Abstract
Although species C human adenoviruses establish persistent infections, the molecular details of this lifestyle remain poorly understood. We previously reported that adenovirus DNA is found in human mucosal T lymphocytes in a noninfectious form (C. T. Garnett, D. Erdman, W. Xu, and L. R. Gooding, J. Virol. 76:10608-10616, 2002). In this study, human tonsil and adenoid tissues were analyzed to determine the dynamics of infection, the rate of clearance of viral DNA, and the possibility of reactivation of virus from these tissues. The presence of viral DNA peaked at 4 years of age and declined thereafter. The average number of viral genomes declined with the age of the donor. The frequency of virus-bearing cells ranged from 3 × 10−7 to 3.4 × 10−4, while the amount of viral DNA per cell varied less, with an average of 280 copies per cell. All species C serotypes were represented in these tissues, although adenovirus type 6 was notably rare. Infectious virus was detected infrequently (13 of 94 of donors tested), even among donors with the highest levels of adenoviral DNA. Adenovirus transcripts were rarely detected in uncultured lymphocytes (2 of 12 donors) but appeared following stimulation and culture (11 of 13 donors). Viral DNA replication could be stimulated in most donor samples by lymphocyte stimulation in culture. New infectious virus was detected in 13 of 15 donors following in vitro stimulation. These data suggest that species C adenoviruses can establish latent infections in mucosal lymphocytes and that stimulation of these cells can cause viral reactivation resulting in RNA transcription, DNA replication, and infectious virus production.
The four species C serotypes (adenovirus type 1 [Ad1], Ad2, Ad5, and Ad6) are the most commonly encountered of the 51 known human adenoviruses. Species C viruses cause roughly 5% of symptomatic upper respiratory tract (18) and 15% of lower respiratory tract (2) infections in children under the age of 5. In a 6-year prospective study, adenoviruses were found in 8% of nasal washings from children with febrile respiratory illness, 81% of which were species C serotypes 1 or 2 (11). Primary infections with these serotypes usually occur within the first few years of life. By comparison, active species C infections were found in only 7 of 1,018 clinical respiratory illnesses in older children and young adults (12-14), and are virtually never seen in military recruits (28).
Species C adenoviruses are normally weakly pathogenic, with a frequency of asymptomatic primary infections of 50 to 90% (3, 11, 18). However, important exceptions exist. Adenovirus infections of very young children can lead to fatal pneumonia (8, 33, 50). Up to half of cases of intussusception (a bowel obstruction commonly caused by lymphoid hyperplasia) in children under the age of 2 are associated with the common adenoviruses (22, 43, 48). Adenoviruses are the most common cause of myocarditis in neonates and infants under 1 year of age (5, 40). Recently we reported a surprising association between prenatal species C adenovirus infection and the development of childhood acute leukemia (23).
Species C adenoviruses also cause significant morbidity and mortality among immunosuppressed individuals, including recipients of bone marrow (4, 16, 59), liver (7, 41), kidney (45), heart (53), and lung (6, 46) transplants, as well as in children with immunodeficiency diseases (24). Among transplant recipients, the incidence of adenoviral disease is much higher in children than in adults (16, 26).
Species C adenoviruses also establish persistent infections characterized by intermittent excretion (19). Although primary infections are respiratory, species C viruses are found in feces months, and even years, after virus is no longer detected in nasopharyngeal washings (18, 19). Restriction analysis of viruses isolated years after initial infection suggests persistent infection, rather than reinfection with the same serotype (1).
Early studies documented the persistence of small amounts of replication-competent species C adenovirus in tonsils and adenoids (14, 30, 58). However, the frequent finding of adenoviral DNA in tonsils that fail to yield infectious virus (44) led investigators to postulate that the virus is latent in these tissues. If indeed these viruses form latent infection in lymphoid tissues as part of their life cycle, one prediction is that quiescent viral DNA should be localized to a unique cell type, as is seen in other latent viruses (15, 49, 57). We have previously reported that adenovirus DNA in human tonsil and adenoid cells is preferentially located in the T-lymphocyte population (20). In the present study, we have expanded our analysis of human tonsil and adenoid tissues harboring species C adenoviruses to determine the dynamics of infection and clearance of viral DNA with age, the unique serotypes involved, the frequency of virus-bearing cells, and the levels of viral gene expression and infectious virus production with and without in vitro activation. These studies suggest that species C adenoviruses form latent infections in children that are generally cleared by the time they become young adults.
MATERIALS AND METHODS
Tonsil and adenoid cell suspensions.
Palatine tonsils or adenoids were obtained from 203 donors undergoing tonsillectomies at Egleston Children's Hospital (Atlanta, GA) following a diagnosis of tonsillar hypertrophy, recurrent bacterial infections, or recurrent otitis media. This study was performed under human investigation approval (no. 666-99) from the Emory University Internal Review Board. Pairs of tonsils from individual donors were pooled. In some cases tonsil and adenoid tissues removed from the same donor were analyzed separately, and in other cases the tonsil and adenoid tissues from the same donor were pooled. In the this paper, clinical samples of separate palatine tonsils or adenoids are designated T or A, respectively. Pooled tonsil and adenoid tissues are designated TA. Surgically removed tissue was placed in sterile HEPES-buffered Hanks' balanced salt solution (HH) containing 5% fetal bovine serum (FBS), 10 mM glutamine, 0.05 mg/ml gentamicin, and Antibiotic-Antimycotic mixture (AA) (Gibco catalog no. 15240-062) (HH5). Tissues were processed into a single-cell suspension as previously described (20). Briefly, tissue was pushed through a stainless steel wire screen, and cells were washed once in HH5 and then stored in aliquots of 1 × 108 to 2 × 108 cells per ml of 90% FBS and 10% dimethyl sulfoxide in liquid nitrogen. For analysis, vials of cells were thawed and viable lymphocytes purified by centrifugation through Ficoll type 400 (Sigma, catalog no. F5415) as described previously (20).
Cell digestion and DNA extraction.
Cells (5 ×106) were washed in phosphate-buffered saline (PBS), suspended in 50 μl of lysis buffer (0.45% NP-40, 0.45% Tween 20, 2 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.5 mg per ml proteinase K), and incubated at 55°C for 24 to 48 h. Tubes were vortexed intermittently to enhance enzymatic digestion. Following the 55°C incubation, proteinase K was inactivated at 95°C for 15 min and the samples stored at −20°C. Samples were thawed and vortexed vigorously before use. Five microliters of each DNA sample was used directly in the PCRs as described below.
Real-time quantitative PCR.
All PCR primers used in this study are described in Table S1 in the supplemental material. Quantitative analysis of species C adenovirus hexon DNA in tonsil and adenoid lymphocytes was performed using real-time PCR as previously described (20). Briefly, PCR amplification was carried out in 50-μl reaction mixtures consisting of Qiagen 1× PCR buffer, 2.25 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (Roche), 2.5 U Qiagen HotStar Taq polymerase, 0.5 μM of each primer, and 0.3 μM of TaqMan probe. Primers P1 and P2 were designed to amplify a conserved region of the species C adenovirus hexon gene, which was detected with the internal TaqMan probe P3. Serial 10-fold dilutions (from 5 × 107 to 5 copies) of Ad2 DNA (Invitrogen) were included with each analysis to generate a standard curve for quantitative assessment of donor adenovirus DNA.
All samples were also tested for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA by real-time PCR using primers P4, P5, and P6. Thermocycling profiles for real-time PCR consisted of 1 cycle of 95°C for 15 min and 50 cycles of 95°C for 15 s, 53°C for 35 s, and 72°C for 30 s in a Bio-Rad iCycler (Bio-Rad). All standard dilutions and samples were run in triplicate or duplicate for hexon or GAPDH, respectively.
To avoid sample-to-sample contamination, different rooms and dedicated equipment were used for DNA purification and processing, PCR setup, and gel analysis. The PCR setup hood was treated with UV light for 15 min prior to setting up any PCR amplification. Positive-displacement pipettes were used for PCR setup, and experimental samples were interspersed with blank or negative samples. In the experiments shown here, no signal was detected in any blank or negative samples.
Nested PCR for adenovirus hexon DNA.
Two different nested PCR assays were used to detect species C adenovirus hexon DNA in clinical samples. Nested PCR assay 1 (nPCR-1) used primers P7 to P10, derived from the Ad2 hexon sequence (GenBank accession no. BK000407), to amplify nucleotides 20721 to 21572, leading to an 852-bp product in the first round and a 286-bp product in the second round. Nested PCR assay 2 (nPCR-2) used primers P11 to P14, designed to amplify a conserved region of the species C adenovirus hexon gene corresponding to nucleotides 18838 to 19205 of Ad2 (GenBank accession no. BK000407). The first-round PCR, with an annealing temperature of 58°C, amplified a 368-bp product, and the second round PCR, with an annealing temperature of 60°C, amplified a 310-bp product from all four species C adenoviruses. Both nested assays routinely detected five copies of Ad1, Ad2, Ad5, and Ad6 and did not amplify representatives from serotypes A, B, D, E, and F (data not shown).
Adenovirus serotype determination by nested PCR product sequencing.
QIAquick (Qiagen) purified nested PCR products from the nPCR-1 assay were sequenced at Agencourt Bioscience Corporation (Beverly, MA). The Lasergene (DNA Star) software package was used for assembly and analysis of the sequences. Virus serotyping was performed by sequence comparison between the PCR product and known nucleotide sequences for species C adenoviruses published in GenBank by the National Center for Biotechnology Information. The predicted nucleotide sequences from the hexon gene were aligned and evaluated empirically by visual inspection for matching with the canonical sequences. Serotype-specific (canonical) bases with nucleotide numbers for Ad1 (nucleotides 21060 to 21345), Ad2 (nucleotides 21049 to 21334), Ad5 (nucleotides 21005 to 21290), and Ad6 (nucleotides 2197 to 2482) were identified by conserved nucleotide base changes within the 286-bp nPCR-1 products.
Species C serotype determination by fiber PCR.
For further evaluation of individual species C serotypes, four different nested PCR assays were developed, each detecting only Ad1, Ad2, Ad5, or Ad6, based on sequences in the fiber gene. Primers P15 to P18 were used to detect the Ad1 fiber gene, using an annealing temperature of 56°C for both rounds. The first round of Ad1 PCR produced a 379-bp product, and the second round produced a product size of 237 bp. Primers P19 to P22 were used to detect the Ad2 fiber gene, with an annealing temperature of 60°C for both first-round and second-round PCR. The Ad2 nested assay produced a first-round PCR product of 331 bp, and the nested round produced a product of 301 bp. The Ad5 fiber gene was detected by primers P23 to P26 used at an annealing temperature of 57°C for both rounds of PCR amplification. The first-round products were of 684 bp, and the nested PCR produced a product of size 347 bp. For Ad6 fiber gene detection, primers P27 to P30 were used at an annealing temperature of 60°C for both rounds of PCR amplification. The sizes of the first-round and second-round products were 308 bp and 303 bp, respectively.
The sensitivity of the assay was determined by performing PCR on 10-fold serial dilution of purified adenovirus DNA. The wild-type Ad1, Ad5, and Ad6 viruses were provided by Dean Erdman, Centers for Disease Control and Prevention, Atlanta, GA, and virus DNA was purified by standard methods (47). Purified Ad2 DNA was purchased from Invitrogen (Carlsbad, CA). For measuring the limits of detection of all of the qualitative and quantitative PCR assays, a known amount of purified Ad2 DNA, from 5 × 106 to 5 copies per reaction mixture, was amplified. The measured sensitivities of serotype assays for all species C were 5 genome copies.
Nested PCR for adenovirus E1A.
The adenovirus E1A genes were detected by nested EIA primers as described by Flomenberg et al. (17). This assay detects most Ad serotypes (species A to F). E1A primers were used at an annealing temperature of 56°C. Following the first round of amplification with primers P31 and P32 using 45 cycles, 5 μl of PCR product was amplified in the second round with primers P33 and P34 for 30 cycles.
Culture assay for infectious virus.
On day −1, 2 × 104 adenovirus-permissive A549 cells were plated per well (24-well plate) with 2 ml Dulbecco's modified Eagle's medium (DME) containing 10% FBS, 0.05 mg/ml AA, and 10 mM glutamine. Cells were left overnight in an 8% CO2 incubator. On day 0, each well was washed once with serum-free DME. A total of 107 Ficoll-purified lymphocytes in 100 μl PBS from adenoids and/or tonsils were freeze-thawed twice and then sonicated for 1 min. Twenty-five microliters of freeze-thawed lysate or control virus (PFU as indicated, usually between 3 and 30 PFU) was then placed onto the previously plated A549 cells in 200 μl of serum-free DME. The plate was incubated at 37°C with 8% CO2 for 2 h with occasional rocking. After 2 h, plates were washed twice, 1.3 ml of DME-2% FBS was added to each well, and the plate was returned to the incubator for 6 days. On day 6, 0.5 ml of DME-2% FBS was added to each well, and the plate was returned to the incubator for an additional 2 days. On day 8, a new 24-well plate was prepared with 1 ml DME-2% FBS per well. On day 8, cells were harvested, and one-fifth of each well volume was passed into the newly prepared 24-well plate. Samples were returned to normal growth conditions for an additional 2 days. On day 10, adherent and nonadherent cells were harvested and the cells processed for intracellular staining of the adenovirus hexon protein or for quantitation of viral DNA by real-time PCR.
Intracellular staining for adenovirus hexon protein.
Intracellular staining for hexon capsid protein was done essentially as described by Weaver and Kadan (61). Briefly, harvested A549 cells were washed in PBS with 2% FBS and adjusted to 1 × 106 cells/ml. Cells were centrifuged and suspended in 1% formaldehyde in PBS at a density of 1 × 106 cells per ml for 30 min at 25°C to fix the cells. Cells were then centrifuged and suspended in PBS with 0.2% Tween 20 at a density of 1 × 106 cells/ml for 15 min at 37°C to permeabilize the cells. Cells were washed twice in PBS with 2% FBS and suspended in the same buffer containing the primary antihexon antibody (Chemicon MAB 8051) at a 1:100 dilution of the 1-mg/ml stock for 30 min at 25°C. Purified mouse immunoglobulin G (Pharmingen 557273) was used as an isotype control for nonspecific staining. After primary antibody staining, cells were washed twice in PBS with 2% FBS and suspended in PBS with 2% FBS containing the secondary antibody, phycoerythrin-conjugated goat F(ab′)2 anti-mouse immunoglobulin G (Southern Biotechnologies no. 1032-09) for 30 min at 25°C. The secondary antibody was used at a 1:100 dilution of a 0.25-mg/ml stock. Cells were then washed in PBS with 2% FBS, resuspended in PBS with 2% FBS, and analyzed on a Becton Dickinson flow cytometer.
RT-PCR.
A total of 5 × 106 to 107 lymphocytes were centrifuged and suspended in 600 μl of RLT buffer from Qiagen. These samples were stored at −80°C until RNA purification. RNA was purified using the Qiagen RNA purification kit. Purified RNA was quantified and subjected to reverse transcription (RT) using the GeneAmp RNA PCR kit from Applied Biosystems (N808-0143). Twenty microliters of purified water was added to each 20-μl RT reaction mixture, and 5 μl of this sample was used directly for E1A, E3, and fiber PCR amplification using the Qiagen HotStar Taq polymerase according to the manufacturer's recommendations. E1A primers P35 and P36 were used at an annealing temperature of 55°C to amplify conserved region 1 of the E1A gene. Fiber was amplified by a seminested approach with primers P37 and P38 for 35 cycles with an annealing temperature of 56°C for the initial reaction. One microliter of this reaction product was amplified with primers P37 and P39 for 25 cycles under the same conditions.
Three sets of primers were used to detect E3 gp19K and E3 14.7K transcripts. Primers P40 and P41 were used to amplify E3 gp19K sequences of all species C adenoviruses. E3 14.7K sequences of Ad1 were amplified by primers P42 and P43, while the same gene from Ad2 was amplified by primers P44 and P45. All E3 primers were used at an annealing temperature of 53°C. All RT-PCR products were visualized on a 1.8% agarose gel stained with ethidium bromide. The gp19K and 14.7K PCR products were 159 bp and 175 bp, respectively. gp19K primers were used to detect each of the species C viruses, while the 14.7K primers could detect serotypes 1 and 2. PCR products were visualized on a 1.8% agarose gel stained with ethidium bromide.
Lymphocyte activation cultures.
Ficoll-purified tonsil and/or adenoid lymphocytes were harvested immediately (“initial”) or cultured at 106 cells/ml in RPMI complete medium (containing 10% FBS, 10 mM HEPES buffer, 10 mM glutamine, AA, and 0.05 mg per ml gentamicin) in a 37°C, 5% CO2 incubator (see Table 3 and Fig. 10B). Lymphocytes were cultured either alone or with phorbol myristate acetate (PMA) (50 ng per ml; Sigma) and ionomycin (1 μM; Calbiochem) (“cultured”) for 48 or 96 h. Samples (equivalent to 5 × 106 to 1 × 107 cells) were tested for adenovirus RNA by RT-PCR. Ficoll-purified tonsil and/or adenoid lymphocytes were either harvested immediately (107 cells, resuspended in DME and frozen at −70°C) (unactivated) or cultured at 3 × 106 cells/ml in RPMI complete medium containing human interleukin-2 (IL-2) (10 μg/ml; eBioscience, catalog no. 14-8029-63), ionomycin (0.25 μg/ml; Sigma, catalog no. I0634), and PMA (33 ng/ml; Fisher, catalog no. PRV1171) in 12-well plates at 1 ml/well in a 37°C, 5% CO2 incubator for 44 h (“activated”) (see Fig. 10C). The plates had been precoated with 1 ml of PBS containing anti-human CD3 (5 μg/ml; BD Pharmingen, catalog no. 555336) and anti-human CD28 (0.5 μg/ml; R&D Systems, catalog no. AF-342-PB) monoclonal antibodies. Following incubation, the activated lymphocytes were harvested, washed twice, and resuspended in DME. Both activated and unactivated lymphocytes were subjected to three cycles of freezing and thawing to release cell-adherent virus, followed by brief sonication and centrifugation. Supernatants were added to A549 cells and incubated at 37°C for 2 hours. Plates were then washed, and DME-2% FBS was added to each well. A549 cells were incubated for 10 days as described above for assay of infectious virus. Cells were then harvested and prepared as described above for real-time PCR quantitation of adenovirus DNA.
TABLE 3.
Virus gene expression by RT-PCR
| Donor status
|
Viral gene expression (no. positive/total)
|
||||||
|---|---|---|---|---|---|---|---|
| E1A
|
E3
|
Fiber
|
|||||
| DNA | Replicating virus | Initial | Cultured | Initial | Cultured | Initial | Cultured |
| Positive | Positive | 1/4 | 3/4 | 1/4 | 2/4 | 0/4 | 3/4 |
| Positive | Negative | 0/8 | 3/9 | 1/8 | 3/9 | 1/8 | 5/9 |
| Negative | Negative | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
FIG. 10.
In vitro lymphocyte activation leads to production of infectious species C adenovirus in tonsil and adenoid tissues. Donors containing moderate to high levels of adenovirus DNA but no detectable infectious virus were chosen for these experiments. (A) Adenovirus DNA levels as determined by real-time PCR in 15 positive donor samples and 1 negative donor sample. (B) Adenovirus DNA levels in lymphocytes from one adenovirus DNA-positive donor (58A) and one negative donor (31A) following activation with and without amplification on permissive A549 cells as described in Materials and Methods. Wild-type (rec700) virus (5 PFU/well) was added to A549 cells, and infected cells were harvested at the indicated times. (C) Adenovirus DNA levels following 10 days of amplification on A549 cells of virus from donor lymphocytes harvested directly (−) or activated for 44 h (+) as described in Materials and Methods.
Statistical analysis.
The number of viral genomes per 107 cells was log transformed for analyses by linear regression and analysis of variance (ANOVA). Pairwise comparison of genome copy numbers from adenoid and tonsil lymphocytes was performed by the Wilcoxon nonparametric rank test. Categorical analysis was performed by Fisher's exact test. P values of less than 0.05 were considered to be statistically significant. Analysis was performed with the suite of tools available as the standard package of the open-source computing environment R (www.R-project.org).
RESULTS
Adenovirus DNA is found in most tonsil and adenoid samples removed by tonsillectomy.
A total of 243 samples, containing Ficoll-purified mononuclear cells from the tonsils and/or adenoids of 203 donors (age 1 to 19 years), were analyzed by a nested PCR assay directed at the hexon region (hexon nPCR-1) for the presence of species C adenovirus DNA. This assay can detect three copies of DNA per reaction from all four species C serotypes, giving a lower limit of sensitivity of 240 or fewer copies in 107 donor lymphocytes. Of these donor samples, 186 (76%) contained detectable levels of the viral DNA. Of the 40 donors whose adenoid and tonsil tissues were assayed separately, 27 contained viral DNA in both tissues, and 2 contained viral DNA in neither tissue. The remaining 11 contained viral DNA in either the tonsil or the adenoid sample, and these 11 positive samples all had numbers of viral DNA copies at the lower limit of detection by the nested PCR assay (see below). In all, viral DNA was detected in tonsil or adenoid tissues from 160 of the 203 donors (78%).
Some samples were also evaluated for species C adenovirus DNA using a different nested PCR assay directed to another region of the hexon gene (hexon nPCR-2) but with a sensitivity similar to that of the first assay (three to five copies). Of 63 samples testing positive for viral DNA by hexon nPCR-1, 60 (95%) were also positive using the second nested PCR assay. Interestingly, the three negative samples contained relatively high levels of viral DNA by quantitative PCR, indicating that the failure to detect them in hexon nPCR-2 was unlikely to be due to sensitivity but rather was due to small sequence variations in the region of nPCR-2. All three negative samples contained species C serotype 5. In addition, of 65 samples from 43 donors that were negative by hexon nPCR-1, viral DNA was detected in only 3 (5%) (from 3 different donors) using either nPCR-2 or yet another assay that detects the E1A region (data not shown).
Figure 1 illustrates the relationship between donor age and the presence of adenovirus DNA in the tonsil and adenoid tissues of the donor population. The fraction of children with detectable adenovirus DNA increased from age 2 to 4. After age 4, this fraction declined with increasing age. The shape of the local regression curve in Fig. 1 suggests that the fraction of young children who contain species C adenovirus DNA in adenoid tissue is roughly 60% at age 2 (the youngest age of donors in this study) and increases until slightly beyond age 4. This analysis leads us to suggest that by age 4 virtually all children undergoing tonsillectomies will have species C adenovirus DNA in lymphoid cells of the tonsils and adenoids. After age 4, viral DNA appears to be lost from some of the individuals, such that approximately half of the donors contained detectable levels by age 15.
FIG. 1.
The fraction of children with adenovirus DNA in lymphoid cells of the adenoids and tonsils peaks at approximately 4 years of age. Adenoid and tonsil samples from 202 donors sorted by age were partitioned into 18 age groups. The percentage of adenovirus DNA-positive samples in each group is plotted as a function of the median age for the group. The solid line is a best-fit second-order local polynomial regression curve.
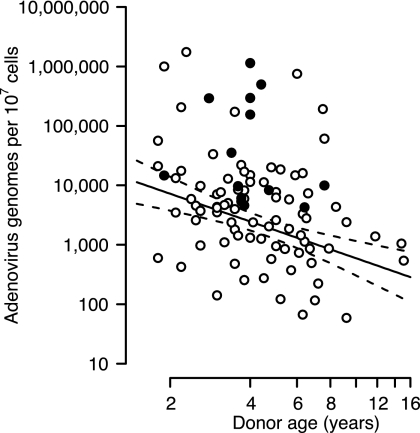
Quantitative real-time PCR was used to determine viral genome copy numbers in donor lymphocytes. Among samples determined to contain viral DNA by nested PCR, most (85%) contained sufficient DNA to be quantifiable by a real-time assay based on the inner primers from the nested assay. This assay (hexon QPCR-1) reliably detects 5 viral genomes per reaction, or 400 per 107 donor lymphocytes, for Ad1, Ad2, and Ad5 and is roughly 4 log units less sensitive to Ad6. Figure 2 depicts the relationship between donor age and amount of viral DNA in tonsil/adenoid lymphocytes, given as virus genome copy numbers per 107 nucleated cells. Although a wide range of values is represented in the 159 nonzero samples shown in Fig. 2, the number of genomes recovered per nucleated cell was inversely correlated with the age of the donor. The linear relationship observed with the log-log plot is consistent with a decline in both the number of viral DNA-containing cells and the amount of viral DNA in each cell. However, it is also possible that the number of viral DNA-positive cells remains constant and the level of viral DNA in each cell declines over time. Under this model, the amount of viral DNA per 107 cells declines with an apparent half-life of 2.6 years (95% confidence interval, 1.7 to 5.0 years). These findings, using nearly six times as many patient samples, confirm our previous suggestion (20) that the youngest donors contain the largest numbers of adenovirus genomes. Separate regression lines for viral DNA obtained from either adenoids or tonsils are offset and parallel, with adenoids containing more viral DNA than tonsils (data not shown), suggesting that similar rates of decline in viral DNA and possibly the number of DNA-containing cells occur in the two tissues.
FIG. 2.
The number of adenoviral genomes per lymphoid cell of the adenoids and tonsils declines with the age of the donor. The viral genome copy number per 107 nucleated cells was determined by quantitative PCR. Open symbols represents values obtained from donors with no replicating virus present in the tissue. Closed symbols represent values from donors with replicating virus present. The solid line represents the best-fit linear regression of log genome number versus log age. Dashed lines represent the 95% confidence interval for the regression line.
Early in these studies, where donor tissue consisted of both palatine tonsils and adenoids, these tissues were analyzed separately by quantitative PCR to identify any systematic difference between them in their adenovirus content. Of the 40 donors for which paired adenoid and tonsils were available, 33 contained more viral DNA (on a per-cell basis) in adenoid than in tonsil samples (P = 0.00014 by the Wilcoxon nonparametric rank test), while only 5 contained more viral DNA in the tonsil than in the adenoid sample. The amount of viral DNA recovered from the paired tissues for which viral DNA was quantifiable in both tissues is depicted in Fig. 3. The amount of viral DNA recovered from the paired tissues ranged from a low of 0.16-fold to a high of 730-fold more DNA being found in adenoid than tonsil, with a median value of 11.5-fold.
FIG. 3.
Lymphoid cells of the adenoid contain more adenovirus DNA than lymphoid cells of the tonsil. Pairwise comparisons of the adenovirus genome content from donors providing both adenoid and tonsil samples are shown. Closed symbols connected by a solid line represent donors with more viral DNA (per nucleated cell) in the adenoid than in the tonsil. Open symbols connected by a dashed line represent donors with less viral DNA in the adenoid than in the tonsil.
Only the four species C serotypes are found in nonreplicating form in tonsil tissues.
Serotype-specific nested PCR assays for the fiber gene were developed to determine which species C serotypes were present in 55 donor samples. Of the 103 species C viruses identified, only three were of serotype 6. The remaining samples were evenly distributed between serotypes 1, 2, and 5 (Fig. 4A). Multiple serotypes were found in nearly half of the donor samples, such that 21 samples (38%) contained two serotypes and 4 (7%) contained three species C serotypes. The distribution seen in Fig. 4B is consistent with a model in which the probability of harboring one species C adenovirus is independent of harboring another species C virus. Over the entire study population, the probability of a sample containing one or more species C virus is 73%. Under this model, the probability of acquiring a single species C serotype is approximately 46%. Neither the age of the donor nor the viral genome level was related to the presence of any specific serotype (P > 0.50 by one-way ANOVA). Similarly, the presence of multiple serotypes was statistically unrelated to the age of the donor (P = 0.07 by one-way ANOVA).
FIG. 4.
(A) Serotype 6 adenovirus is uncommon in lymphoid cells of the adenoids and tonsils compared to serotypes1, 2, and 5. Fiber-specific PCR was used to determine the serotype of virus present in samples obtained from 55 donors. The number of donors containing the indicated virus serotype is shown. (B) Multiple adenovirus serotypes are present in the adenoids and tonsils of nearly half of children undergoing tonsillectomy.
The PCR product amplified using hexon nPCR-1 (nucleotides 21060 to 21345 of Ad1) contains 21 nucleotides that differ among the four species C serotypes. In an earlier study, these sequence differences were used to determine which species C serotype was dominant in tonsil samples (20). Sequences in this area of the hexon gene were compared to sequences in GenBank for a total of 21 patient samples. Four each of Ad1, Ad2, and Ad6 samples contained identical nucleotides at the 21 variant residues. Seven of nine Ad5 samples were identical to published Ad5 sequence, but two of the Ad5 samples contained multiple nucleotide substitutions or deletions (see Fig. S1 in the supplemental material).
The PCR assays used to detect hexon and fiber sequences in this study were all designed and confirmed to be specific for the species C serotypes only. To determine whether any of the other subspecies of adenoviruses could be detected, 62 samples from 42 individual donors that lacked species C hexon DNA were tested for the presence of other adenovirus DNA using a nested PCR assay for the E1A region that detects representatives of all species of adenovirus (17). All samples were negative, suggesting that in this age group, only species C virus DNA is present in nonreplicating form in these mucosal lymphoid tissues.
Replicating virus is rare in tonsil tissues.
Early studies on the epidemiology of adenoviruses found that recurrent shedding of virus in stools of young children is the likely reservoir of infectious virus in the population (19). This suggests that reactivation of latent virus in mucosal lymphoid tissues maintains the life cycles of these endemic viruses. These early studies also suggested that, because individual infants could go long periods of time without shedding detectable virus and then have brief periods of virus shedding, only a minority of viral DNA-bearing donors should contain infectious virus at any given time. In our previous study (20) we reported finding that only 1 of 16 samples tested contained infectious virus as measured by coculture of tonsil lymphocytes with permissive cells and monitoring of cytopathology. Initial attempts to use plaque assays to detect infectious virus in tonsil samples revealed few, small plaques from some samples, suggesting that the persistent virus in these tissues may replicate and/or spread more slowly than the tissue culture-adapted “wild-type” strains to which they were being compared (not shown). An infectivity assay was developed that permitted longer incubation of virus with permissive cells followed by virus detection using intracellular staining for hexon and flow cytometry to confirm infection with adenovirus. Figure 5 illustrates the application of this assay using a small inoculum of Ad2 (5 PFU) compared to representative positive and negative patient samples. The infectivity assay was compared directly with the plaque assay on eight DNA-positive donor samples (Table 1). Of these samples, three (45A, 30A, and 15A) produced small numbers of plaques. All three plaque-positive samples were also positive in the infectivity assay. However, the infectivity assay detected virus growth in two samples (43T and 51A) that did not produce detectable plaques, making the infectivity assay a more sensitive measure of the presence of replicating adenovirus than the traditional plaque assay.
FIG. 5.
Flow cytometry analysis of adenovirus infectivity of clinical samples following 10 days of incubation on permissive (A549) cells. Lysates of donor lymphocytes were used to inoculate A549 cells, followed by a 10-day incubation. Cells were harvested and stained for intracellular hexon protein as described in Materials and Methods. Controls were uninfected A549 cells or A549 cells infected with 5 PFU Ad2. Patient samples contained the following numbers of species C adenovirus genomes/107 cells: 45A, 1,140,000; 36A, 292,000; 15A, 35,000; 31A, none detected; 39A, none detected.
TABLE 1.
Quantitation of infectious virus in tonsil and adenoid lymphocytes
| Donor sample | Genotype | Genomes per 107 cells | Plaques per 107 cells | Hexon-positive cells (%) after 10 daysa |
|---|---|---|---|---|
| 45A | Ad1 | 1,140,000 | 35 | 35, 64, 61 |
| 30A | Ad1 | 1,000,000 | 0 | 1, 0 |
| 36A | Ad1 | 292,000 | 5 | 6, 21 |
| 43T | Ad1 | 295,000 | 0 | 47, 49 |
| 15A | Ad1 | 35,000 | 5 | 8, 14 |
| 43A | Ad1 | 11,400 | 0 | 1, 0, 1 |
| 51A | Ad2 | 10,000 | 0 | 37, 33 |
| 44A | Ad2 | 11,400 | 0 | 0, 0 |
Multiple independent determinations.
The infectivity assay was used to test tissue samples from 90 DNA-positive and 4 DNA-negative donors, in which cells were thawed from liquid nitrogen and Ficoll purified to remove dead cells and debris and the remaining cells lysed to release virus. Replication-competent virus was identified in 11 of 90 DNA-positive donors (12.2%) and in none of the 4 DNA-negative donors. Eight donors with both adenoid and tonsil samples were included in this analysis. The status of replication-competent virus was identical in seven of the eight paired samples, with the exception being a donor with replicating virus in tonsil but not adenoid tissue. This donor was unusual in that the viral DNA load was substantially higher in the tonsils (295,000 genomes per 107 cells) than in the adenoids (15,000 genomes per 107 cells), perhaps due to an active adenovirus infection in tonsils at the time of tissue removal. Samples containing the highest levels of viral DNA (median, 22,000; range, 9,680 to 1.75 × 106 genomes per 107 cells) were more likely to contain replication-competent virus than samples with low levels of viral DNA (Fig. 6A). Nonetheless, most samples with substantial amounts of viral DNA (median, 2,800; range, 0 to 106 genomes per 107 cells) did not contain replication-competent virus, consistent with the notion that most virus present in the lymphocytes was not replicating at the time of tissue removal.
FIG. 6.
(A) Replication-competent virus is more frequently found in adenoid and tonsil tissue with high levels of viral genome. Adenoid and tonsil samples (n = 96) from 92 Ad-positive donors were partitioned in three groups based on adenovirus DNA content, and the fraction of samples containing replication-competent virus in each group is shown. (B) Replication-competent virus is found in adenoid and tonsil tissue over a wide range of ages. The samples analyzed for panel A were partitioned into four age groups, and the fraction of samples containing replication-competent virus in each group is shown.
Curiously, although younger donors tended to have more viral DNA, the greatest fraction of replication-competent virus was found in donors with a median age of 3.7 (Fig. 6B). Although this distribution was not statistically significant (P = 0.055, Fisher's exact test), the trend suggested by these results indicates that although replication-competent virus was most likely to be found in donors with high viral genome levels, this may be more likely in patients between 3 and 4 years of age. This relationship can also be seen in Fig. 2, where donor samples found to have replicating virus are seen above the regression line and clustered about age 4.
The failure of some samples to produce plaques when replicating virus was present coupled with the sometimes less robust replication in the 10-day-long infectivity assay (Fig. 5) led us to compare directly the kinetics of DNA replication between patient-derived and laboratory-adapted virus. For this experiment, A549 cells were inoculated with either the wild-type serotype 2 virus or lysates from five donor samples determined to contain replication-competent virus and from one negative sample. Viral DNA levels were measured in the A549 cells shortly after adsorption and 2, 5, and 10 days postinfection. As seen in Fig. 7A, levels of the laboratory-adapted virus DNA rose by approximately 4 orders of magnitude after only 2 days. In contrast, none of the patient-derived samples showed a substantial increase in viral DNA at this time (Fig. 7B). An increase in viral DNA was observed for three of the five positive samples after 5 days, but a significant change was not observed until 10 days postinfection. Note that by 10 days, viral DNA from some patient samples had amplified to the same level as the laboratory-adapted virus. The basis for these differences remains unknown; however, it seems unlikely that heritable differences in the viral genome contribute to this effect because with subsequent passages, the patient-derived virus replicated with the same kinetics as the laboratory-adapted virus (data not shown). Adenovirus DNA integrated into the cellular chromatin becomes methylated, which in turn diminishes transcription from viral promoters (9). Perhaps similar epigenetic modifications occur to viral DNA carried in a latently infected cell.
FIG. 7.
Replication-competent virus recovered from donor samples displays a lower initial rate of replication than laboratory-adapted virus. (A) Either 3 or 30 PFU of laboratory-adapted serotype 2 adenovirus was used to infect 2 × 104 A549 cells, and the amount of viral DNA in the culture was determined at the times indicated. (B) Freeze-thaw lysates of Ficoll-purified lymphocytes from the indicated donor samples were used to infect 2 × 104 A549 cells. The amount of viral DNA in the culture was determined at the times indicated. Open symbols represent donor samples from which replication-competent virus was detected. Closed symbols represent a negative control from an adenovirus DNA-positive sample with no replication-competent virus.
Following the 10-day incubation used to determine the presence of replicating virus, samples were collected and the serotype of the infectious virus was determined by fiber PCR (Table 2). In each of the nine samples tested, only a single serotype of virus was replicating, even in samples containing more than one serotype at the beginning of culture. In 5/9 samples (56%), the replicating virus was Ad1. Ad5 and Ad2 were found in 2/9 (22%) and 1/9 (11%), respectively. A single donor, 141T, contained replicating virus by hexon staining, but no DNA could be detected by fiber or hexon PCR assays, both of which are specific to species C adenoviruses. The replicating virus in 141T was tested by Dean Erdman at the Centers for Disease Control and Prevention and found to be Ad3, a relatively common species B serotype. This is the only time an adenovirus other than the species C viruses was detected in these tissues. Although serotypes 1, 2, and 5 were equally represented among donor samples, this limited survey identifies Ad1 as the replication-competent virus in the largest fraction of samples.
TABLE 2.
Replicating virus in donor samples
| Donor sample | Genomes per 107 cells | Serotype by PCR | Replicating virus serotype |
|---|---|---|---|
| 23T | 5,400 | 1 | 1 |
| 43T | 295,000 | 1 | 1 |
| 45T | 155,000 | 1 | 1 |
| 54A | 4,260 | 1, 2 | 1 |
| 141T | 1,752,000 | 1 | 3 |
| 144TA | 498,000 | 1, 5 | 1 |
| 171T | 14,600 | 1, 2, 5 | 5 |
| 182T | 9,700 | 2 | 2 |
| 205T | 8,300 | 5 | 5 |
Frequency of virus-containing cells.
A limiting-dilution assay was used to determine the frequency of tonsil lymphocytes that contained adenovirus DNA. For this assay, fivefold dilutions of Ficoll-purified donor lymphocytes were diluted with noninfected lymphocytic cells. Cells from each dilution were lysed and tested for the presence of adenovirus DNA using a nested PCR assay able to detect as few as three copies of the viral genome. The fraction of viral DNA-positive samples is plotted as a function of the log10 of the dilution in Fig. 8A. A nonlinear mixed-effects model was used to fit a two-parameter logistic equation to the data, in which the slope parameter was assumed to be constant between donors, while the inflection point was allowed to vary between donors. The x value at the point where the best-fit curve is 63% corresponds to one DNA-positive cell per well. This value, shown in each panel of Fig. 8A, represents the number of DNA-positive cells per 107 lymphocytes. For the samples analyzed here, this value ranged from 3 to 3,400 cells per 107 lymphocytes, for frequencies ranging from 3 × 10−7 to 3.4 × 10−4.
FIG. 8.
The frequency of adenovirus DNA-positive lymphocytes varies among donors and is correlated with the viral DNA load. (A) Fivefold dilutions of Ficoll-purified donor lymphocytes were analyzed by nested PCR to determine the frequency of cells with adenovirus DNA. The solid line represents a best-fit logistic regression line for each sample. The dashed lines show the x value where the regression line equals 63%, corresponding to one DNA-positive cell per well. The value above the dashed line is the number of DNA-positive cells per 107 lymphocytes. (B) The frequency of DNA-positive cells is correlated with the total number of genomes per 107 cells. The regression line describing the log frequency versus the log number of genomes per 107 cells is shown by the solid line. Dashed lines represent the 95% confidence interval for the regression line. The intersection of these lines with the y axis represents the estimated number of genomes per infected cell.
A plot of the frequency of DNA-positive cells as a function of the number of genomes per 107 cells is shown in Fig. 8B. The data can be described by a straight line. The projection of this regression line intersects the y axis when the x value corresponds to one adenovirus genome per cell. The reciprocal of this number is the average number of genomes per infected cell. The nine samples analyzed here yield an estimated value of 280 genomes per infected cell with a 95% confidence interval of 15 to 5,000 genomes per infected cell.
While measuring the number of viral genomes per cell and the frequency of infected cells, we noted substantial variation within some samples. To identify the source and nature of this variation, we performed repeated measures on replicate aliquots from nine donor samples. Between 9 and 18 replicate aliquots of 5 × 106 Ficoll-purified cells were separately lysed, and the number of adenovirus genomes present in each lysate was determined by quantitative PCR. Each value plotted in Fig. 9 is an average of three to five measurements from each replicate aliquot. The solid line shows the robust mean for all replicates from a single donor. This was determined by Huber's M-estimator, which diminishes the influence of outlying values on the mean. This analysis reveals at least two sources of heterogeneity. Variation around the mean was observed for all samples. In donor samples 43T, 45T, and 54A all measured values were within 6 standard errors of the mean (SEM) (dotted line). This heterogeneity among aliquots of 5 × 106 cells is consistent with the limited number of cells within each aliquot that are expected to contain adenovirus DNA. The Poisson distribution would govern the frequency of adenovirus-positive cells in each aliquot.
FIG. 9.
Repeated determinations of the number of viral genomes per lymphocyte reveal at least two sources of heterogeneity. The amount of adenoviral DNA in 9 to 18 replicate aliquots of 5 × 106 Ficoll-purified cells from nine donors was determined three to five times. The mean value for each replicate is plotted on a linear scale. The solid line shows the robust mean for all replicates from a single donor. The dashed line shows an upper limit corresponding to 6 SEM above the mean. Open symbols identify replicates that fall at or below the 6-SEM upper limit. Closed symbols identify replicate values above this limit. The donor number and tissue (A for adenoid and T for tonsil) is identified above each panel.
The second source of heterogeneity is apparent in the remaining samples analyzed in Fig. 9 and is reflected by values falling beyond the 6-SEM limit. It seems likely that these values arise from rare cells with substantially more viral genomes than the average value. Because typically only one or two replicates contained DNA at levels significantly above the 6-SEM limit, it seems likely that these replicates contained a single cell with approximately 3-to 10-fold more viral DNA than the typical infected cell. From Poisson considerations, the frequency of these cells is approximately 1 in 5 × 107, which is on the order of 3- to 3,000-fold less frequent than the typical adenovirus-positive cell.
Latent species C adenovirus DNA in tonsil and adenoid tissues.
In order for the nonreplicating adenovirus DNA detected in tonsil tissues to represent a stage in the life cycle of the virus, it must be capable of replicating and forming infectious virus when appropriate signals are delivered to the T cells that carry it. The capacity of tonsil-associated viral genomes to transcribe viral early and late genes was tested by incubating Ficoll-purified tonsil lymphocytes in vitro with and without PMA and ionomycin to simulate T-cell receptor signaling and testing for the presence of several virus transcripts by RT-PCR. The results are summarized in Table 3. At the initiation of culture, only 2 of 12 DNA-positive samples (17%) contained detectable viral RNA. Following in vitro culture, 11 of 13 (85%) were transcribing viral early and/or late genes, suggesting that some activation signals are indeed capable of stimulating viral transcription. The experiments summarized in Table 3 involved time course analysis of viral RNA over the period of from 8 to 96 h in culture. The donor was considered positive for the mRNA in question if it could be detected at least one of the time points. Most, but not all, samples were positive at multiple time points. Interestingly, the classical progression of immediate-early to early to late genes was not observed in these experiments, probably due to heterogeneity within the cell population being analyzed (data not shown).
If the quiescent adenovirus DNA in tonsil tissues is the reservoir for virus persistence in the population, then appropriate activation signals should also lead to viral DNA replication and infectious virus production. To test this prediction, lymphocytes from several DNA-positive, replicating-virus-negative donors were cultured for up to 4 days with or without PMA and ionomycin, and samples were removed at intervals for quantitation of adenoviral DNA. Samples from half the donors (10/20) contained higher levels of viral DNA following culture than uncultured samples (data not shown). To determine whether these increases in viral DNA involved production of infectious virus, lysates of cultured lymphocytes were added to permissive A549 cells and viral DNA was quantified after 10 days of culture. Most donors showed evidence of infectious virus production as monitored by the presence of replicating viral DNA following the 10-day culture on permissive cells (Fig. 10). Figure 10B illustrates the results of these experiments for two donors, 58A (viral DNA positive) and 31A (viral DNA negative). This approach was extended to a panel of donors, all of whom had moderate to high levels of viral DNA (Fig. 10A) and no evidence of infectious adenovirus at the time of tissue removal. Ficoll-purified lymphocytes were cultured for 44 h in medium containing PMA/ionomycin along with IL-2 and antibodies to CD3 and CD28 to maximize activation of T cells, followed by coculture of lysates on permissive A549 cells. After the 10-day coculture, A549 cells were harvested and tested for adenovirus DNA by real-time PCR. Figure 10C illustrates the results of this approach with 14 viral DNA-positive donors and one viral DNA-negative donor (31A). Including the findings with donor 58A, 13 of 15 adenovirus DNA-positive donors (87%) released infectious virus during the activation period. Serotype analysis revealed that all three of the common species C serotypes (Ad1, Ad2, and Ad5) were activated from latency in these experiments (not shown).
DISCUSSION
This study establishes that most (here 78%) pediatric donors carry species C adenovirus DNA in their nasopharyngeal lymphoid tissues, with the frequency of adenovirus-positive tissues peaking at around age 4 and then decreasing with age of the donor. This tempo of seeding of the tonsil tissues with adenoviruses parallels early studies that described the rapid rise in the development of anti-species C adenovirus antibodies during the first 5 years of life (29).
Among donor samples where tonsil (palatine tonsil) and adenoid (pharyngeal tonsil) samples were analyzed separately, significantly more virus was present in the adenoid than in the tonsil from the same donor (on a per-cell basis). A similar distinction between virus levels in tonsils versus adenoids was observed, with the same set of samples, for human bocavirus (38). In addition, early studies on outgrowth of virus from tissues explanted to culture found more adenovirus in adenoid than in tonsil tissues (30). The basis for this difference is unknown but may relate to differences in the epithelia of these organs and their susceptibility to infection by these viruses. Adenoids lie under a mucosa of pseudo-stratified columnar epithelium similar to the rest of the respiratory tract, while the tonsils are covered by stratified squamous epithelium (10).
Although all four species C serotypes were identified in tonsil tissues, serotypes 1, 2, and 5 each represented about a third of the total, with type 6 appearing only rarely. This distribution was somewhat surprising, given that serotypes 1 and 2 are encountered significantly more often in respiratory diseases than is type 5 (29, 52, 60). It is possible that type 5 is less pathogenic than types 1 and 2 and hence more likely to lead to asymptomatic infections but is equally prevalent in the population and in its ability to infect tonsil tissue long-term. This is supported by observations that the prevalence of antibodies against type 5 is only slightly less than that against types 1 and 2 in children (55). Nearly half of donor samples tested contained more than one species C serotype, and statistical analysis indicates that prior infection with one type has no effect on establishment of infection with another type. This suggests that cell-mediated immunity, which is cross-reactive among the species C serotypes (34, 54), has little or no impact on the establishment of persistent infection in lymphoid tissues. Thus, sequential infections with species C serotypes early in life lead to establishment of multiple persistent infections of the mucosa-associated lymphoid tissues.
Of the 21 samples analyzed by sequencing of the nPCR-1 hexon product, only two were different from the prototype sequences in GenBank. Four each of Ad1, Ad2, and Ad6 had sequences identical to those of published stains, but two of nine Ad5 isolates contained multiple substitutions or deletions compared to the GenBank sequence. Sequence variants in the hypervariable regions of Ad5 are encountered more frequently than with the other species C viruses in isolates from acute disease, which suggests that there is more diversity in the Ad5 population generally (37).
In the present study, fewer than 15% of samples containing adenoviral DNA also contained infectious virus. Quantitative analysis suggests that only a small amount of viral DNA is present as infectious virus, even in samples with large amounts of viral DNA. This finding is in sharp contrast to early descriptions of adenoviruses in tonsil and adenoid tissues, where most tissues explanted to culture yielded infectious virus. One key difference between the present study and early investigations is that the latter identified adenoviruses in tonsil and adenoid tissues by explanting pieces of these tissues to organ culture and observing over weeks or months for virus-specific cytopathic effects (14, 29, 30, 51, 58). The organ culture approach presumably included both viral DNA-containing lymphocytes and fully permissive fibroblast and epithelial cells and permitted small quantities of infectious virus to spread in the culture over time. In most cases, culture was necessary to detect infectious virus (58). In the present study, time in culture also appears to activate latent virus in the tissues, which was detected by transferring “activated” lymphocyte-derived virus onto permissive A549 cells instead of relying on permissive cells in the tonsil/adenoid tissue itself. Thus, the numbers of donors from which live virus can be recovered following culture approach the high levels noted in the early studies (14, 29, 30, 58). Reports of infection of laboratory workers in the early adenovirus groups suggest that some exogenous contamination might have elevated the frequency with which live virus was found in these studies (27, 29).
Among the samples investigated here, only species C viruses were detected, with one exception. A single donor sample contained replicating Ad3, a common species B serotype. No evidence of nonreplicating adenoviral DNA from any other species was identified in these samples. It remains possible that other serotypes of adenovirus form latent or persistent infections in other tissues, such as species B serotypes in kidney (25, 56, 60), lung (35), or possibly brain (32), but because of different tissue tropism, these viruses were not detected in tonsil and adenoid tissue.
In this study, a limiting-dilution approach was used to determine the frequency of cells containing species C adenovirus DNA in tonsil and adenoid tissues. A wide range of frequencies, from 3 to 3,400 DNA-containing cells per 107 total lymphocytes, was determined for eight donors, with a median frequency of 150 DNA-containing cells per 107 lymphocytes. These donors were chosen because of their relatively high levels of viral DNA, so this frequency range is likely higher than the overall average in the general population. However, it lies within the range encountered in Epstein-Barr virus latency, where healthy donors contain between 5 and 500 virus-bearing cells per 107 B lymphocytes in peripheral blood (31, 42).
While working with tonsil cells frozen in multiple aliquots in liquid nitrogen, marked sample-to-sample variation was observed. The analysis illustrated in Fig. 9 revealed that many donors contain a rare cell population that harbors large quantities of adenovirus DNA. At present the identity of this rare cell is not known. Its presence has so far defeated attempts to identify the T-cell subpopulation in which adenovirus is latent. It will be necessary to remove these cells prior to any analysis that relies on enrichment or depletion of viral DNA to identify the relevant cell subpopulations.
Two approaches were taken to determine whether the adenoviral DNA found in tonsil tissues represents a true latent state for the virus. First, the presence of virus transcripts was determined by RT-PCR before and after in vitro incubation under conditions that lead to generalized cellular activation. Although initial cultures rarely showed evidence of viral transcription, nearly all samples (85%) became transcriptionally active following culture. To determine whether DNA-containing samples could be induced to produce infectious virus, tonsil/adenoid cells were incubated with a cocktail of T-cell activation stimuli (PMA, ionomycin, IL-2, anti-CD3, and anti-CD28), and then lysates of the activated cells were placed on permissive cells and monitored for evidence of infection. Most (87%) showed evidence that activation does indeed lead to production of infectious virus. Although at present the natural ligands that trigger virus-containing T cells to initiate virus replication are unknown, it is clear that this pathway is available to the virus and represents the likely reservoir of species C adenoviruses in the human population (19). Furthermore, like all DNA viruses that form latent or persistent infections (21), human species C adenoviruses encode a variety of gene products, primarily within the E3 transcription unit, that function to counteract host antiviral defense mechanisms (36). We have previously reported that the E3 promoter is upregulated when cells are exposed to signals that activate T lymphocytes (39). Hence, it appears likely that the immune evasion strategies of these viruses are directed toward protecting the T lymphocyte from destruction during the period of viral activation from latency.
Supplementary Material
Acknowledgments
The assistance of Dean Erdman (CDC) in serotyping non-species C adenovirus is gratefully acknowledged.
We acknowledge NIH grants R01-AI052280 (to L.R.G.), T32-AI007610 (to G.T.), and RO1-CA077342 (to D.A.O.).
Footnotes
Published ahead of print on 24 December 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adrian, T., G. Schafer, M. K. Cooney, J. P. Fox, and R. Wigand. 1988. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol. Infect. 101503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avila, M. M., G. Carballal, H. Rovaletti, B. Ebekian, M. Cusminsky, and M. Weissenbacher. 1989. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 140634-637. [DOI] [PubMed] [Google Scholar]
- 3.Bell, J. A., R. J. Huebner, R. S. Paffenbarger, Jr., W. P. Rowe, R. G. Suskind, and T. G. Ward. 1956. Studies of adenoviruses (APC) in volunteers. Am. J. Public Health Nations Health 461130-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanke, C., C. Clark, E. R. Broun, G. Tricot, I. Cunningham, K. Cornetta, A. Hedderman, and R. Hromas. 1995. Evolving pathogens in allogeneic bone marrow transplantation: increased fatal adenoviral infections. Am. J. Med. 99326-328. [DOI] [PubMed] [Google Scholar]
- 5.Bowles, N. E., J. Ni, D. L. Kearney, M. Pauschinger, H. P. Schultheiss, R. McCarthy, J. Hare, J. T. Bricker, K. R. Bowles, and J. A. Towbin. 2003. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J. Am. Coll. Cardiol. 42466-472. [DOI] [PubMed] [Google Scholar]
- 6.Bridges, N. D., T. L. Spray, M. H. Collins, N. E. Bowles, and J. A. Towbin. 1998. Adenovirus infection in the lung results in graft failure after lung transplantation. J. Thorac. Cardiovasc. Surg. 116617-623. [DOI] [PubMed] [Google Scholar]
- 7.Cames, B., J. Rahier, G. Burtomboy, J. de Ville de Goyet, R. Reding, M. Lamy, J. B. Otte, and E. M. Sokal. 1992. Acute adenovirus hepatitis in liver transplant recipients. J. Pediatr. 12033-37. [DOI] [PubMed] [Google Scholar]
- 8.Deinhardt, F., V. V. Bergs, G. Henle, and W. Henle. 1958. Studies on persistent infections of tissue cultures. III. Some quantitative aspects of host cell-virus interactions. J. Exp. Med. 108573-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doerfler, W. 2008. In pursuit of the first recognized epigenetic signal—DNA methylation: a 1976 to 2008 synopsis. Epigenetics 3125-133. [DOI] [PubMed] [Google Scholar]
- 10.Dolen, W. K., B. Spofford, and J. C. Selner. 1990. The hidden tonsils of Waldeyer's ring. Ann. Allergy 65244-248. [PubMed] [Google Scholar]
- 11.Edwards, K. M., J. Thompson, J. Paolini, and P. F. Wright. 1985. Adenovirus infections in young children. Pediatrics 76420-424. [PubMed] [Google Scholar]
- 12.Evans, A. S. 1957. Acute respiratory disease in University of Wisconsin students. N. Engl. J. Med. 256377-384. [DOI] [PubMed] [Google Scholar]
- 13.Evans, A. S. 1957. Establishment of human adult tonsil cells in continuous culture and their virus susceptibilities. Proc. Soc. Exp. Biol. Med. 96752-757. [DOI] [PubMed] [Google Scholar]
- 14.Evans, A. S. 1958. Latent adenovirus infections of the human respiratory tract. Am. J. Hyg. 67256-266. [DOI] [PubMed] [Google Scholar]
- 15.Faulkner, G. C., A. S. Krajewski, and D. H. Crawford. 2000. The ins and outs of EBV infection. Trends Microbiol. 8185-189. [DOI] [PubMed] [Google Scholar]
- 16.Flomenberg, P., J. Babbitt, W. R. Drobyski, R. C. Ash, D. R. Carrigan, G. V. Sedmak, T. McAuliffe, B. Camitta, M. H. Horowitz, N. Bunin, and J. T. Casper. 1994. Increasing incidence of adenovirus disease in bone-marrow transplant recipients. J. Infect. Dis. 169775-781. [DOI] [PubMed] [Google Scholar]
- 17.Flomenberg, P., E. Gutierrez, V. Piaskowski, and J. T. Casper. 1997. Detection of adenovirus DNA in peripheral blood mononuclear cells by polymerase chain reaction assay. J. Med. Virol. 51182-188. [DOI] [PubMed] [Google Scholar]
- 18.Fox, J. P., C. D. Brandt, F. E. Wassermann, C. E. Hall, I. Spigland, A. Kogon, and L. R. Elveback. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am. J. Epidemiol. 8925-50. [DOI] [PubMed] [Google Scholar]
- 19.Fox, J. P., C. E. Hall, and M. K. Cooney. 1977. The Seattle virus watch. VII. Observations of adenovirus infections. Am. J. Epidemiol. 105362-386. [DOI] [PubMed] [Google Scholar]
- 20.Garnett, C. T., D. Erdman, W. Xu, and L. R. Gooding. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J. Virol. 7610608-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gooding, L. R. 1992. Virus proteins that counteract host immune defenses. Cell 715-7. [DOI] [PubMed] [Google Scholar]
- 22.Guarner, J., B. de Leon-Bojorge, E. Lopez-Corella, T. Ferebee-Harris, L. Gooding, C. T. Garnett, W. J. Shieh, J. Dawson, D. Erdman, and S. R. Zaki. 2003. Intestinal intussusception associated with adenovirus infection in Mexican children. Am. J. Clin. Pathol. 120845-850. [DOI] [PubMed] [Google Scholar]
- 23.Gustafsson, B., W. Huang, G. Bogdanovic, F. Gauffin, A. Nordgren, G. Talekar, D. A. Ornelles, and L. R. Gooding. 2007. Adenovirus DNA is detected at increased frequency in Guthrie cards from children who develop acute lymphoblastic leukaemia. Br. J. Cancer 97992-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hierholzer, J. C. 1992. Adenoviruses in the immunocompromised host. Clin. Microbiol. Rev. 5262-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hierholzer, J. C., N. O. Atuk, and J. M. Gwaltney, Jr. 1975. New human adenovirus isolated from a renal transplant recipient: description and characterization of candiate adenovirus type 34. J. Clin. Microbiol. 1366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard, D. S., I. G. Phillips, D. E. Reece, R. K. Munn, J. Henslee-Downey, M. Pittard, M. Barker, and C. Pomeroy. 1999. Adenovirus infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 291494-1501. [DOI] [PubMed] [Google Scholar]
- 27.Huebner, R. J., and W. P. Rowe. 1957. Adenoviruses as etiologic agents in conjunctivitis and keratoconjunctivitis. Am. J. Ophthalmol. 4320-25. [DOI] [PubMed] [Google Scholar]
- 28.Huebner, R. J., W. P. Rowe, J. R. Seal, H. C. Turner, J. E. Whiteside, and R. L. Woolridge. 1956. A study of the role of adenoviruses in acute respiratory infections in a navy recruit population. Am. J. Hyg. 64211-219. [DOI] [PubMed] [Google Scholar]
- 29.Huebner, R. J., W. P. Rowe, T. G. Ward, R. H. Parrott, and J. A. Bell. 1954. Adenoidal-pharyngeal-conjunctival agents: a newly recognized group of common viruses of the respiratory system. N. Engl. J. Med. 2511077-1086. [DOI] [PubMed] [Google Scholar]
- 30.Israel, M. 1962. The viral flora of enlarged tonsils and adenoids. J. Pathol. Bacteriol. 84169-176. [Google Scholar]
- 31.Joseph, A. M., G. J. Babcock, and D. A. Thorley-Lawson. 2000. EBV persistence involves strict selection of latently infected B cells. J. Immunol. 1652975-2981. [DOI] [PubMed] [Google Scholar]
- 32.Kosulin, K., C. Haberler, J. A. Hainfellner, G. Amann, S. Lang, and T. Lion. 2007. Investigation of adenovirus occurrence in pediatric tumor entities. J. Virol. 817629-7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladenheim, H. S., A. S. Mistchenko, and R. Drut. 1995. Expression of early and late adenoviral proteins in fatal adenovirus bronchopneumonia. Pediatr. Pathol. Lab. Med. 15291-298. [DOI] [PubMed] [Google Scholar]
- 34.Leen, A. M., U. Sili, E. F. Vanin, A. M. Jewell, W. Xie, D. Vignali, P. A. Piedra, M. K. Brenner, and C. M. Rooney. 2004. Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells. Blood 1042432-2440. [DOI] [PubMed] [Google Scholar]
- 35.Leung, A. Y., M. Chan, V. C. Cheng, K. Y. Yuen, and Y. L. Kwong. 2005. Quantification of adenovirus in the lower respiratory tract of patients without clinical adenovirus-related respiratory disease. Clin. Infect. Dis. 401541-1544. [DOI] [PubMed] [Google Scholar]
- 36.Lichtenstein, D. L., K. Toth, K. Doronin, A. E. Tollefson, and W. S. Wold. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 2375-111. [DOI] [PubMed] [Google Scholar]
- 37.Lu, X., and D. D. Erdman. 2006. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch. Virol. 1511587-1602. [DOI] [PubMed] [Google Scholar]
- 38.Lu, X., L. R. Gooding, and D. Erdman. 2008. Human bocavirus in tonsillar lymphocytes. Emerg Infect. Dis. 141332-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahr, J. A., J. M. Boss, and L. R. Gooding. 2003. The adenovirus E3 promoter is sensitive to activation signals in human T cells. J. Virol. 771112-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, A. B., S. Webber, F. J. Fricker, R. Jaffe, G. Demmler, D. Kearney, Y. H. Zhang, J. Bodurtha, B. Gelb, J. Ni, et al. 1994. Acute myocarditis. Rapid diagnosis by PCR in children. Circulation 90330-339. [DOI] [PubMed] [Google Scholar]
- 41.Michaels, M. G., M. Green, E. R. Wald, and T. E. Starzl. 1992. Adenovirus infection in pediatric liver transplant recipients. J. Infect. Dis. 165170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyashita, E. M., B. Yang, K. M. Lam, D. H. Crawford, and D. A. Thorley-Lawson. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80593-601. [DOI] [PubMed] [Google Scholar]
- 43.Montgomery, E. A., and E. J. Popek. 1994. Intussusception, adenovirus, and children: a brief reaffirmation. Hum. Pathol. 25169-174. [DOI] [PubMed] [Google Scholar]
- 44.Neumann, R., E. Genersch, and H. J. Eggers. 1987. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 793-97. [DOI] [PubMed] [Google Scholar]
- 45.Norris, S. H., T. C. Butler, N. Glass, and R. Tran. 1989. Fatal hepatic necrosis caused by disseminated type 5 adenovirus infection in a renal transplant recipient. Am. J. Nephrol. 9101-105. [DOI] [PubMed] [Google Scholar]
- 46.Ohori, N. P., M. G. Michaels, R. Jaffe, P. Williams, and S. A. Yousem. 1995. Adenovirus pneumonia in lung transplant recipients. Hum. Pathol. 261073-1079. [DOI] [PubMed] [Google Scholar]
- 47.Ornelles, D. A., R. N. Broughton-Shepard, and F. D. Goodrum. 2007. Analysis of adenovirus infections in synchronized cells. Methods Mol. Med. 13183-101. [DOI] [PubMed] [Google Scholar]
- 48.Porter, H. J., C. J. Padfield, L. C. Peres, L. Hirschowitz, and P. J. Berry. 1993. Adenovirus and intranuclear inclusions in appendices in intussusception. J. Clin. Pathol. 46154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 1024140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosman, F. C., A. S. Mistchenko, H. S. Ladenheim, J. P. do Nascimento, H. N. Outani, K. Madi, and H. L. Lenzi. 1996. Acute and chronic human adenovirus pneumonia: cellular and extracellular matrix components. Pediatr. Pathol. Lab. Med. 16521-541. [DOI] [PubMed] [Google Scholar]
- 51.Rowe, W., R. J. Huebner, and L. K. Gillmore. 1953. Isolation of a cytopathic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84570-573. [DOI] [PubMed] [Google Scholar]
- 52.Schmitz, H., R. Wigand, and W. Heinrich. 1983. Worldwide epidemiology of human adenovirus infections. Am. J. Epidemiol. 117455-466. [DOI] [PubMed] [Google Scholar]
- 53.Shirali, G. S., J. Ni, R. E. Chinnock, J. K. Johnston, G. L. Rosenthal, N. E. Bowles, and J. A. Towbin. 2001. Association of viral genome with graft loss in children after cardiac transplantation. N. Engl. J. Med. 3441498-1503. [DOI] [PubMed] [Google Scholar]
- 54.Smith, C. A., L. S. Woodruff, C. Rooney, and G. R. Kitchingman. 1998. Extensive cross-reactivity of adenovirus-specific cytotoxic T cells. Hum. Gene Ther. 91419-1427. [DOI] [PubMed] [Google Scholar]
- 55.Sterner, G. 1962. Adenovirus infection in childhood. An epidemiological and clinical survey among Swedish children. Acta Paediatr. Suppl. 1421-30. [PubMed] [Google Scholar]
- 56.Takayama, R., N. Hatakeyama, N. Suzuki, M. Yamamoto, T. Hayashi, Y. Ikeda, H. Ikeda, H. Nagano, T. Ishida, and H. Tsutsumi. 2007. Quantification of adenovirus species B and C viremia by real-time PCR in adults and children undergoing stem cell transplantation. J. Med. Virol. 79278-284. [DOI] [PubMed] [Google Scholar]
- 57.Taylor-Wiedeman, J., J. G. Sissons, L. K. Borysiewicz, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 722059-2064. [DOI] [PubMed] [Google Scholar]
- 58.van der Veen, J., and M. Lambriex. 1973. Relationship of adenovirus to lymphocytes in naturally infected human tonsils and adenoids. Infect. Immun. 7604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venard, V., A. Carret, D. Corsaro, P. Bordigoni, and A. Le Faou. 2000. Genotyping of adenoviruses isolated in an outbreak in a bone marrow transplant unit shows that diverse strains are involved. J. Hosp. Infect. 4471-74. [DOI] [PubMed] [Google Scholar]
- 60.Wadell, G. 1984. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110191-220. [DOI] [PubMed] [Google Scholar]
- 61.Weaver, L. S., and M. J. Kadan. 2000. Evaluation of adenoviral vectors by flow cytometry. Methods 21297-312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.