the relative growth rates are indicated. +++, normal growth; ++, slow growth; +, very slow growth; −, no growth. For calcofluor white, a B indicates that cells were blue, while a W indicates that cells were white. For invasive growth assays, a Y indicates that cells were competent for invasive growth, while an N indicates that the cells were not. The percentage of RACK1 associated with ribosomes in each mutant is included for reference. Representative images of the calcofluor white staining assay are included for each mutant to illustrate the blue and white phenotypes.
Calcofluor white is a blue, fluorescent dye that binds to chitin in the yeast cell wall (12). When grown on synthetic medium containing calcofluor white, wild-type cells appear white while cells with elevated levels of chitin become blue. RACK1 is implicated in cell wall maintenance in yeast, showing a physical interaction with the cell wall remodeling protein Knr4p (5) as well as genetic interactions with a number of chitin synthase genes (40). Consequently, a RACK1Δ strain exhibits elevated levels of chitin (23), and thus these cells appear blue when grown in such media (Fig. 4). We observed that cells harboring the RACK1 mutant with the strongest binding defect (R38D K40E) appeared as brilliant blue as RACK1Δ cells and that those expressing mutants with intermediate binding defects (K87A R90A and K62A) were also blue. In contrast, cells expressing the R102A mutant, which had the weakest 40S subunit binding defect, appeared as white as the wild type. Similarly, cells expressing the four other RACK1 mutants, none of which displayed a ribosome binding defect, also produced colonies of similar color to wild-type cells. Thus, the calcofluor white phenotype associated with RACK1Δ depends on proper localization of RACK1 at the ribosome.
We also wanted to test the proposed function of RACK1 as a ribosomal scaffold by directly testing its hypothetical role in the delivery of the yeast vigilin homologue Scp160 to actively translating ribosomes (Fig. 5). Previous studies had shown that Scp160 associates with polysomes in close physical proximity to RACK1 and that this association is compromised in RACK1Δ strains (6). However, whether this association actually depends on RACK1's localization at ribosomes or is a consequence of downstream effects resulting from the deletion of the protein has not been formally addressed. To distinguish between these two possibilities, we examined the polysome association of Scp160 in a strain of yeast carrying the RACK1 R38D K40E double mutation, which had the most substantial ribosome binding defect that we observed. In wild-type cells, Scp160 was found to be ∼100% associated with polyribosomes as detected by Western blotting (Fig. 5). In contrast, cells expressing the RACK1 R38D K40E mutant protein contained a small but significant (∼25%) fraction of Scp160 in nonribosome-associated fractions. As ∼13% of RACK1 is ribosome associated in the R38D K40E mutant (Fig. 3B) and RACK1 is estimated to be present in an 8- to 15-fold excess over Scp160 in cells (27), we had anticipated that most of the Scp160 would still be able to associate with polysomes and only a minority would appear in the lighter fractions. Thus, the shift we observed in strains harboring the RACK1 R38D K40E mutant is entirely consistent with the hypothesized role of RACK1 as a scaffold for Scp160 at the ribosome.
FIG. 5.
Effect of RACK1's ribosome localization on Scp160 association with polysomes. Upper panel, cartoon of the hypothesized RACK1/Scp160 scaffold at the ribosome; direct interaction could deliver specific mRNAs to the ribosome. Lower panel, polysome Western blot assay for Scp160 present in samples from wild-type and RACK1 R38D K40E mutant yeast strains.
DISCUSSION
The direct modulation of protein synthesis in response to environmental signals is central to cellular regulation, yet the mechanisms by which external signals are transmitted to the translation machinery are only beginning to emerge. More than a decade of research has established RACK1 as a key player in multiple signaling pathways; its recently discovered role as a core component of the 40S ribosomal subunit has raised the exciting possibility that RACK1 connects the signaling and translation machinery in the cell. However, the majority of research on RACK1 to date has addressed either its function in signaling or its potential activities as a ribosomal protein, but not both.
In this investigation, we aimed to determine the function of RACK1 localization at ribosomes directly. Aided by the high-resolution crystal structure of RACK1 obtained in this study, we constructed strains of yeast in which RACK1 binding to ribosomes was compromised. Our discovery that some, but not all, of the phenotypes associated with RACK1 deletion were reproduced in strains defective for the RACK1-ribosome association suggests that RACK1 plays specific, rather than general, roles in translational control. These findings underscore the importance of considering RACK1's ribosome localization when investigating its function in signaling pathways.
RACK1 structural analysis reveals a conserved surface for ribosome binding.
The crystal structure of RACK1 showed that its overall seven-bladed β-propeller architecture is markedly asymmetric and deviates substantially from the prototypical Gβ-like structure. These asymmetries arise from an unusually intimate and extended closure of the propeller between the N and C termini as well as a large, structured insertion in the sixth blade. Additionally, the loop insertion between blades III and IV, which is disordered in our structure, likely contributes additional asymmetry to RACK1. In β-propeller proteins, accessory loops or unusual surfaces often serve as platforms for interaction with other proteins (11, 30, 38). Thus, the identification of such features may prove helpful in predicting binding sites for RACK1 interaction partners.
Our own modeling efforts combining the crystal structure with a prior cryo-EM model of RACK1 on the 40S ribosomal subunit showed that a cluster of positively charged and conserved RACK1 residues positioned near the junction of helices 39 and 40 of the 18S rRNA creates a high-affinity platform for the phosphate backbone of the rRNA. Although mutational analysis implicates these positive charges as critical for RACK1 binding to ribosomes in vivo, we anticipate there are other features of the interaction as well. There is likely a nonionic component of the interaction, as RACK1 binding is resistant to high-salt washes in vitro (19). Additionally, it is not yet clear how RACK1 specifically recognizes helices 39 and 40 of the 18S rRNA, as opposed to other RNAs.
Other conserved features, like the large structured knob (Fig. 1D), did not contribute to RACK1's affinity for ribosomes but did lie along the RACK1-40S interface. This could indicate that the knob is a platform for recruiting specific proteins to this particular position on the ribosome or that there may be a function of this structure unrelated to the ribosome or translation that is sequestered when RACK1 is bound to the ribosome.
How might RACK1 localization at ribosomes contribute to its function in vivo?
Using the mutants created in this study, we identified a subset of RACK1's functions for which ribosome localization was critical. The sensitivity of yeast to staining by calcofluor white and the ability of Scp160 to associate with polysomes both exhibited a clear dependence on RACK1's positioning at ribosomes, while sensitivity to other toxins like rapamycin and potassium disulfite appeared to be a consequence of particular RACK1 mutations rather than of RACK1's ribosome localization.
There are two plausible, but very different, explanations for a ribosome localization-dependent function of RACK1. At one extreme, RACK1 might not have any function at the ribosome directly. Instead, the ribosome serves as a place to sequester RACK1 in a nonfunctional state and then release it at specific times to perform specific duties. Indeed, as RACK1 is substantially more abundant than the proteins with which it is known to interact (10-fold greater than Scp160 and 100-fold greater than Gpa2p, Pkc1p, or Knr4p [27]), its localization at ribosomes could temper this stoichiometric overabundance by decreasing the effective concentration of RACK1 available for interaction in the cytosol. Disruption of RACK1's position at the ribosome would increase the effective concentration of free RACK1 in the cell, in turn perturbing pathways in which RACK1 is involved. At the other extreme, RACK1 might perform its functions on the 40S subunit itself, allowing kinases, signaling proteins, or other interacting partners to directly engage with the ribosome to regulate and affect cellular processes.
This first scenario would suggest that ribosome-bound RACK1 is not competent for some of its cellular functions and that there is a means of regulating the portion of free and ribosome-bound pools of RACK1 in the cell, both of which are supported by previous research. As an example of the former, RACK1 was recently shown to play the role of the β-subunit of the heterotrimeric G-protein coupled to the glucose-sensing G-protein-coupled receptor Gpa2 in yeast (44). In this case, homology to other heterotrimeric G-proteins would place the Gα-anchoring partner to RACK1 on the ribosome-facing side of the protein (18, 22). However, our own modeling suggests that the possibility of these three interacting simultaneously would result in steric clashes (data not shown). Furthermore, such an interaction would orient the ribosome so as to collide with the cell membrane. Thus, it seems likely that RACK1's Gβ function occurs when it is not associated with the ribosome, and thus some functions of RACK1 are restricted to the free form of the protein. There is also evidence in yeast that cells may have the ability to alter the distribution of free and ribosome-associated RACK1 in specific situations. For example, during stationary phase in yeast, RACK1 shifts out of ribosomes and into the cytosol (6). Although the precise mechanism of this shift is unknown, the binding data we have presented show that the placement of two negative charges along the binding platform is sufficient to shift the majority of the protein into the cytosol in vivo. This raises the exciting possibility that phosphorylation or other modifications of the protein in this region could provide a means of modulating the ratio of free and ribosome-bound pools of RACK1 in the cells in response to environmental cues.
However, there are also compelling data that RACK1 has functions in translational control, providing evidence in favor of the second proposed mechanism, in which RACK1 functions directly on the 40S subunit. RACK1 has been suggested to play a role in 40S and 60S subunit joining in vivo by facilitating the phosphorylation of the 60S-associated eukaryotic initiation factor eIF6 by protein kinase C (8). RACK1 depletion experiments hint at a role for the protein in recruiting specific messages to ribosomes in both Schizosaccharomyces pombe (29, 36) and mammalian cells (31). Furthermore, RACK1 is thought to regulate the translation of specific mRNAs in S. cerevisiae by recruiting the mRNA binding protein Scp160 and its associated messages to the ribosome (6). Finally, the fact that RACK1 interacts with intermembrane integrin-β receptors (9) could allow for the positioning of ribosomes at specific sites within the cell for localized translation.
In light of these mechanistic possibilities, how do we interpret the phenotypes we have observed in this study? The ribosome localization-specific sensitivity to calcofluor white, indicative of a defect in cell wall integrity, is most consistent with a direct role for RACK1 at the ribosome. In this case, RACK1 might play a role in regulating the translation of genes involved in cell wall upkeep, and so removal of RACK1 from its position at the ribosome would then result in the failure of RACK1 to regulate translation of these genes. Similarly, the importance of RACK1 localization for the association of Scp160 with polysomes also points to a direct role for RACK1 on the ribosome. Our data establish that Scp160 associates with ribosomes more effectively when RACK1 is present at ribosomes than when it is not, implying that RACK1 acts as a scaffold for the interaction between ribosomes and Scp160.
Given the diversity and sheer volume of functions for RACK1 in the cell, the majority of research concerning RACK1 to date has considered its functions within distinct categories that involve signaling or the ribosome exclusively. However, the duality of RACK1 as both a signaling scaffold and a ribosomal protein requires us to consider the functional interplay between its two roles. The additional dimension of RACK1 functionality we have established here brings these two disparate aspects of RACK1 function together and lays a precedent for the importance of determining the ribosome localization dependency of RACK1 function in all its roles in the cell.
Acknowledgments
We gratefully acknowledge members of the Doudna laboratory for discussions and comments on the manuscript. In particular we thank Ian MacRae and Martin Jinek for help with diffraction data collection and RACK1 structure determination. We thank Matthias Seedorf for providing us with antibodies against Scp160p.
This work was supported by a grant from the NIH and a gift from Gilead Inc. to J.A.D.; W.V.G. is supported by a K99 award from the NIH.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 581948-1954. [DOI] [PubMed] [Google Scholar]
- 3.Armon, A., D. Graur, and N. Ben-Tal. 2001. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 307447-463. [DOI] [PubMed] [Google Scholar]
- 4.Barton, M. C., M. F. Hoekstra, and B. M. Emerson. 1990. Site-directed, recombination-mediated mutagenesis of a complex gene locus. Nucleic Acids Res. 187349-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basmaji, F., H. Martin-Yken, F. Durand, A. Dagkessamanskaia, C. Pichereaux, M. Rossignol, and J. Francois. 2006. The ‘interactome’ of the Knr4/Smi1, a protein implicated in coordinating cell wall synthesis with bud emergence in Saccharomyces cerevisiae. Mol. Genet. Genomics 275217-230. [DOI] [PubMed] [Google Scholar]
- 6.Baum, S., M. Bittins, S. Frey, and M. Seedorf. 2004. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 380823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ceci, M., C. Gaviraghi, C. Gorrini, L. A. Sala, N. Offenhauser, P. C. Marchisio, and S. Biffo. 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426579-584. [DOI] [PubMed] [Google Scholar]
- 9.Chang, B. Y., K. B. Conroy, E. M. Machleder, and C. A. Cartwright. 1998. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol. Cell. Biol. 183245-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y.-W. E., and H. A. Traugh. 1997. Phosphorylation of elongation factor 1 and ribosomal protein S6 by multipotential S6 kinase and insulin stimulation of translational elongation. J. Biol. Chem. 27228252-28257. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri, I., J. Soding, and A. N. Lupas. 2008. Evolution of the beta-propeller fold. Proteins 71795-803. [DOI] [PubMed] [Google Scholar]
- 12.Darken, M. A. 1962. Absorption and transport of fluorescent brighteners by microorganisms. Appl. Microbiol. 10387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 14.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59407-450. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, B., H. U. Mosch, E. Sattlegger, I. B. Barthelmess, A. Hinnebusch, and G. H. Braus. 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol. Microbiol. 31807-822. [DOI] [PubMed] [Google Scholar]
- 18.Hurowitz, E. H., J. M. Melnyk, Y. J. Chen, H. Kouros-Mehr, M. I. Simon, and H. Shizuya. 2000. Genomic characterization of the human heterotrimeric G protein alpha, beta, and gamma subunit genes. DNA Res. 7111-120. [DOI] [PubMed] [Google Scholar]
- 19.Inada, T., E. Winstall, S. Z. Tarun, Jr., J. R. Yates III, D. Schieltz, and A. B. Sachs. 2002. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji, H., C. S. Fraser, Y. Yu, J. Leary, and J. A. Doudna. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. USA 10116990-16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 212916-924. [Google Scholar]
- 22.Lambright, D. G., J. Sondek, A. Bohm, N. P. Skiba, H. E. Hamm, and P. B. Sigler. 1996. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature 379311-319. [DOI] [PubMed] [Google Scholar]
- 23.Lesage, G., J. Shapiro, C. A. Specht, A. M. Sdicu, P. Menard, S. Hussein, A. H. Tong, C. Boone, and H. Bussey. 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link, J. A., and J. R. Yates. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17676-682. [DOI] [PubMed] [Google Scholar]
- 25.Longtine, M. S., and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 26.Lu, H. C., E. C. Swindell, W. D. Sierralta, G. Eichele, and C. Thaller. 2001. Evidence for a role of protein kinase C in FGF signal transduction in the developing chick limb bud. Development 1282451-2460. [DOI] [PubMed] [Google Scholar]
- 27.Lu, P., C. Vogel, R. Wang, X. Yao, and E. M. Marcotte. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25117-124. [DOI] [PubMed] [Google Scholar]
- 28.McCahill, A., J. Warwicker, G. B. Bolger, M. D. Houslay, and S. J. Yarwood. 2002. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 621261-1273. [DOI] [PubMed] [Google Scholar]
- 29.McLeod, M., B. Shor, A. Caporaso, W. Wang, H. Chen, and L. Hu. 2000. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell. Biol. 204016-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neer, E. J., C. J. Schmidt, R. Nambudripad, and T. F. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371297-300. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, J., J. Sengupta, J. Frank, and P. Nissen. 2004. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 51137-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, M. M., D. Ron, K. Touhara, C. H. Chen, and D. Mochly-Rosen. 1999. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry 3813787-13794. [DOI] [PubMed] [Google Scholar]
- 33.Ron, D., and D. Mochly-Rosen. 1994. Agonists and antagonists of protein kinase C function, derived from its binding proteins. J. Biol. Chem. 26921395-21398. [PubMed] [Google Scholar]
- 34.Rupp, S., and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 181257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sengupta, J., J. Nilsson, R. Gursky, C. M. Spahn, P. Nissen, and J. Frank. 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat. Struct. Mol. Biol. 11957-962. [DOI] [PubMed] [Google Scholar]
- 36.Shor, B., and M. McLeod. 2003. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 27849119-49128. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24181-185. [DOI] [PubMed] [Google Scholar]
- 39.Steele, M. R., A. McCahill, D. S. Thompson, C. MacKenzie, N. W. Isaacs, M. D. Houslay, and G. B. Bolger. 2001. Identification of a surface on the beta-propeller protein RACK1 that interacts with the cAMP-specific phosphodiesterase PDE4D5. Cell. Signal. 13507-513. [DOI] [PubMed] [Google Scholar]
- 40.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303808-813. [DOI] [PubMed] [Google Scholar]
- 41.Ullah, H., E. L. Scappini, A. F. Moon, L. V. Williams, D. L. Armstrong, and L. C. Pedersen. 2008. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci. 171771-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagin, A. T., A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 301022-1025. [Google Scholar]
- 43.Valerius, O., M. Kleinschmidt, N. Rachfall, F. Schulze, S. Lopez Marin, M. Hoppert, K. Streckfuss-Bomeke, C. Fischer, and G. H. Braus. 2007. The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11-dependent adhesive growth and dimorphism. Mol. Cell Proteomics 61968-1979. [DOI] [PubMed] [Google Scholar]
- 44.Zeller, C. E., S. C. Parnell, and H. G. Dohlman. 2007. The RACK1 ortholog Asc1 functions as a G-protein beta subunit coupled to glucose responsiveness in yeast. J. Biol. Chem. 28225168-25176. [DOI] [PubMed] [Google Scholar]