Abstract
Human immunodeficiency virus type 1 (HIV-1) elite controllers (EC) maintain viremia below the limit of commercial assay detection (<50 RNA copies/ml) in the absence of antiviral therapy, but the mechanisms of control remain unclear. HLA-B57 and the closely related allele B*5801 are particularly associated with enhanced control and recognize the same Gag240-249 TW10 epitope. The typical escape mutation (T242N) within this epitope diminishes viral replication capacity in chronically infected persons; however, little is known about TW10 epitope sequences in residual replicating viruses in B57/B*5801 EC and the extent to which mutations within this epitope may influence steady-state viremia. Here we analyzed TW10 in a total of 50 B57/B*5801-positive subjects (23 EC and 27 viremic subjects). Autologous plasma viral sequences from both EC and viremic subjects frequently harbored the typical cytotoxic T-lymphocyte (CTL)-selected mutation T242N (15/23 sequences [65.2%] versus 23/27 sequences [85.1%], respectively; P = 0.18). However, other unique mutants were identified in HIV controllers, both within and flanking TW10, that were associated with an even greater reduction in viral replication capacity in vitro. In addition, strong CTL responses to many of these unique TW10 variants were detected by gamma interferon-specific enzyme-linked immunospot assay. These data suggest a dual mechanism for durable control of HIV replication, consisting of viral fitness loss resulting from CTL escape mutations together with strong CD8 T-cell immune responses to the arising variant epitopes.
A subset of human immunodeficiency virus type 1 (HIV-1)-infected persons who control viremia to below the limit of detection (<50 RNA copies/ml plasma) without antiviral therapy has been termed elite controllers/suppressors (EC) (2, 3, 6, 13, 32). Some of these individuals have been infected in excess of 30 years, indicating prolonged containment of HIV replication, but the mechanisms associated with this extreme viremia control remain elusive (13). Among EC, certain HLA class I alleles are overrepresented, in particular HLA-B57, strongly suggesting that HIV-1-specific cytotoxic T-lymphocyte (CTL) responses restricted by these alleles may be crucial for viremia control (16, 29, 32). However, to date, there has been no clear explanation as to why some subjects can control viremia but others cannot, even when carrying the same allegedly protective HLA alleles. Moreover, the characteristics of virus-specific immune responses as well as the impact of viral escape mutations on in vitro replicative fitness in persons with different disease outcomes remain unclear.
Growing numbers of studies suggest that CTL targeting Gag, particularly the p24 capsid protein, play an important role in controlling viremia (7, 15, 22, 26, 32, 33, 38). Indeed, the most protective HLA class I allele, B57, which is present in over 40% of EC (32), restricts four immunodominant CTL epitopes in the p24 capsid protein. Previous studies have failed to find differences in the recognition of Gag epitopes or in gamma interferon (IFN-γ) responses to HIV proteins between B57-positive (B57+) long-term nonprogressors and B57+ progressors (28). Other studies have shown differences in the frequency of polyfunctional CD8+ T cells between B57+ EC and B57+ progressors (5); likewise, differences in the frequency of IFN-γ/interleukin-2-producing CD8+ T cells between controllers and progressors with protective HLA alleles were reported (16). Recently, Bailey et al. reported that plasma viruses in B57+ EC can harbor CTL escape mutations in the Gag protein, and in some cases these autologous variants were recognized by CTL (3). However, since there were no comparisons to progressors, it is unclear whether the viral variants that were detected or the apparent de novo CTL responses to the variant viruses are characteristic features among B57+ persons who maintain persistent control.
Of the four immunodominant Gag CTL epitopes restricted by HLA-B57, TW10 (TSTLQEQIGW [Gag residues 240 to 249]) is known to be the earliest target in acute infection (1, 11, 36), therefore likely playing an important role in defining the plasma viral load set point. This epitope is also known to be presented by the closely related B*5801 allele, which is also associated with viral control (21). One of the most frequently detected mutations within this epitope, T242N, is known to occur rapidly and almost universally after acute infection in persons expressing HLA-B57/B*5801 (11, 17, 23). The same mutation has been shown to have a negative impact on viral replication capacity (VRC) by both clinical observation and in vitro experiments (8, 23, 25). Moreover, as plasma viral load increases, compensatory mutations accumulate, restoring VRC to some extent (8). Additional studies, predominantly with children, indicated that some TW10 escape variants may be targeted by specific immune responses (17). Together, these data suggest a hypothesis to explain the diverse disease courses among B57+ subjects, namely, that a combination of fitness cost by CTL escape from the TW10 response, variable accumulation of compensatory mutations, and variable generation of specific CTL responses to the new variant influence plasma viral loads.
In this study, we investigated plasma viral sequences and IFN-γ-specific enzyme-linked immunospot (ELISPOT) assay responses to autologous Gag TW10 sequences in HLA-B57/B*5801-positive EC and compared these data to those obtained from persons with detectable viremia. Our results indicate that the TW10 T242N mutation does not differentiate HLA-B57/B*5801 EC from those with viremia and that CTL responses to this variant epitope are frequently detected in both viremic and aviremic subjects. However, some rare variants within and flanking this epitope were observed exclusively in HIV controllers, most of which not only reduced VRC but also were recognized by specific CTL at a high magnitude. These data suggest that the additive effects of both CTL-mediated selection for less fit viral variants and CD8 T-cell responses to the variant viruses contribute to strict viremia control in HLA-B57/B*5801-positive controllers.
MATERIALS AND METHODS
Study subjects.
A total of 50 B57/B*5801-positive subjects were involved in this study. Twenty-three were EC, 15 were viremic controllers (VC) (50 to 2,000 RNA copies/ml), and 12 were chronically infected untreated subjects with higher plasma viral loads (chronic progressors [CP]) (median virus load, 14,500 RNA copies/ml; range, 3,460 to 604,000 copies/ml). The definitions of EC and VC are described in detail elsewhere (32). Briefly, controllers are defined as subjects infected with HIV for at least a year and maintaining <50 (EC) or 50 to 2,000 (VC) RNA copies/ml without antiretroviral treatments. CP were defined as subjects infected with HIV for more than a year, with plasma viral loads of >2,000 RNA copies/ml without treatment. Written informed consent was obtained from all participants. Plasma and peripheral blood mononuclear cells (PBMC) were obtained by standard procedures and stored at −80C° and in liquid nitrogen, respectively, until use.
Viral RNA isolation, genomic DNA isolation, PCR, and sequencing.
Viral RNA isolation from plasma and genomic DNA isolation were performed as described elsewhere (30). Reverse transcription-PCR and DNA PCR were also performed as described before (30). Multiple alignments of viral sequences were made with ClustalW. Phylogenetic trees were drawn by the maximum likelihood method (DNAml of PHYLIP, included in Bioedit software). Proviral sequences which carried premature stop codons were eliminated from the analysis.
IFN-γ ELISPOT assay.
PBMC were plated in RPMI 1640 medium containing 10% serum at 50,000 to 100,000 cells/well. Tenfold serial dilutions of peptide (range, 0.0143 to 14.3 μg/ml) were added to cells. Wash and detection steps were performed by standard methods. Results were calculated as the number of spot-forming cells (SFC) per million input cells after subtraction of the background. For the experiments at a fixed peptide concentration, 14.3 μg/ml of peptide was used. A positive response was defined as over 50 SFC/million cells after subtracting the background. The background was the average for three negative control wells in each experiment and never exceeded 30 SFC/million cells.
Mutagenesis and VRC assay.
Mutant NL4-3 viruses were constructed by site-directed mutagenesis, using a method described elsewhere (30). Briefly, mutagenesis reactions were performed on a pUC19 plasmid backbone carrying the SacI-SbfI fragment of NL4-3 (2,353 bp containing the entire gag gene). Mutations were confirmed by sequencing the entire fragment. Fragments were then ligated back into full-length pNL4-3. Viruses were obtained by transfecting HEK293T cells with plasmid variants and were titrated using a Tat-driven green fluorescent protein (GFP) reporter T-cell line (GXR cells) (9). Replication capacity assays were performed in monoculture by infecting GXR cells with virus (multiplicity of infection [MOI] = 0.002), and the proportion of GFP-positive (GFP+) cells was measured up to day 8. The natural log slope of each replication curve was calculated using the data from days 2 to 6, during which viruses grew exponentially. All experiments were performed in duplicate or triplicate.
Viral competition assay.
Viral competition assays were performed by coinfecting mutant virus with NL4-3 vif synonymous mutant C, which carries synonymous mutations in the vif gene (5321-C-T-C-G-C-5336 [HXB2 numbering]). These mutations do not change the expressed protein but allow the use of a distinct probe to distinguish this virus in competition assays. Two viruses were used to infect GXR cells (0.25 million) in 100 μl of R10 Plus medium (RPMI 1640 medium [Sigma-Aldrich] containing 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% [vol/vol] bovine calf serum [Atlantic Biologicals]) in round-bottomed 96-well plates at a 1:1 ratio, maintaining a total MOI of 0.002 on day 2. After overnight infection, cells were washed twice with R10 Plus, resuspended in 200 μl of R10 Plus, and cultured in new 96-well plates in order to remove extra HIV RNA in the inoculum. Seventy microliters of supernatant was harvested on days 2, 4, 6, and 8 and stored at −80°C. Viral RNA was extracted using 96-well-format Charge-Switch (Invitrogen) following the manufacturer's instructions. cDNA was synthesized using an AffinityScript cDNA synthesis kit (Stratagene) and random primers in 20 μl of reaction mix. Real-time PCR was performed by adding 25 μl quantitative PCR mix (12.5 μl of 2× Brilliant multiplex quantitative PCR master mix [Stratagene], a 2 μM final concentration of forward primer qVif F [GAAAGAGACTGGCATTTGGGTCAGGG] and reverse primer qVif R [GATGAATTAGTTGGTCTGCTAGGTCAGGG], 200 nM [final concentration] of TaqMan MGB probe, and reference dye) to 96-well plates containing Charge-Switch. For each sample, two wells were used to detect mutant NL4-3 and vif synonymous mutant virus separately. TaqMan MGB probe sequences were as follows: 6-carboxyfluorescein-ATAGCACACAAG for mutant NL4-3 and HEX-ACAGTACCCAGGTCGA for vif synonymous NL4-3. All samples from the same subjects were always run on the same plate. Standard curves were drawn in each experiment, using known amounts of pNL4-3 wild type and vif synonymous mutant proviral plasmid.
Statistical analysis.
All of the comparisons for independent continuous variables were performed by the Mann-Whitney U test. All of the comparisons for paired continuous variables were performed by the Wilcoxon matched-pair test. Categorical data were compared using Fisher's exact test. For all comparisons, P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
Some sequences from this study were submitted to GenBank previously (30, 31), and the remainder were added under accession numbers FJ387525 to FJ387541.
RESULTS
T242N escape mutation is frequently seen within the TW10 Gag CTL epitope in HLA-B57/B*5801-positive subjects.
The dominant early CTL response in persons expressing the protective allele HLA-B57 is directed against the Gag TW10 epitope, and the earliest escape mutation arising in persons expressing B57 is a T242N mutation within this epitope (11). This mutation confers a fitness cost, as wild-type virus has been shown to outcompete the mutant after 10 to 20 days in culture competition assays performed in vitro (25). The same epitope is targeted by persons expressing HLA-B*5801, and the identical T242N mutation arises in those individuals. In order to determine how frequently the T242N mutation is seen over the range of viral loads in persons expressing HLA-B57/B*5801, we obtained plasma virus Gag sequences for 48 B57- and 2 B*5801-positive subjects. We included 23 EC and 27 subjects with detectable viremia (15 VC and 12 CP). Since it is known that there are discrepancies between plasma and proviral sequences in subjects with low plasma viral loads (3; our unpublished data), we used only plasma viral sequences for all EC. Plasma virus sequences were also used for viremic subjects, except for four CP and two VC for whom only proviral sequences were available (see Materials and Methods).
As shown in Table 1, we found that 83% (10/12 sequences) of CP and 87% (13/15 sequences) of VC viruses exhibited the T242N escape mutation, whereas only 65% (15/23 sequences) of EC viruses had this mutation. Although EC viruses harbored the T242N mutation less frequently, this did not reach statistical significance (P = 0.183), despite >1,000-fold differences in viral loads. We also examined each of the subjects for evidence of three known upstream compensatory mutations associated with the T242N mutation (H219Q, I223V, and M228I), which restore viral fitness and are associated with an increase in plasma viral load (8). Among those who expressed the T242N mutation, there was a progressive increase in these compensatory mutations with increasing plasma viral loads (4/15 EC viruses, 6/13 VC viruses, and 6/10 CP viruses), but these differences also did not reach statistical significance (P = 0.238; chi-square test). Moreover, grouping all viremic patients together (VC and CP combined) and comparing them to EC also did not show statistical significance (12/23 sequences versus 4/15 sequences; P = 0.182). However, among HIV controllers not expressing the T242N mutation, we noted additional mutations compared to the consensus B sequence within and surrounding this epitope that were not observed in persons with chronic progressive infection.
TABLE 1.
TW10 epitope and its flanking sequences in B57/B*5801-positive subjects
| Category | Subjectc | Difference from consensus B sequencea | No. of sequencesb |
|---|---|---|---|
| Consensus B | DRLHPV HAGPIAPGQM REPRGSDIAGT TSTLQEQIGW MTNNPP | ||
| CP | PARC_CP002 | -----A Q----P---I ----------- --N-----T- ------ | Pop |
| BCP_001 | N----- ----V----- ----------- --N------- ------ | Pop | |
| PARC_CP001 | --T--X ----V----- ----------- --N------- --S--- | Pop | |
| PARC_CP006 | ------ ----P----L ----------- --N------- ------ | Pop | |
| PARC_CP019 | --M--- ---------- ----------- --N------- --S--- | Pop | |
| PARC_CP021 | ------ Q---V----L ----------- --N-----A- --H--- | Pop | |
| PARC_CP025 | -----X ----X----- ----------- --N------- --S--- | Pop | |
| PRLS12 | ------ ----V----- ----------- --N------- ------ | Pop | |
| F7165 | ------ ---------- ----------- --N-----A- ------ | Pop | |
| F719 | ------ ---------- ----------- --N------- ------ | Pop | |
| BCP_002 | ------ ---------L ----------- -------VQ- I----- | Pop | |
| PARC_CP038 | ------ ----V----L ----------S ---------- --S--- | Pop | |
| VC | BVC_002 | ------ ----V----- ----------- --N-----A- ------ | Pop |
| BVC_012 | ------ Q---V----- ----------- --N------- --H--- | Pop | |
| BVC_004 | ------ ---------- ----------- --N------- --S--- | Pop | |
| BVC_013 | ------ P--------- ----------- --N------A ------ | 3/11 clones | |
| ------ P--------I ----------- --N-----TR ------ | 8/11 clones | ||
| BVC_005 | --V--- Q---V----I ----------- --N-----A- ------ | Pop | |
| BVC_006 | --V--- ---------- ----------- --------D- --H--- | Pop | |
| BVC_007 | ------ ---------- ----------- --N-----A- ------ | Pop | |
| BVC_008 | ------ ---------- ----------- --N-----A- I-S--- | Pop | |
| BVC_009 | ------ Q---V----- ----------- --N------- --H--- | Pop | |
| BVC_014 | ------ Q--------- ----------- --N------- --H--- | Pop | |
| BVC_010 | ------ ---------- ----------- --N------- ------ | Pop | |
| BVC_015 | ------ ---------L ----------- --N-----A- --H--- | Pop | |
| BEC 147 | ------ ---------- ----------- --------D- --H--- | Pop | |
| BVC_003 | --M--- ---------- ----------- --N-----A- ------ | Pop | |
| BVC_011 | ------ ---------- ----------- --N------- --H--- | Pop | |
| EC | BEC 5 | ------ ---------L ----------- -------MA- I-H--- | Pop |
| BEC 13 | ------ ---------- ----------- --N-----A- ------ | Pop | |
| BEC 15 | ------ ---------- ----------- --N------- --H--- | Pop | |
| BEC 24 | ------ ----V----- ----------- --N-----T- ------ | Pop | |
| BEC 33 | ------ ---------- ----------- --------A- I----- | 3/5 clones | |
| ------ Q--------- ----------- -------MA- I----- | 1/5 clones | ||
| ------ Q--------- -G--------- -------MA- V----- | 1/5 clones | ||
| BEC 36 | ------ ---------- ----------- --N------- --S--- | Pop | |
| BEC 43 | ------ ---------- ----------- --N-----A- ------ | Pop | |
| BEC 53 | ------ ---------- ----------- -------LN- I----- | 2/9 clones | |
| ------ ---------- ----------- --------N- I----- | 7/9 clones | ||
| BEC 59 | ------ ---------- ----------- --N------- ------ | Pop | |
| BEC 60 | --M--- ---------- ----------- --N------- ------ | Pop | |
| BEC 62 | ------ ---------- ----------V --N------- ------ | Pop | |
| BEC 63 | ------ ---------- ----------- ---------- ------ | Pop | |
| BEC 65 | -X---- ----X----- ----------- --N-----A- --H--- | Pop | |
| BEC 71 | ------ ----V----- ----------- --N-----N- --G--- | Pop | |
| BEC 72 | ------ Q--------L ----------- -------MT- I----- | Pop | |
| BEC 78 | ------ P---V----L ----------- --N-----A- --H--- | Pop | |
| BEC 82 | ------ ---------- ----------- --------D- --H--- | Pop | |
| BEC 90 | ------ ---------- ----------- --------D- --R--- | Pop | |
| BEC 106 | ------ ---------- ----------- --N-----A- ------ | Pop | |
| BEC 110 | ------ ---------- ----------- --N------- ------ | Pop | |
| BEC 121 | ------ ---------- ----------- --N------- ------ | Pop | |
| BEC 149 | ------ ---------- ----------- --N------- --H--- | Pop | |
| BEC 105 | ------ ----N----- ----------- --------A- --G--A | Pop |
X, mixed amino acids (223I/A/V in PARC_CP025 and 223I/V in BEC65).
Pop, population sequencing.
The relatively high prevalence of TW10 mutations in EC, VC, and CP indicate that there is significant pressure on the TW10 epitope in persons expressing HLA-B57/B*5801, regardless of plasma viral load. These data also indicate that the T242N mutation and its compensatory mutations alone cannot account for the differences in plasma viral load among HLA-B57/B*5801-positive persons.
CD8 T-cell recognition of the autologous TW10 epitope.
Given that there was no significant difference between EC and viremic subjects in the frequency of the T242N mutation, we hypothesized that there might be differences in recognition of the T242N mutant by CD8 T-cell responses. Indeed, the T242N mutation has been shown to frequently be targeted in children (17), indicating that this variant may be presented effectively by the HLA class I allele. IFN-γ ELISPOT responses to wild-type and T242N mutant epitopes were measured, and responses in EC were compared to those in viremic subjects (VC and CP combined) (Fig. 1A). Limited numbers of subjects tested responded to the wild-type TW10 peptide in either group, and there was no statistically significant difference in the magnitude or frequency of positive responses between the groups (P = 0.766) (Fig. 1A and data not shown [for frequency]). Since the lack of responses might be due to the lack of the wild-type TW10 sequence in vivo, we also tested for recognition of the Gag KF11 peptide as a control, since this epitope has been noted to have few variations (12, 37). Here we saw significant differences in the magnitude and frequency of the IFN-γ response to wild-type KF11, namely, viremic subjects showed stronger responses than did EC (P = 0.0011) (Fig. 1B and data not shown [for frequency]).
FIG. 1.
Comparison of IFN-γ ELISPOT responses to wild-type and variant TW10 in EC and viremic subjects (VC and CP). The magnitudes of IFN-γ ELISPOT responses for randomly selected subjects are indicated in SFC per 106 PBMC. (A) Comparison of responses to wild-type TW10 peptide between EC (n = 33) and viremic subjects (n = 28). (B) Comparison of responses to wild-type KF11 peptide between EC (n = 29) and viremic subjects (n = 25). (C) Pairwise comparison of responses to wild-type TW10 peptide and autologous variant peptides. (D) Comparison of responses to autologous variant TW10 between EC (n = 19) and viremic subjects (n = 19). Only subjects with available sequences were involved in the analysis for panels C and D.
We next compared IFN-γ ELISPOT responses to wild-type TW10 peptides and autologous variant TW10 peptides in a total of 38 subjects for whom autologous viral sequences, PBMC, and corresponding variant peptides were available (Table 2). For subjects from whom we obtained multiple sequences (BEC33 and BEC53) (Table 1), variants with the largest numbers of mutations within TW10 were tested. On average, responses to autologous variants were of significantly greater magnitude than those to wild-type TW10 (P = 0.0179) (Fig. 1C). Although we failed to see a statistically significant difference in either the magnitude or the frequency between EC and viremic subjects (Fig. 1D) (P = 0.0675 for the magnitude and P = 0.515 for the frequency), there was a trend toward stronger responses to autologous variants in viremic subjects, similar to what was observed for responses to the highly conserved KF11 epitope.
TABLE 2.
Autologous TW10 variants used for IFN-γ ELISPOT assay
| Subject(s) | Difference from TSTLQEQIGW |
|---|---|
| 15 subjects | --N------- |
| 12 subjects | --N-----A- |
| BEC105 | --------A- |
| BEC82, BEC90, BEC147, and BVC006 | --------D- |
| BEC33 | -------MA- |
| BEC53 | -------LN- |
| BEC71 | --N-----N- |
| BEC24 | --N-----T- |
These results indicate that CTL recognition of the autologous escape variants within the TW10 epitope occurs frequently in HLA-B57/*5801-positive subjects but that simple recognition of the autologous virus by CD8 T cells does not explain the enhanced control of viremia.
Rare variants in the TW10 Gag CTL epitope are associated with fitness defects and strong virus-specific CD8 T-cell responses.
Although the T242N mutation and its associated compensatory mutations were not significantly more frequent in progressive infection, many of the HIV controllers studied here had other mutations both within and downstream of the TW10 epitope. A number of these mutants were seen only in the absence of the T242N mutant, in particular a G248D mutant that was seen only in two EC and two VC and was associated with mutations at codon 252, where the consensus B sequence has an asparagine (N). Other mutations rarely seen in persons not expressing HLA-B57/*5801 or in CP included dual mutations at positions 247 and 248, together with mutations at position 250 and, on occasion, position 252 (Table 1). Those that were unique to EC and VC particularly involved positions 247 and 248 within the epitope and residues 250 and 252 downstream of the epitope (Table 3).
TABLE 3.
TW10 sequences in 134 B57/58-negative subjectsa
| Codon | Substitution,b no. (%) of subjects
|
|||||
|---|---|---|---|---|---|---|
| 242 | 247 | 248 | 249 | 250 | 252 | |
| 1 | T, 129 (96.3) | I, 128 (95.5) | G, 106 (79.1) | W, 134 (100) | M, 133 (99.3) | N, 76 (56.7) |
| 2 | N, 4 (3.0) | V, 6 (4.4) | A, 21 (15.6) | R, 0 (0.0) | I, 1 (0.75) | H, 28 (20.9) |
| 3 | S, 1 (0.75) | M, 0 (0.0) | T, 6 (4.4) | V, 0 (0.0) | S, 28 (20.9) | |
| 4 | L, 0 (0.0) | D, 1 (0.75) | Q, 1 (0.75) | |||
| 5 | N, 0 (0.0) | G, 1 (0.75) | ||||
| 6 | R, 0 (0.0) | |||||
Thirty-one EC, 18 VC, and 85 CP.
Boldface type indicates rare variants that were observed in HIV-1 controllers.
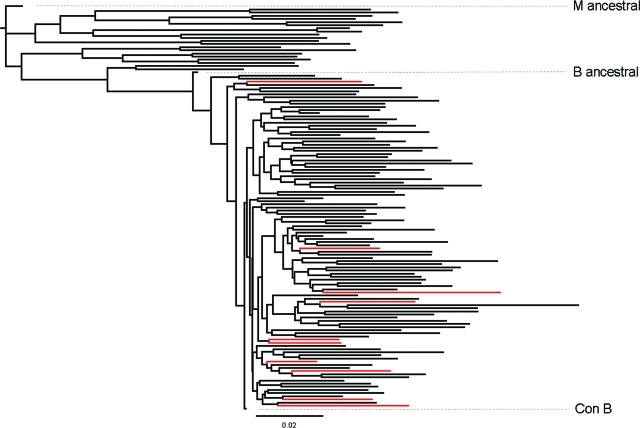
Since these mutations were rarely or never seen in the non-B57/58 population or in persons with chronic progressive infection, we first examined whether they shared common ancestors. A phylogenetic tree was constructed based on the entire gag sequence. As shown in Fig. 2, the viruses expressing rare mutations (shown in red) were scattered throughout the tree, indicating that these did not arise from a common ancestor. The frequencies of these mutations in the setting of HLA-B57/B*5801 and the fact that these mutations were rarely or never seen in CP expressing HLA-B57/B*5801 or in persons who did not express these alleles (Table 3) make it most likely that these variants were selected in vivo after transmission. These data indicate strong selection pressure and that this pressure may force the virus down different mutational pathways in controllers from those in CP.
FIG. 2.
Phylogenetic analysis of HIV-1 gag sequences in B57/B*5801 subjects. A maximum likelihood tree was drawn using a total of 129 entire gag sequences, including consensus B, M ancestral, B ancestral, and some reference sequences for each clade. Red branches indicate 10 B57/B*5801-positive subjects with rare mutations in TW10. Consensus B, M ancestral, and B ancestral viruses are indicated in the figure.
We next undertook to define the consequences of these relatively unique mutations on VRC, as well as their recognition by autologous CD8 T cells. Mutant NL4-3 viruses were generated by mutagenesis based on autologous viral sequences, including those in the C-terminal flanking region of TW10. We chose for further analysis a subset of those viruses that had rare mutations that were preferentially seen in the context of HIV control. We compared VRC by measuring the slope of the natural log of the percentage of GFP expression from days 2 to 6, as previously described (Fig. 3A) (8, 9, 30, 34). Because NL4-3 differs from the current consensus B sequence at position 252, namely, consensus B has 252N but NL4-3 has 252H, we also engineered these mutations as indicated. NL4-3 with 252N displayed a slightly reduced replication capacity compared to that of wild-type NL4-3, which is consistent with a previous study (8) (Fig. 3A). We also used an IFN-γ ELISPOT assay and limiting peptide concentrations to determine the effects of these mutations on T-cell recognition.
FIG. 3.
VRC of mutant NL4-3. The slope (days 2 to 6) of the natural log of the GFP expression percentage was calculated, and relative values compared to wild-type NL4-3 are shown (means and standard errors of the means). Experiments were performed in either duplicate or triplicate. (A) Relative VRC of mutant NL4-3 viruses compared to that of the wild type. (B) Compensation of fitness cost due to G248D mutation by 252H/R substitution.
(i) G248D and 252H/R.
One of the mutations within the TW10 epitope, G248D, was observed only in EC (n = 2) and VC (n = 2), not in any of the B57/B*5801-positive CP examined here, and it was seen in only 1 of 134 sequences from non-HLA-B57/58 subjects in our cohort and in only 1 of 530 sequences from non-HLA-B57 subjects in a cohort with progressive infection from British Columbia, Canada (10). In each of the subjects in whom we observed the G248D substitution, it was seen in the absence of the T242N mutation (Table 1) and was always accompanied by either an H or R at position 252, downstream of the epitope. When the G248D/252H mutants seen in an EC and two VC were constructed and tested, they displayed 54% VRC compared to that of wild-type NL4-3. In contrast, the same virus with T242N/G248A, a common variant selected in persons with B57/B*5801 (23), did not differ in VRC from the wild-type virus. Although the single T242N mutant was not included in the current experiments, the VRC of the T242N mutant was similar to that of the T242N/G248A mutant in the exact same experimental setting (8). The mutant with the G248D and 252R mutations, which was seen in a single EC, displayed 74% VRC compared to wild-type NL4-3 (Fig. 3A). These findings were confirmed by a viral competition assay where the replication capacities of mutant viruses were compared directly to that of wild-type NL4-3 in a head-to-head manner (Fig. 4A and B).
FIG. 4.
Viral competition assay. Relative replication capacities of mutant NL4-3 viruses were measured by competition assay (A, B, and C). NL4-3 mutants and vif synonymous NL4-3 (containing synonymous mutations in the vif gene that allowed PCR-based detection of the reference virus [see Materials and Methods]) were used at a 1:1 ratio to infect GXR cells. (D) Comparable replication capacities of vif synonymous NL4-3 and wild-type NL4-3 were confirmed. Representative data from duplicate experiments are shown.
Since the G248D mutation was always associated with an additional position 252 mutation, we also made a mutant NL4-3 virus with G248D mutation together with 252N mutation, a combination that was never seen in vivo. As shown in Fig. 3B, this mutant displayed only 24% of the replication capacity of wild-type NL4-3, suggesting that 252H and 252R act as compensatory mutations for the G248D mutation.
Unlike the G248T, G248A, and G248N mutations, none of the subjects with the G248D mutation had the T242N escape mutation. Therefore, we also made a mutant with T242N, G248D, and 252H mutations, as well as one with the consensus B amino acid residue 252N. Although all viruses were used at the same MOI, these two mutants were extremely defective in the replication capacity assay (Fig. 3B). These data suggest that there may be mutually exclusive pathways to immune escape in HLA-B57/B*5801-positive subjects, in that the G248D mutation not only affects VRC but also may be incompatible with the typical T242N escape mutation in B57/B*5801-positive subjects.
We next examined the effects of these mutations on IFN-γ ELISPOT responses. Strong responses to the G248D TW10 variant peptide were observed in all four subjects in whom this mutation was detected, and in each case these were stronger than those to the wild-type TW10 peptide (Fig. 5A to D), though the differences did not reach statistical significance with this number of subjects (P = 0.125) (Fig. 6A). We also tested responses to this variant peptide in other B57/B*5801-positive subjects whose autologous viral sequences did not include the G248D mutation (n = 26) (Fig. 6B), using the same concentration of peptide. Only one subject showed a strong response to the G248D variant as well as to wild-type TW10, but the others showed low responses to the G248D variant as well as to wild-type TW10 (P = 0.820).
FIG. 5.
IFN-γ ELISPOT responses to rare variant TW10 in HIV controllers. IFN-γ ELISPOT responses to TW10 variant peptides in individual subjects are shown. Backgrounds were subtracted. TW10 variants in boxes indicate autologous viral sequences. (A through D) Results for subjects with G248D mutations. (E through H) Results for subjects with other rare mutations.
FIG. 6.
IFN-γ response to G248D variant TW10 in B57/B*5801-positive subjects. IFN-γ ELISPOT assay was performed with a peptide concentration of 14.3 μg/ml. (A) IFN-γ responses in the four subjects whose autologous viral sequences contained the G248D mutation. (B) IFN-γ responses in the 26 subjects whose autologous viral sequences did not contain the G248D mutation.
Together, these data indicate that the G248D mutation affects viral load by two mechanisms, namely, a reduction in viral replicative capacity through the induced mutation and a strong CTL response to the mutated epitope. Moreover, the data suggest that the responses to the G248D mutation are not simply due to cross-reactivity of previously existing TW10 responses but rather represent a type-specific response to the G248D variant.
(ii) I247L/M, G248A/T/N, M250I/V, and 252N/H.
A second group of unusual mutations that were repeatedly detected in EC but were not observed in progressors involved mutations within the TW10 epitope at positions 247 and/or 248, together with positions 250 and 252 in the epitope flanking region. The I247M/G248T/M250I/252N mutant (from subject BEC72) displayed a lower replication capacity than did G248D variants (Fig. 3A), and a viral competition assay strongly supported this finding (Fig. 4C). Two subjects had I247M/G248A/M250I mutants with a 252N/H variation (BEC33 and BEC5). Since bulk Gag sequences from patient BEC33 showed mixed amino acids in this region, we obtained clonal sequences (Table 1). This revealed three different populations, corresponding to I247M/G248A/M250I/252N, G248A/M250I/252N, and I247M/G248A/M250V/252N mutants (Table 1). The I247M/G248A/M250V/252N mutant was extremely attenuated, since we could not obtain viral stocks with a high enough titer for use in the VRC assay, despite the fact that transfection of the proviral plasmid was performed under exactly the same conditions as those used for the other mutants. As shown in Fig. 3A, both the I247M/G248A/M250I/252N and G248A/M250I/252N mutants displayed reduced replication capacities (71% and 62% of wild-type capacity, respectively). Therefore, all three mutants for BEC33 were attenuated compared to the consensus B sequence and wild-type NL4-3, and all of these mutants resulted in a greater loss of VRC than did the typical T242N/G248A mutations (Fig. 3A). The I247M/G248A/M250I/252H mutant for patient BEC5, which had 252H instead of 252N, displayed an improved replication capacity (Fig. 3A) but still had a slightly lower VRC than the wild type (91%), supporting the compensatory effect of the N252H mutation. Adding the T242N mutation to the I247M/G248A/M250I/252N and G248A/M250I/252N mutants made them more defective, and we could not obtain high enough titers for use in the VRC assay (data not shown), suggesting that this combination of mutations is incompatible with effective viral replication, as shown for the G248D mutation.
We next examined IFN-γ responses to these autologous variant peptides by ELISPOT assay, as described for the G248D mutants. As shown in Fig. 5E, BEC33 showed stronger responses to the G248A variant of TW10 than to wild-type TW10 but failed to respond to the I247M/G248A variant. Therefore, the I247M and G248A mutations likely represent mutations that lead to escape from CD8 T-cell recognition but that induce a considerable fitness defect with M250I and 252N mutations. BEC5 also carried this variant (I247M/G248A), but the PBMC from this subject had such high background levels that valid data could not be obtained despite repeated experiments using samples from different time points, so whether these mutations also led to escape in subject BEC5 could not be determined. However, summarizing the variants seen in BEC33, one of the TW10 variants (G248A) with M250I/252N mutations displayed reduced VRC and was also recognized by CTL, and the other two variants (I247M/G248A with M250I/V and 252N) appeared to escape from CTL responses but had reduced VRC as well. However, the mechanism of strict viremia control in BEC5 was unclear. Unfortunately, PBMC for BEC72 (I247M/G248T) were not available for testing, but likewise the virus had an extremely reduced VRC.
The I247L/G248N/M250I/252N and G248N/M250I/252N mutants from patient BEC53 also displayed reduced replication capacities (79% and 35% of wild-type capacity, respectively) (Fig. 3A). We observed positive IFN-γ responses to the G248N mutation alone or to the G248N mutation in combination with the I247L mutation, and these responses were stronger than those to wild-type TW10 (Fig. 5F). The introduction of the T242N mutation rendered the viruses further defective, as reflected in the fact that insufficient titers of these viruses could be obtained (data not shown). Therefore, both of the TW10 variants for BEC53 not only displayed reduced VRC but also were recognized by type-specific CTL.
(iii) T242N and G248T/N.
Autologous viral sequence from BEC71 contained T242N, G248N, and 252G mutations (Table 1). The presence of these mutations in an NL4-3 backbone gave a comparable to slightly reduced replication capacity compared to that of the wild type (85%) (Fig. 3A). Mutants carrying the G248N/252G mutations and 252G substitution alone displayed 89% and 72% of the replication capacity of the wild type, respectively (data not shown). Although we did not make a mutant with the G248N mutation alone, unlike the mutations already discussed, the combination of this mutation with the T242N mutation did not severely affect VRC, which is probably the reason that this combination was seen in vivo. Moderate IFN-γ responses to the G248N variant, which were stronger than those to wild-type TW10, but weak responses to the T242N/G248N variant peptide were observed (Fig. 5G). Therefore, the mechanism of strict viremia control in this case was unclear.
Autologous viral sequence from subject BEC24 had T242N, G248T, and 252N mutations. The G248T mutation was seen in 4.4% of non-B57/58 subjects and in a single case of a B57/B*5801-positive CP (Tables 1 and 3). Therefore, it was not necessarily specific for controllers. The T242N/G248T/252N mutant in BEC24 was slightly less robust in replication capacity (92% of wild-type capacity) (Fig. 3A). However, stronger IFN-γ responses to variant peptide (T242N/G248T) than to wild-type TW10 were observed (Fig. 5H). Interestingly, all of the variant peptides (T242N/G248X) were recognized in this subject (Fig. 5H). We then examined whether this feature characterizes CTL responses in B57/B*5801-positive EC. We expanded the experiment to include 10 subjects (4 EC, 3 VC, and 3 CP) who showed strong responses (>300 SFC/million PBMC) to the T242N variant of TW10. All of them showed similar responses to the T242N/G248X variant peptides (data not shown). Therefore, the mechanism of viremia control involving the variant in BEC24 is unlikely to be explained by differences in recognition of autologous virus. One possibility is that functional differences in variant-specific CTLs might exist between controllers and progressors.
(iv) T242N, G248T, and W249R.
Autologous viral sequence from subject BVC013 (VC) contained T242N, G248T, and W249R mutations. Codon 249 is the anchor position of TW10. None of the viral sequences from other subjects had the W249R mutation at this codon, regardless of HLA type (Tables 1 and 3). Although we made a mutant proviral NL4-3 incorporating these mutations, we could not obtain sufficient viral stock for the subsequent experiments, which indicated that the virus containing these mutations was extremely defective. Since PBMC for this subject were not available, we were not able to perform IFN-γ ELISPOT assays to test variant recognition.
In summary, many viruses from controllers, particularly EC, carried rare variants of TW10 which compromised VRC and/or were targeted by variant-specific CTL responses. However, although these mechanisms contributing to control were frequently observed among HLA-B57/B*5801-positive controllers, there was considerable heterogeneity in the fitness of viruses evolving in vivo and the host responses to these variants, indicating a complex interplay between these factors among individual subjects.
DISCUSSION
One of the strongest associations with durable “elite” control of HIV identified to date is expression of HLA-B57, but the mechanisms of this control remain unclear. To address this, we analyzed viral sequences within and flanking the immunodominant and earliest targeted Gag CTL epitope, TW10, in a large cohort of B57/B*5801+ subjects, as well as immune responses to the autologous virus epitopes. Our results show that the commonly selected mutation within this epitope that impairs viral fitness, T242N, is more frequently observed in persons with progressive infection. When T242N is the sole mutation within TW10, compensatory flanking mutations that restore VRC are frequently observed in viremic subjects but are rare in EC. When the T242N mutation is absent in EC, rare variants both within and flanking the TW10 epitope are detected that result in a greater impairment of VRC and are also frequently associated with strong type-specific CD8 T-cell responses. These data suggest a dual mechanism of immune control in a subset of EC, involving CD8 T-cell-induced defects in VRC together with strong CTL responses to the variant viruses evolving in vivo.
The remarkable frequency of plasma virus mutations within the TW10 epitope in viruses from EC reported here provides clear evidence of immune selection pressures in EC viruses, though it was unknown whether they had been exerted during primary infection or were ongoing, as seen for neutralizing antibodies in EC (24). Many of the mutations both within and flanking the TW10 epitope were observed exclusively in B57/B*5801+ HIV controllers, and most of them have never been reported. These mutations were also not seen in our local cohort of HLA-B57/58-negative subjects, providing robust evidence that these are being selected in the context of these alleles. The exact pathways leading to these mutations are not known. For example, codon usage for the G248D mutation was not caused by hypermutation by APOBEC3G/F (data not shown). Whether EC might have a distinct T-cell receptor repertoire that accommodates this mutation will require additional study. Importantly, these rare variants not only affected VRC but also were frequently recognized by variant-specific CD8 T-cell responses. The potential impact of functional differences in these responses to the different variants in terms of cytokine secretion and ability to inhibit virus replication in vitro should be addressed in future studies.
Our data also suggest that immune responses to and immune escape from TW10 may play a dominant role in determining set point viral loads in B57/B*5801 EC, in that we saw far fewer distinct associations among three other Gag epitope sequences in these B57-positive subjects and viral control (data not shown). The A146P mutation immediately preceding ISPRTLNAW (ISW9; Gag147 to 155) is known to act as an antigen-processing mutation but not to affect VRC (14). In the present study, we observed no significant difference in the frequency of A146P mutation between EC and viremic subjects (19/26 versus 14/22 subjects; P = 0.543). KAFSPEVIPMF (KF11; Gag162 to 172) is known to be targeted during chronic infection (20, 21, 27, 29). Only three viremic subjects and four EC showed variation within KF11, again with no significant difference between aviremic and viremic subjects. Escape mutations within a third Gag epitope, QW9 (QASQEVKNW; Gag308 to 316), have never been reported, although E312D mutation tends to occur in B*5701-positive subjects (28). In our data set, there was no statistically significant difference in the frequency of this mutation between viremic subjects and EC (6/26 versus 5/22 subjects; P = 1.0).
Among the rare TW10 variants identified in this study, the G248D variant was particularly interesting in that it was observed only in the setting of in vivo HIV control and had already been reported for an untreated B57/B*5801-positive pediatric patient with a low plasma viral load (1,100 RNA copies/ml) (17). Bailey et al. recently reported a B57+ EC in whom the G248D variant arose as plasma viral loads increased, but the G248D variant was not recognized by CTL in that subject (4). This case is of importance for two reasons. First, the study confirms that this rare mutation can occur in the setting of B57 in another cohort, and second, it shows a higher level of viremia when the G248D variant is not targeted by the autologous CTL response. On the other hand, we observed strong IFN-γ responses to this variant of TW10 in all of four B57/B*5801+ controllers presenting this mutation who were suppressing viremia effectively. Taken together, the data suggest that B57/B*5801+ subjects who mount CTL responses to this rare variant peptide can control viremia. Thus, although this mutation reduced VRC in vitro, virus-specific CTL toward this variant likely play an important role in controlling viremia in vivo.
For other rare variants we observed in the present study, the relative contributions of defective VRC, CD8 T-cell targeting of the variant virus, and other potential mechanisms are not as clear as for the G248D mutation. Mutations seen in two EC (BEC33 and BEC53) affected VRC; specific CTL responses to two of the three variant peptides for BEC33 and to both variants for BEC53 were observed, as for the G248D variant. However, the relative contributions to control of the variant-specific CTL response to the mutant viruses or the in vitro viral fitness cost itself are unclear. Although IFNγ ELISPOT data could not be obtained for BEC72 and BVC013, the replication capacity of mutant NL4-3 containing these autologous TW10 variants was profoundly compromised. It is possible that variant-specific CTL responses are detected in these persons, but we were not able to confirm this experimentally due to a lack of appropriate samples. Note that, as for mutations seen in BEC71 and BEC5, not all rare variants in TW10 affected VRC. Thus, despite these data indicating a dual mechanism of control involving a combination of CTL-induced escape mutations that impair VRC together with a response to the variant virus, there clearly are additional factors that are likely to be involved in the heterogeneous outcome of infection.
The mechanisms by which the mutations described here impact VRC might be mediated through interactions with cyclophilin A (8, 34). The T242N escape mutation in TW10 was reported to make the virus more dependent on cyclophilin A (8). In addition, compensatory mutations in the cyclophilin A binding loop, which is just upstream of TW10 (18, 19), were shown to recover the loss of replication capacity by making the viral capsid independent of cyclophilin A (8). Therefore, atypical mutations in the TW10 epitope observed in the present study may affect the interaction between cyclophilin A and capsid as well. Indeed, some of the variants were accompanied by H219Q mutation, which is one of the compensatory mutations for T242N escape (Table 1) (8). Looking at the downstream region of TW10, an amino acid change at codon 252 seems to act as a compensatory mutation for escape mutations in TW10, which was also suggested in a previous study (8). In the current study, we clearly show that 252H/R mutations improve VRC loss by the G248D mutation. Although we do not know the mechanism for the loss of VRC by G248D mutation or the compensation by 252H/R, there is an interesting study regarding HIV-2 and cynomolgus monkey Trim5α. A single amino acid change at codon 120 of the HIV-2 capsid, which corresponds to codon 253 of HIV-1 Gag, determines the sensitivity to cynomolgus monkey Trim5α (35). Therefore, the in vitro fitness cost of escape by G248D mutation and compensation by 252H/R could be associated with the sensitivity of HIV-1 capsid to human Trim5α. This hypothesis could also apply to the in vitro fitness cost of one of the mutants in BEC33 (I247M/G248A/M250I/252N) and its recovery by 252H mutation (BEC5) as well. Further studies are warranted to reveal the mechanism of fitness cost and compensation for these rare mutations seen in B*57/5801-positive EC.
The presence of replication-impaired viruses in plasmas of HIV controllers raises an important additional question, i.e., why do such viruses not mutate back to a wild type that is better fit? There are several feasible explanations. One is that wild-type virus may be controlled more strongly by wild-type-specific CTL than variant viruses are controlled by variant-specific CTL. IFN-γ ELISPOT assay also does not measure the strength of individual clonal populations of CTL but merely quantitates the number of memory cells. As shown in a previous study, the magnitudes of responses measured by IFN-γ ELISPOT assay are likely driven by in vivo antigen loads (32). Therefore, stronger IFN-γ ELISPOT responses to variant peptides do not necessarily mean that variant-specific CTL suppress viral replication in vivo better than do CTL specific for the wild-type virus. Another possible explanation is that there might be few or no actively replicating wild-type viruses in these EC during the chronic phase of infection, but rather these might be archived completely after acute infection. Moreover, the extremely low viral loads in EC reduce the chance that variant viruses will acquire additional mutations required to revert to the wild type or develop compensatory mutations to enhance viral replication.
In conclusion, a large fraction of B57/B*5801+ HIV-1 controllers carry rare unique mutations in the TW10 Gag CTL epitope, affecting both VRC and inducing specific CTL responses. These dual mechanisms of immunologically selected mutations that impair in vitro virus fitness, which are also targeted by variant-specific CTL responses, likely contribute to strict viremia control in these cases. Despite these findings shining light on durable immune control, much remains unclear. Although these differences were seen in the majority of subjects examined, they clearly are not necessary for control, as some B57/B*5801-expressing EC had entirely wild-type viruses at TW10. Moreover, the mechanisms of control in persons with other HLA types have yet to be defined, indicating that further studies will be necessary to define additional determinants accounting for durable containment of HIV.
Acknowledgments
We thank the patients, investigators, and clinical staff at Massachusetts General Hospital for their essential contributions to this research study. We also thank the members of the International HIV Controllers Consortium (www.hivcontrollers.org), who are contributing additional subjects to the cohort used here in order to facilitate broader genetic studies.
This work was supported by grants AI028568 and AI030914 from the NIAID/NIH, by the Howard Hughes Medical Institute, by the Harvard University Center for AIDS Research (HU CFAR), by the Mark and Lisa Schwartz Foundation, by the IAVI, and by the Bill and Melinda Gates Foundation. Z.L.B. is supported by a postdoctoral fellowship from the Canadian Institutes for Health Research (CIHR).
The ideas and opinions expressed in the manuscript are solely the responsibility of the authors and are not necessarily shared by the NIH or other funding sources, Massachusetts General Hospital, or its affiliates. We declare no conflicts of interest related to this study.
Footnotes
Published ahead of print on 30 December 2008.
REFERENCES
- 1.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, J. R., K. G. Lassen, H. C. Yang, T. C. Quinn, S. C. Ray, J. N. Blankson, and R. F. Siliciano. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 804758-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 2031357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, J. R., H. Zhang, B. W. Wegweiser, H. C. Yang, L. Herrera, A. Ahonkhai, T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2007. Evolution of HIV-1 in an HLA-B*57-positive patient during virologic escape. J. Infect. Dis. 19650-55. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankson, J. N., J. R. Bailey, S. Thayil, H. C. Yang, K. Lassen, J. Lai, S. K. Gandhi, J. D. Siliciano, T. M. Williams, and R. F. Siliciano. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J. Virol. 812508-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borghans, J. A., A. Molgaard, R. J. de Boer, and C. Kesmir. 2007. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS ONE 2e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 8112608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockman, M. A., G. O. Tanzi, B. D. Walker, and T. M. Allen. 2006. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J. Virol. Methods 131134-142. [DOI] [PubMed] [Google Scholar]
- 10.Brumme, Z. L., I. Tao, S. Szeto, C. J. Brumme, J. M. Carlson, D. Chan, C. Kadie, N. Frahm, C. Brander, B. Walker, D. Heckerman, and P. R. Harrigan. 2008. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS 221277-1286. [DOI] [PubMed] [Google Scholar]
- 11.Brumme, Z. L., C. J. Brumme, J. Carlson, H. Streeck, M. John, Q. Eichbaum, B. L. Block, B. Baker, C. Kadie, M. Markowitz, H. Jessen, A. D. Kelleher, E. Rosenberg, J. Kaldor, Y. Yuki, M. Carrington, T. M. Allen, S. Mallal, M. Altfeld, D. Heckerman, and B. D. Walker. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 829216-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crawford, H., J. G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, J. I. Mullins, R. Kaslow, P. Goepfert, S. Allen, E. Hunter, J. Mulenga, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 818346-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deeks, S. G., and B. D. Walker. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27406-416. [DOI] [PubMed] [Google Scholar]
- 14.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 762298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emu, B., E. Sinclair, H. Hatano, A. Ferre, B. Shacklett, J. N. Martin, J. M. McCune, and S. G. Deeks. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 825398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feeney, M. E., Y. Tang, K. Pfafferott, K. A. Roosevelt, R. Draenert, A. Trocha, X. G. Yu, C. Verrill, T. Allen, C. Moore, S. Mallal, S. Burchett, K. McIntosh, S. I. Pelton, M. A. St John, R. Hazra, P. Klenerman, M. Altfeld, B. D. Walker, and P. J. Goulder. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 1747524-7530. [DOI] [PubMed] [Google Scholar]
- 18.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 871285-1294. [DOI] [PubMed] [Google Scholar]
- 19.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273231-235. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retrovir. 121691-1698. [DOI] [PubMed] [Google Scholar]
- 22.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 23.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. St John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10282-289. [DOI] [PubMed] [Google Scholar]
- 24.Mahalanabis, M., P. Jayaraman, T. Miura, F. Pereyra, E. M. Chester, B. Richardson, B. Walker, and N. L. Haigwood. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 803617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews, P. C., A. Prendergast, A. Leslie, H. Crawford, R. Payne, C. Rousseau, M. Rolland, I. Honeyborne, J. Carlson, C. Kadie, C. Brander, K. Bishop, N. Mlotshwa, J. D. Mullins, H. Coovadia, T. Ndung'u, B. D. Walker, D. Heckerman, and P. J. Goulder. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 828548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Migueles, S. A., and M. Connors. 2001. Frequency and function of HIV-specific CD8(+) T cells. Immunol. Lett. 79141-150. [DOI] [PubMed] [Google Scholar]
- 28.Migueles, S. A., A. C. Laborico, H. Imamichi, W. L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C. W. Hallahan, and M. Connors. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral Gag sequences. J. Virol. 776889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura, T., M. A. Brockman, C. J. Brumme, Z. L. Brumme, J. M. Carlson, F. Pereyra, A. Trocha, M. M. Addo, B. L. Block, A. C. Rothchild, B. M. Baker, T. Flynn, A. Schneidewind, B. Li, Y. E. Wang, D. Heckerman, T. M. Allen, and B. D. Walker. 2008. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J. Virol. 828422-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura, T., M. A. Brockman, Z. L. Brumme, C. J. Brumme, F. Pereyra, A. Trocha, B. L. Block, A. Schneidewind, T. M. Allen, D. Heckerman, and B. D. Walker. 2009. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J. Virol. 83140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereyra, F., M. M. Addo, D. E. Kaufmann, Y. Liu, T. Miura, A. Rathod, B. Baker, A. Trocha, R. Rosenberg, E. Mackey, P. Ueda, Z. Lu, D. Cohen, T. Wrin, C. J. Petropoulos, E. S. Rosenberg, and B. D. Walker. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197563-571. [DOI] [PubMed] [Google Scholar]
- 33.Rolland, M., D. Heckerman, W. Deng, C. M. Rousseau, H. Coovadia, K. Bishop, P. J. Goulder, B. D. Walker, C. Brander, and J. I. Mullins. 2008. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS ONE 3e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. Labute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 8112382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, H., E. E. Nakayama, M. Yokoyama, H. Sato, J. A. Levy, and T. Shioda. 2007. A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J. Virol. 817280-7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 Gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 817725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, X. G., M. Lichterfeld, S. Chetty, K. L. Williams, S. K. Mui, T. Miura, N. Frahm, M. E. Feeney, Y. Tang, F. Pereyra, M. X. Labute, K. Pfafferott, A. Leslie, H. Crawford, R. Allgaier, W. Hildebrand, R. Kaslow, C. Brander, T. M. Allen, E. S. Rosenberg, P. Kiepiela, M. Vajpayee, P. A. Goepfert, M. Altfeld, P. J. Goulder, and B. D. Walker. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol. 811619-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuniga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 803122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]