Abstract
A recombinant vesicular stomatitis virus (VSV-PeGFP-M-MmRFP) encoding enhanced green fluorescent protein fused in frame with P (PeGFP) in place of P and a fusion matrix protein (monomeric red fluorescent protein fused in frame at the carboxy terminus of M [MmRFP]) at the G-L gene junction, in addition to wild-type (wt) M protein in its normal location, was recovered, but the MmRFP was not incorporated into the virions. Subsequently, we generated recombinant viruses (VSV-PeGFP-ΔM-Mtc and VSV-ΔM-Mtc) encoding M protein with a carboxy-terminal tetracysteine tag (Mtc) in place of the M protein. These recombinant viruses incorporated Mtc at levels similar to M in wt VSV, demonstrating recovery of infectious rhabdoviruses encoding and incorporating a tagged M protein. Virions released from cells infected with VSV-PeGFP-ΔM-Mtc and labeled with the biarsenical red dye (ReAsH) were dually fluorescent, fluorescing green due to incorporation of PeGFP in the nucleocapsids and red due to incorporation of ReAsH-labeled Mtc in the viral envelope. Transport and subsequent association of M protein with the plasma membrane were shown to be independent of microtubules. Sequential labeling of VSV-ΔM-Mtc-infected cells with the biarsenical dyes ReAsH and FlAsH (green) revealed that newly synthesized M protein reaches the plasma membrane in less than 30 min and continues to accumulate there for up to 2 1/2 hours. Using dually fluorescent VSV, we determined that following adsorption at the plasma membrane, the time taken by one-half of the virus particles to enter cells and to uncoat their nucleocapsids in the cytoplasm is approximately 28 min.
Vesicular stomatitis virus (VSV), the prototypic rhabdovirus within the family Rhabdoviridae and the order Mononegavirales, is an enveloped virus with a negative-stranded RNA genome of 11,161 nucleotides. The viral genome encodes five proteins, namely, the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large polymerase protein (L) (35). The genome is present within the virion core as a ribonucleoprotein (RNP) or nucleocapsid (NC) complex tightly encapsidated by the N protein and associated with the viral RNA-dependent RNA polymerase, a multiprotein complex of the viral L and P proteins. The G protein forms spikes on the viral envelope, binds to cell surface receptors, and plays a role in entry of virus into susceptible cells. The M protein is multifunctional; it plays a role in virus assembly and is responsible for cytopathogenesis observed in virus-infected cells (6).
Studies on viral protein transport and virus motility in infected cells have been facilitated by imaging of fluorescent proteins fused to viral structural proteins (for reviews, see references 8 and 23). Recent advances in imaging techniques, coupled with the ability to genetically tag viral structural proteins with fluorescent proteins or to label viral membranes with lipophilic dyes, have allowed studies of the dynamic events of virus entry as well as of virus-cell interactions (17, 27-29, 31, 34, 40, 43, 51, 53). For enveloped viruses, the hallmark event of infection is the fusion of the viral envelope and the release of the NC (or RNP) into the cytoplasm. To examine the infection process by fluorescence microscopy and to distinguish between the enveloped virion and the uncoated NC, it is essential to differentially label the viral envelope and the NC core. Dually fluorescent viruses in which the viral core component, such as the NC or the RNP, is labeled with one fluorescent color and the envelope component is labeled with another color are thus powerful reagents for studies of virus entry and NC uncoating during early stages of infection as well as for studies of virus assembly during late stages of infection. Recently, dually fluorescent rabies virus (27) and human immunodeficiency virus (HIV) (9, 30) have been generated successfully. Using the dually fluorescent rabies virus, it was demonstrated that complete virus particles are transported in a retrograde manner (27).
Successful recovery of a recombinant VSV encoding the P protein fused in frame with enhanced green fluorescent protein (PeGFP) allowed us to track the intracellular transport of viral NCs by live-cell imaging (14). This study demonstrated that microtubules were involved in viral NC transport toward the cell periphery (14), presumably to the plasma membrane for virus assembly. Whether the M protein interacts with the viral NCs before transport to the plasma membrane or at the plasma membrane prior to virus assembly remains a fundamental question in VSV assembly. Previous studies have shown that the M protein and the NCs do interact in vitro and in vivo (11, 12, 26), although more recent studies suggest that such interactions may occur only at the plasma membrane (18, 54). To examine the events of virus entry, uncoating, and also assembly, we wanted to generate a dually fluorescent VSV encoding PeGFP and monomeric red fluorescent protein fused in frame with the M protein at its carboxy terminus (MmRFP). Although repeated attempts to recover VSV with the fluorescently tagged M protein in place of wild-type (wt) M were unsuccessful, viruses encoding the fluorescently tagged M protein could be recovered when it was inserted as an extra cistron at the G-L gene junction. Further use of this virus for studies of virus entry, uncoating, and egress was limited because the MmRFP fusion protein was not incorporated into the virions.
Recently, a new method of genetic tagging of proteins for fluorescence imaging was developed wherein the protein is tagged with a relatively smaller tetracysteine (tc) motif (CCPGCC). This motif can be recognized specifically by membrane-permeable biarsenical dyes that fluoresce when covalently bound to the cysteine pairs in the tc motif (1, 24, 39). Such a small tag can be fused to the protein of interest with minimal disruption of protein function. This is a powerful approach for real-time visualization of nascent protein synthesis and trafficking, as the existing and newly synthesized pools of proteins can be labeled differentially with the two fluorescent biarsenical dyes, FlAsH (green) and ReAsH (red) (38, 48). Using a tc-tagged M protein (Mtc) encoded in place of the wt M protein in the VSV genome, we rescued recombinant viruses (VSV-PeGFP-ΔM-Mtc and VSV-ΔM-Mtc) and demonstrated that the Mtc was incorporated into infectious virions in amounts similar to that observed for M protein in wt VSV. Moreover, dynamic imaging of newly synthesized M protein by sequential labeling with the two biarsenical dyes revealed that the M protein is transported from the site of synthesis inside the cytoplasm to the plasma membrane in less than 30 min. We have also shown that the M protein reaches the plasma membrane independent of NCs and the microtubules. Additionally, our results show that following adsorption, entry and uncoating of VSV in the infected cells occur with a half-life of approximately 28 min.
MATERIALS AND METHODS
Cell culture, viruses, antibodies, and reagents.
Baby hamster kidney (BHK-21) cells were grown and maintained as described before (16). Stocks of vaccinia virus vTF7-3 (19) were prepared and titrated in BHK-21 cells as described previously (25). VSV (Indiana serotype) was grown and titrated in BHK-21 cells. Mouse anti-VSV antibody has been described previously (15). Anti-M monoclonal antibody (MAb)-producing 23H12 hybridoma cells were kindly provided by Douglas Lyles (32). Supernatant from the hybridoma culture was used as the source of MAb. Secondary antibodies were obtained from Molecular Probes. The biarsenical dyes, ReAsH and FlAsH, were obtained from Invitrogen. Nocodazole (NOC), colcemid, and cycloheximide were obtained from Sigma-Aldrich and were used at concentrations of 10 μg/ml, 10 μg/ml, and 100 μg/ml of growth medium, respectively.
Plasmid construction, transfection, and virus recovery.
The plasmid pGEM-M encodes wt M protein of VSV under the control of the T7 RNA polymerase promoter. The coding region of mRFP was amplified from the plasmid pRSET-mRFP, kindly provided by Roger Tsien (10), using primers RFPBamSnF and RFPBsiSphR (Table 1), and cloned into pGEM-3 vector under the control of the T7 RNA polymerase promoter, resulting in the plasmid pGEM-mRFP. The pGEM-MmRFP and pGEM-mRFPM plasmids encode MmRFP and mRFPM fusion proteins, with mRFP fused in frame at the carboxy and amino termini of the M protein, respectively. To construct the pGEM-MmRFP plasmid, a two-step PCR procedure was used. The sequences of the primers used to construct these fusion proteins are shown in Table 1. In the first PCR, a forward primer (MBamSnF) containing the ATG start codon and the downstream 12 nucleotides from the 5′ coding region of VSV M protein and a reverse primer (M-RFPR) containing the 3′ end of the M coding region and the 5′ end of the mRFP coding region were used to amplify the entire M coding region. Another PCR was performed with a forward primer (M-RFPF) comprising the 3′ end of the M coding region and the 5′ end of the mRFP coding region and a reverse primer (RFPBsiSphR) containing the 3′ end of the mRFP coding region to amplify the entire mRFP coding region. In the second-round PCR, which combined the M and mRFP coding regions, the above two PCR products were annealed together and amplified by PCR, using the MBamSnF and RFPBsiSphR primers. The PCR product was digested with BamHI and SphI and cloned into pGEM vector that had been digested with the same enzymes. The plasmid pGEM-mRFPM was also generated using a similar PCR approach, with the primers RFPBamSnF, RFP-MR, RFP-MF, and MBsiSphR, described in Table 1. To construct the plasmid coding for the M protein fused with a tc motif at the carboxy-terminal end, the forward primer MBamSnF was used in conjunction with a reverse primer (MtcBsiSphR) constituting the 3′ end of the M coding sequence and the coding region of the tc tag (FLNCCPGCCMEP) (39). The PCR product was then digested with BamHI and SphI and cloned into pGEM vector, and the plasmid was designated pGEM-Mtc. The coding regions of MmRFP, mRFPM, and Mtc in the constructs were checked for accuracy by nucleotide sequencing of the clones.
TABLE 1.
Primers used in this study
| Primer | Nucleotide sequence (5′-3′)a |
|---|---|
| VSVXbaI2070F | ATATATCTCATCTAGAGGAGAGTTCATCT |
| VSVSnaBI2240R | CTTTAAGGAACTCATGGTACGTAGATTGGGATAACAC |
| VSVMEco47III2378R | ATATATGGAGCGCTCGGAGCATACTCCATGCT |
| VSVGMluIR | GCACTTCATAGTGACGCGTAAACAGATCGATCTCT |
| VSVMEco47IIIF | GAGTATGCTCCGAGCGCTCCAATTGACA |
| VSVMBsiWI2943R | GTTCAGAAGCTAGAAGTCGTACGAGCTCATTTGAAGTGG |
| MBamSnF | ATATATGGATCCTACGTACCATGAGTTCCTTAAAG |
| RFPBamSnF | ATATATGGATCCTACGTACCATGGCCTCCTCCG |
| MBsiSphR | ATATATGCATGCCGTACGAGCTCATTTGAAGTGGCTGATAG |
| RFPBsiSphR | ATATATGCATGCCGTACGTTAGGCGCCGGTGGAGTG |
| M-RFPF | GATTCTATCAGCCACTTCAAAATGGCCTCCTCCGAGGACGTC |
| M-RFPR | GACGTCCTCGGAGGAGGCCATTTTGAAGTGGCTGATAGAATC |
| RFP-MF | CGCCACTCCACCGGCGCCATGAGTTCCTTAAAGAAG |
| RFP-MR | CTTCTTTAAGGAACTCATGGCGCCGGTGGAGTGGCG |
| MBamBsiF | ATATATGGATCCCGTACGGCCGCCACCATGAGTTCCTTAAAG |
| MtcBsiSphR | ATATATGCATGCCGTACGCTAcggttccatgcaacatccgggacaacagttgaggaa TTTGAAGTGGCTGATAGAATC |
Restriction enzyme sites are shown in bold. Sequences corresponding to the tc tag are shown with lowercase letters.
To insert the coding region for the MmRFP fusion protein as an extra cistron at the G-L gene junction in the full-length VSV genome, the plasmid FLVSVBsiWI (14) was used. The MmRFP region was PCR amplified from the plasmid pGEM-MmRFP, using primers MBamBsiF and RFPBsiSphR (Table 1), digested with the enzyme BsiWI, and cloned into plasmid FLVSVBsiWI that had been digested with the same enzyme. The resulting plasmid was designated pVSV-M-MmRFP. A full-length genome plasmid encoding both PeGFP and MmRFP was then generated by fragment exchanges between plasmids pVSV-MmRFP and pVSV-PeGFP, using the restriction enzymes BstZ17I and MluI, and the resulting plasmid was designated plasmid pVSV-PeGFP-M-MmRFP.
To introduce the MmRFP or Mtc coding sequence into the full-length VSV genome in place of the wt M gene, two unique restriction sites were engineered, one (SnaBI) before the start codon and the other (BsiWI) after the stop codon of the M gene in the plasmid pVSV-PeGFP (14), using the megaprimer method (52) of PCR, to give rise to the plasmid pVSV-PeGFPSB. For insertion of the SnaBI site at the 5′ noncoding region of the M gene, a PCR was performed using a forward primer (VSVXbaI2070F) spanning the XbaI site, located in the P gene, in combination with a reverse primer (VSVSnaBI2240R) containing the SnaBI site, which had been placed upstream of the ATG start codon of the M gene. This PCR product was used as a megaprimer for another round of PCR with a reverse primer (VSVMEco47III2378R) spanning the Eco47III site, located inside the M coding region. For both PCRs, plasmid pVSV-PeGFP was used as the DNA template. The final PCR product was then digested with the enzymes XbaI and Eco47III and cloned into pVSV-PeGFP plasmid that had been digested with the same enzymes. The resulting plasmid, which contained the SnaBI site, was designated pVSV-PeGFPSnaBI. To insert the BsiWI site at the 3′ noncoding region of the M gene, a forward primer (VSVMEco47IIIF) spanning the Eco47III site and a reverse primer (VSVMBsiWI2943R) containing a BsiWI site placed downstream of the stop codon of the M gene were used to generate a megaprimer, using plasmid pVSV-PeGFP as the template. For the second round of PCR, the megaprimer was used in conjunction with a reverse primer (VSVGMluIR) spanning the MluI site, located at the 5′ noncoding region of the G gene, with plasmid pVSV-PeGFP as the template. The second PCR product was digested with the enzymes Eco47III and MluI and replaced in the corresponding region of plasmid pVSV-PeGFPSnaBI that had been digested with the same enzymes, resulting in the plasmid pVSV-PeGFPSB. These two restriction sites were also inserted in the corresponding region of plasmid pFLVSV to give rise to plasmid pFLVSVSB. The plasmid pVSV-PeGFPSB was then used to replace the coding sequence of the wt M protein with that of either MmRFP or Mtc to give rise to the plasmid pVSV-PeGFP-ΔM-MmRFP or pVSV-PeGFP-ΔM-Mtc, respectively. The MmRFP or Mtc coding region was also inserted into plasmid pFLVSVSB in place of wt M, resulting in the plasmid pVSV-ΔM-MmRFP or pVSV-ΔM-Mtc, respectively. All clones were sequenced to ensure that only plasmids with the correct coding sequences were used for further studies. Virus recovery and identification of recovered viruses were performed as described previously (15). The recovered viruses were designated VSV-ΔM-Mtc and VSV-PeGFP-ΔM-Mtc and encoded Mtc in place of M.
Metabolic labeling and analysis of proteins in transfected and infected cells.
Metabolic labeling of proteins in transfected or virus-infected cells or of proteins in virions, immunoprecipitation using anti-VSV antibody (1:100) or anti-M antibody (1:100), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detection of proteins by fluorography were carried out as described previously (15). Proteins incorporated into extracellular virions were examined by radiolabeling as described previously (14).
Determination of single-step growth kinetics.
Single-step growth kinetics of wt and mutant viruses in BHK-21 cells were determined as described previously (15).
Biarsenical labeling of transfected or virus-infected cells.
Biarsenical labeling with FlAsH and ReAsH was performed per the manufacturer's recommendations provided in the tc tag detection kit (Invitrogen). Briefly, BHK-21 cells were grown on glass coverslips in 12-well cell culture plates. At 4 h postinfection (hpi) or posttransfection (hpt), cells were washed once with Opti-MEM (Invitrogen) and labeled with ReAsH dye (1.25 μM to 2.5 μM) in 400 μl of Opti-MEM for 30 min at room temperature. Supernatant was then discarded, and cells were washed in BAL (2,3-dimercapto-1-propanol) wash buffer (supplied in the kit; Invitrogen) in Hanks' balanced salt solution three times at room temperature. Subsequently, the cells were fixed in 4% paraformaldehyde and processed for staining with anti-M antibody as described previously (14). For dual biarsenical labeling, cells infected with VSV-Mtc or transfected with pGEM-Mtc were treated with ReAsH for 30 min at 3 hpi or 4 hpt, respectively, as described above. Supernatant was discarded, and cells were washed in BAL wash buffer and either labeled immediately with the second biarsenical dye, FlAsH (1.25 μM to 2.5 μM), in Opti-MEM or incubated at 37°C in complete growth medium for 1 or 2 h before FlAsH labeling. For the later time period analyses, cells were again rinsed with Opti-MEM and labeled with FlAsH (1.25 μM to 2.5 μM) for 30 min at room temperature. After FlAsH labeling, cells were washed thoroughly in BAL wash buffer in Hanks' balanced salt solution as described above and processed for staining with anti-M antibody. Cells were then visualized under an inverted confocal microscope. For ReAsH labeling of extracellular virus particles, infected cells at 4 hpi were treated with ReAsH as described above for 12 h. The culture supernatant containing the dye-labeled virus was collected and clarified. The virus particles were purified by sucrose density gradient centrifugation and used for further studies.
Fluorescence microscopy.
For epifluorescence and differential interference contrast microscopy, BHK-21 cells were grown in 35-mm culture dishes, with or without glass coverslips, and were either fixed with 4% paraformaldehyde or visualized directly under an Olympus FV500/IX81 inverted laser scanning confocal microscope as described previously (14). Immunofluorescence staining was performed with anti-M MAb (1:100) as described previously (14). For measurement of fluorescence intensities of virus particles, images of particles were collected with an Olympus FV500 confocal laser scanning microscope, using a dual-laser line and sequential image collection program (488-nm excitation and 522-nm emission for GFP; 565-nm excitation and 595-nm emission for ReAsH) with the same confocal settings for laser power, gain, photomultiplier tube, and offset levels. The mean fluorescence intensities of viral particles were obtained with the same defined threshold and intensity measurement for green and red panels, using AnalySIS-Opti image analysis software (Soft Imaging System Co., Lakewood, CO). The relative fluorescence intensities were calculated with the green fluorescence intensity set arbitrarily at 100. For measurement of the relative levels of ReAsH-labeled Mtc on the membranes of cells treated with or without NOC, fluorescence intensities on the membrane regions of the cells infected with VSV-PeGFP-ΔM-Mtc were quantitated using the software FV500, version 4.3, with an FV500/IX81 inverted confocal microscope (Olympus). Twelve randomly selected membrane areas of equal size were chosen per cell, and eight cells from each of the two independent experiments, treated with or without the drug, were used for data collection. Total fluorescence intensities were determined in arbitrary units and analyzed statistically as described below.
Statistical analysis.
Differences in total fluorescence intensity on membranes in BHK-21 cells after VSV-PeGFP-ΔM-Mtc virus infection for drug-treated and untreated groups were analyzed by the t test, using SAS software, version 9.1 (SAS Institute Inc., Cary, NC). P values of >0.05 were considered insignificant.
RESULTS
Dually fluorescent VSV expressing PeGFP and MmRFP fusion proteins.
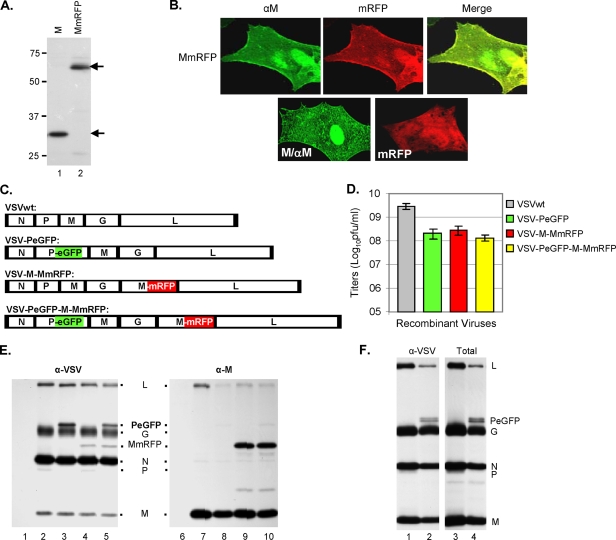
Previously, we reported the recovery of a fluorescent virus (VSV-PeGFP) encoding PeGFP, a fusion protein with EGFP inserted in frame within the P protein (14). In the present study, we attempted to generate a dually fluorescent recombinant VSV encoding mRFP fused in frame with the matrix protein (M) in the genomic background of VSV-PeGFP. The use of such a dually fluorescent VSV will be invaluable in examining virus uncoating during the initial stages of the infection cycle and virus assembly at the late stages of infection. To generate the dually fluorescent VSV, we first constructed a fusion protein (MmRFP) in which the entire coding region of mRFP was fused in frame at the carboxy terminus of the M protein. In cells transfected with the plasmid encoding MmRFP, the fusion protein was expressed with the predicted molecular mass of ∼65 kDa (Fig. 1A), at levels similar to that of the M protein, and was also found to be as stable as the M protein (data not shown). Unlike mRFP, which was localized in the cytoplasm (Fig. 1B, lower right panel), the MmRFP fusion protein was found to be associated with the plasma membrane (Fig. 1B, upper panels) in a pattern that is typical of the M protein (Fig. 1B, lower left panel) (12).
FIG. 1.
Recovery and characterization of recombinant VSV encoding fluorescent fusion proteins. (A) Expression of M and MmRFP fusion proteins. BHK-21 cells infected with vTF7-3 and transfected with a plasmid encoding M (lane 1) or MmRFP (lane 2) were radiolabeled with Expre35S35S label for 2 h at 16 hpt. The radiolabeled proteins were immunoprecipitated with anti-VSV antibody, separated by SDS-PAGE, and detected by fluorography. Size markers in kDa are shown on the left. The M and MmRFP proteins are identified with arrows on the right. (B) Epifluorescence and immunofluorescence staining of cells expressing the MmRFP, M, or mRFP protein. (C) Recombinant VSV genome plasmids. VSVwt, the wt VSV genome, with the N, P, M, G, and L genes shown in rectangular boxes; VSV-PeGFP, genome encoding PeGFP in place of wt P; VSV-M-MmRFP, genome encoding MmRFP at the G-L gene junction; VSV-PeGFP-M-MmRFP, genome encoding MmRFP at the G-L gene junction of the VSV-PeGFP genome. Intergenic regions as well as 3′-leader-gene and 5′-trailer sequences are shown in black boxes. (D) Growth of recombinant viruses. BHK-21 cells were infected with various recombinant VSVs at an MOI of 5. Growth of each recombinant virus was determined by plaque assay of culture supernatants at 16 hpi. Virus titers shown are averages from three independent experiments, with the standard deviations indicated by error bars. (E) Analysis of viral proteins in cells infected with recombinant VSVs. BHK-21 cells were infected with wt VSV (lanes 2 and 7), VSV-PeGFP (lanes 3 and 8), VSV-M-MmRFP (lanes 4 and 9), or VSV-PeGFP-M-MmRFP (lanes 5 and 10) at an MOI of 10 or were left uninfected (lanes 1 and 6). The proteins were radiolabeled for 1 h at 4 hpi, immunoprecipitated with anti-VSV antibody (lanes 1 to 5) or with anti-M antibody (lanes 6 to 10), separated by SDS-PAGE, and detected by fluorography. The viral proteins along with the fusion proteins are identified in the middle. (F) Examination of proteins incorporated into extracellular virions. The viral proteins in cells infected with VSV or VSV-PeGFP-M-MmRFP were radiolabeled with Expre35S35S label for 12 h at 4 hpi, and the proteins incorporated into the purified virions (VSV, lanes 1 and 3; and VSV-PeGFP-M-MmRFP, lanes 2 and 4) were examined by immunoprecipitation with anti-VSV antibody (lanes 1 and 2) or as total proteins (lanes 3 and 4). The positions of the viral proteins are shown on the right.
Several attempts to recover viruses encoding MmRFP in place of wt M were unsuccessful, perhaps indicating that the MmRFP fusion protein is not competent to support infectious virus assembly and budding. Subsequently, plasmids carrying recombinant VSV genomes constructed by inserting the MmRFP coding sequence between the G-L gene junction in the wt VSV or VSV-PeGFP genome (Fig. 1C) led to recovery of the recombinant viruses (VSV-M-MmRFP and VSV-PeGFP-M-MmRFP). Examination of single-step and multistep growth kinetics revealed no apparent differences between the viruses (data not shown), although the overall yields of the viruses were lower than that of wt VSV (Fig. 1D). The VSV-PeGFP-M-MmRFP virus grew to a final titer that was 25- to 50-fold less than that of wt VSV, whereas the VSV-M-MmRFP titer was about 10-fold less than that of wt VSV. Examination of the viral proteins synthesized in cells infected with these recombinant viruses suggested that both PeGFP and MmRFP fusion proteins of predicted molecular masses were detected in appropriate recombinant VSV-infected cells (Fig. 1E). In most cells infected with VSV-PeGFP-M-MmRFP, both green and red fluorescence was readily observed, and these cells appeared yellow when observed simultaneously under both laser lights (data not shown). However, a small number of patches of cells infected with VSV-PeGFP-M-MmRFP appeared exclusively green or red (data not shown). Further examination indicated that these cells were infected with viruses that expressed only green or red fluorescence. On average, the green- or red-only viruses represented <3% of the total virus population. The molecular basis of the phenotype of these viruses is currently being examined and will be reported elsewhere.
MmRFP fusion protein is not incorporated into VSV-PeGFP-M-MmRFP virions.
Since our goal is to use dually fluorescent viruses for studies of virus entry, NC uncoating, assembly, and egress, we wanted to determine whether both PeGFP and MmRFP fusion proteins were incorporated into extracellular virions. Accordingly, radiolabeled virions from the supernatants of cells infected with VSV-PeGFP-M-MmRFP were concentrated by ultracentrifugation, and the viral proteins were analyzed for the presence of the two fluorescent fusion proteins by electrophoretic analyses. Although the PeGFP fusion protein could readily be detected in the virions, the MmRFP fusion protein was not detectable by examining the immunoprecipitated or total proteins from the virions (Fig. 1F, lanes 2 and 4). Repeated experiments with increasing amounts of total viral proteins from purified virions failed to demonstrate the presence of MmRFP. Similar studies with mRFPM fusion protein also failed to demonstrate incorporation of the fusion protein into the extracellular virions (data not shown). These results indicate that the MmRFP or mRFPM fusion protein is not competent to be incorporated into virions.
Mtc supports assembly of infectious VSV and is incorporated into virions.
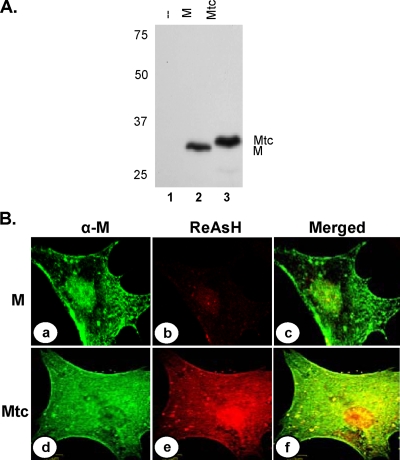
We rationalized that the inability of MmRFP to be incorporated into virions might be due to its increased size or the fact that it is not competent to support virus assembly. To address the potential size constraint, we examined if a smaller tag could be used at the carboxy terminus of the M protein to generate the recombinant virus. tc motifs usually form hairpin structures and bind specifically to biarsenical dyes through covalent interactions, causing fluorescence and allowing live imaging of cells expressing the tc-tagged proteins (24). Depending on the biarsenical dyes used, the proteins can be visualized as either green (labeled with FlAsH) or red (labeled with ReAsH) under a fluorescence microscope. To examine if a tc motif could be fused with M protein without adversely affecting its functions, we fused a tc motif of 12 amino acids (FLNCCPGCCMEP) (39) at the carboxy terminus of the M protein. Mtc was expressed in transfected cells (Fig. 2A) and was associated with the plasma membrane (Fig. 2B, panel d) in a manner similar to that of the wt M protein (Fig. 2B, panel a), as judged by immunofluorescence staining of the transfected cells. To examine if the Mtc protein could be labeled specifically with ReAsH, cells transfected with the plasmid encoding Mtc were incubated with the biarsenical dye for 30 min at 4 hpt. Under a fluorescence microscope, Mtc (red fluorescence) was seen to be distributed throughout the cytoplasm and also localized to the plasma membrane (Fig. 2B, panel e). Immunofluorescence staining of the ReAsH-labeled cells with anti-M MAb showed the localization of the Mtc (Fig. 2B, panel d) in these transfected cells. The merged image (Fig. 2B, panel f) showed that the two signals (red and green) overlapped, demonstrating that the ReAsH dye specifically labeled the Mtc protein. Additionally, because of the close overlap of the ReAsH staining and immunostaining with anti-M antibody (Fig. 2B, panel f), it appears that similar pools of M proteins are detected by the two methods. A low level of nonspecific background staining with ReAsH was observed in the nuclei of cells transfected with the wt M protein (Fig. 2B, panel b), which could easily be distinguished from the authentic M signal by anti-M staining (Fig. 2B, panel a). This type of nonspecific staining with ReAsH has also been observed by others (48). Incubation of cells with ReAsH did not have any adverse effect on the localization of M or on the morphology of the treated cells. These data indicate that biarsenical labeling of tc tags can authentically detect M inside cells. Overall, the results demonstrate that wt M and Mtc localize to the same membranous areas in the cell, indicating that the tc tag does not affect the membrane localization properties of the protein.
FIG. 2.
Expression of Mtc fusion protein in transfected cells. (A) Cells transfected with a plasmid encoding M (lane 2) or Mtc (lane 3) protein or mock-transfected cells (lane 1) were radiolabeled with Expre35S35S label for 2 h at 16 hpt. The radiolabeled proteins were immunoprecipitated with anti-VSV antibody, analyzed by SDS-PAGE, and detected by fluorography. Size markers in kDa are shown on the left. The M and Mtc proteins are identified on the right. (B) Biarsenical dye ReAsH labeling of cells expressing the M (a, b, and c) or the Mtc (d, e, and f) protein. Transfected cells were treated with ReAsH as described in Materials and Methods. Following washing of the dye, the cells were fixed and subjected to immunofluorescence staining with anti-M antibody and anti-mouse Alexa 488-conjugated secondary antibody. ReAsH staining (b and e), anti-M staining (a and d), and merged images of staining of the same cells (c and f) are shown.
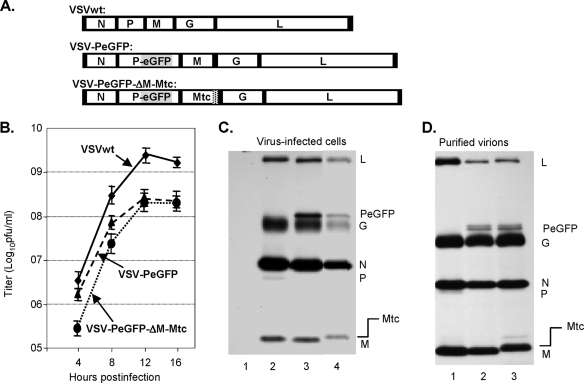
In order to recover a recombinant VSV that could express and incorporate both Mtc and PeGFP fusion proteins, we constructed the plasmid VSV-PeGFP-ΔM-Mtc by replacing the coding region of M with that of Mtc in the plasmid VSV-PeGFP (Fig. 3A) and subsequently recovered the virus. Cytopathic effects specific for VSV were observed in infected cells, which also exhibited green fluorescence due to the expression of the PeGFP fusion protein. However, the cytopathic effects in these cells were less pronounced than those observed for VSV-PeGFP or wt VSV, indicating that the recovered virus might be less cytopathic (data not shown). VSV-PeGFP-ΔM-Mtc grew at a lower rate than VSVwt and VSV-PeGFP did, with a maximum titer of approximately 2 × 108 PFU/ml (Fig. 3B). Analysis of viral proteins in infected cells showed expression of Mtc with a slightly slower electrophoretic mobility consistent in size with the fusion of the tc tag with M (Fig. 3C). In addition, the Mtc protein of correct size was incorporated into virions at levels similar to that of wt M (Fig. 3D). These results demonstrate that Mtc is competent to support VSV assembly and is incorporated into infectious VSV particles.
FIG. 3.
Recovery of VSV encoding Mtc. (A) Genome organization of wt VSV, VSV-PeGFP, and VSV-PeGFP-ΔM-Mtc. (B) Single-step growth kinetics of recombinant viruses. Virus titers shown are averages from three independent experiments, with the standard deviations indicated by error bars. (C) Analysis of viral proteins in cells infected with recombinant VSVs. BHK-21 cells were mock infected (lane 1) or infected with wt VSV (lane 2), VSV-PeGFP (lane 3), or VSV-PeGFP-ΔM-Mtc (lane 4) at an MOI of 10. The proteins were radiolabeled for 1 h at 4 hpi, immunoprecipitated with anti-VSV antibody, separated by SDS-PAGE, and detected by fluorography. The viral proteins along with the fusion proteins are identified on the right. (D) Examination of proteins incorporated into purified virions. Viral proteins in cells infected with VSV, VSV-PeGFP, or VSV-PeGFP-ΔM-Mtc were radiolabeled as described in the legend to Fig. 1F, and the proteins incorporated into purified virions (VSV, lane 1; VSV-PeGFP, lane 2; and VSV-PeGFP-ΔM-Mtc, lane 3) were examined. The positions of various proteins are shown on the right.
Biarsenical labeling with ReAsH allows detection of dually fluorescent virions.
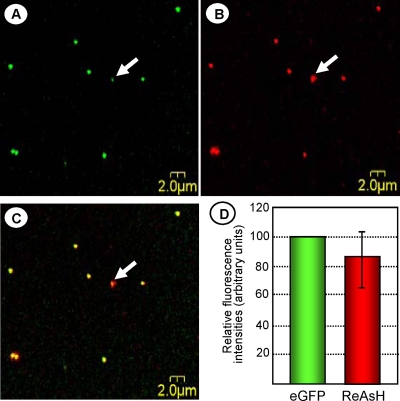
In order to produce dually fluorescent VSV particles, we incubated VSV-PeGFP-ΔM-Mtc-infected cells at 4 hpi with ReAsH for 12 h. Labeled virus particles present in the clarified culture supernatant were placed on a microscope slide and examined under a confocal laser scanning fluorescence microscope. Double-labeled particles with perfect colocalization of EGFP and ReAsH fluorescence were readily detectable (Fig. 4A and B) as yellow particles (Fig. 4C), indicating that both the fusion proteins were incorporated into the virions. We determined the relative fluorescence intensities of 50 individual dually fluorescent virus particles and found that the relative red fluorescence intensities of the particles were, on average, about 85% of the green fluorescence intensities (Fig. 4D). However, some of the particles (as an example, the one identified by the white arrows in the panels) possessed a green fluorescence intensity considerably lower than the average intensity displayed by most particles. In addition, a small number of particles (<5%) with either green or red fluorescence were also detected (data not shown). The red-only particles were, on average, smaller in size than the yellow or green-only particles and could potentially represent empty membrane vesicles containing Mtc, whereas the green-only signals could be due to uneven labeling of the particles with ReAsH.
FIG. 4.
VSV-PeGFP-Mtc virions exhibit dual fluorescence. Cells infected with VSV-PeGFP-ΔM-Mtc were treated with ReAsH for 12 h at 4 hpi. Samples (1 to 2 μl) of the clarified culture supernatant were placed on glass coverslips and visualized under an inverted microscope. High-resolution images of the viral particles were collected sequentially, using a 488-nm laser line for eGFP (A) and a 596-nm laser line for ReAsH (B), and then displayed using the simultaneous dual-channel mode (C). Arrows in the panels show one virus particle with a significantly reduced eGFP signal compared to those of the other particles. (D) Quantitation of fluorescence intensities of a total of 50 dually fluorescent particles. Histograms show average relative intensities for green and red fluorescence, with the error bar showing the standard deviation.
Plasma membrane localization of M protein is not affected by inhibitors of microtubule polymerization.
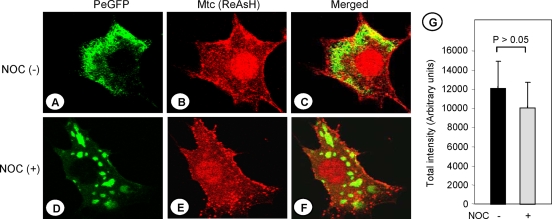
We had previously shown that in cells infected with VSV, newly synthesized NCs are transported from the sites of synthesis to the cell periphery by a microtubule-dependent mechanism (14). The M protein, synthesized in the cytoplasm, is also localized to the plasma membrane for virus assembly. The mechanism by which the M protein is localized to the plasma membrane is not known. It was previously shown that the VSV M protein physically associates with tubulin (42). Therefore, we rationalized that the M protein distribution in the cytoplasm as well as its membrane association may be mediated by microtubules. To examine if microtubules have any effect on the intracellular distribution or plasma membrane localization of M protein in virus-infected cells, we pretreated the cells with NOC, a drug known to inhibit microtubule polymerization. Under these conditions, complete depolymerization of microtubules has been observed (14). Subsequently, the cells were infected with VSV-PeGFP-ΔM-Mtc. At 4 hpi, the infected cells were treated with ReAsH for 30 min to label the Mtc protein. The cells were washed, fixed, and then observed under a fluorescence microscope. The Mtc protein (stained red) was seen to be localized to the plasma membranes of cells that were treated with NOC (Fig. 5E), in a manner similar to that in cells not treated with the drug (Fig. 5B). On the other hand, the distribution of viral NCs was significantly altered in the presence of the drug (Fig. 5D), as we demonstrated previously (14). We then quantitated the accumulation of the Mtc protein in the membranes of both NOC-treated and untreated cells, using the software FV500 that was provided with the confocal microscopy system. Our data show that there was no statistically significant difference in the membrane accumulation of Mtc protein in both drug-treated and untreated cells (Fig. 5G). Colcemid, another drug that is known to inhibit microtubule polymerization, also did not have any effect on M protein distribution (data not shown). Together, these results indicate that microtubules do not play any role in intracellular distribution or plasma membrane localization of the M protein. Previous studies have shown that the M protein associates with NCs, both in vitro and in vivo (11, 12, 26), albeit with a low affinity (18). However, a recent study showed no direct colocalization of M and the NCs in the cytoplasm or at sites on the plasma membrane other than virus budding sites (54). Our data shown in Fig. 5 clearly reveal a lack of colocalization of the M protein with the viral NCs in the cytoplasm or at the plasma membrane, thus confirming the results of the recent study (54).
FIG. 5.
Plasma membrane association of the M protein is not affected by microtubule depolymerization. BHK-21 cells were left untreated (A, B, and C) or treated with 10 μg per ml of NOC (D, E, and F) for 3 h prior to infection with VSV-PeGFP-ΔM-Mtc at an MOI of 10. The cells were incubated in medium lacking or containing NOC for 4 h. The infected cells were then treated with ReAsH for 30 min. After being washed, the cells were fixed and examined for epifluorescence under a fluorescence confocal microscope. NOC (-), NOC-untreated cells; NOC (+), NOC-treated cells. Merged PeGFP (A and D) and ReAsH (B and E) fluorescent images of cells are shown in the right panels (C and F). (G) Quantitation of the amount of Mtc accumulated in the plasma membranes of infected cells in the presence (+) or absence (−) of NOC. Total fluorescence intensities (in arbitrary units) were determined as described in Materials and Methods and then plotted as histograms, with error bars indicating standard deviations. The P value for the difference was >0.05.
Dynamic imaging of newly synthesized and existing pools of M protein in virus-infected cells by dual biarsenical labeling.
To study the dynamics and destination of newly synthesized M protein, we employed the procedure of dual biarsenical labeling. Currently, biarsenical labeling is the only available method to visualize newly synthesized proteins by imaging (20, 38, 46, 48). With this method, one biarsenical dye is used to label the existing pool (steady-state level) of protein. Once the existing pool of proteins is labeled, the unbound dye is washed out of the cells and the other dye is added to the cells to label the newly synthesized proteins. Since the dye binding is irreversible, the existing pool of proteins is labeled with one dye and the newly synthesized pool of proteins is labeled with the other dye, and both pools of proteins can be visualized simultaneously by live- or fixed-cell imaging.
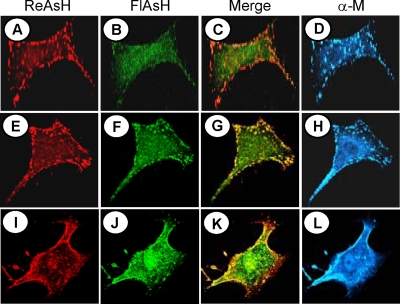
To examine the existing pool of M proteins as well as the pool of newly synthesized proteins in VSV-infected cells, we first recovered a recombinant VSV (VSV-ΔM-Mtc) encoding the Mtc protein in place of the wt M protein, which grew to titers similar to those of wt VSV (data not shown). BHK-21 cells were then infected with VSV-ΔM-Mtc. At 4 hpi, cells were labeled with ReAsH for 30 min. Following washing away of the dye, the cells were exposed immediately or after 1 or 2 h to FlAsH for an additional 30 min and then washed, fixed, and examined under a fluorescence microscope. As shown in Fig. 6A, the existing pool of M protein, labeled red, was seen to be localized to the plasma membrane, with some M protein present inside the cytoplasm. The newly made M protein (labeled green) synthesized immediately after ReAsH labeling was seen to be localized mostly in the cytoplasm, with a small fraction detected at the plasma membrane (Fig. 6B). This result indicates that newly synthesized M protein reaches the plasma membrane less than 30 min after synthesis. Accumulation of the existing pool of M protein (labeled red) in the plasma membrane continued to increase with time, with a concomitant decrease in the cytoplasm (Fig. 6E and I). Data from two similar but independent labeling studies suggested that the existing pool of M protein shows a maximum plasma membrane association within 2 1/2 h after synthesis (that is, at 2 h post-ReAsH labeling). The level of plasma membrane association of newly synthesized M protein as well as the M protein synthesized post-ReAsH labeling also increased steadily with time (Fig. 6F and J) and colocalized with the already existing pool of M protein at the plasma membrane (Fig. 6G and K). By 2 h post-ReAsH labeling, most of the newly synthesized M protein (labeled green) was localized to the plasma membrane (Fig. 6J and K), with a fraction of the protein still in the cytoplasm. At this time point, both red and green fluorescence showed maximum colocalization (Fig. 6K) as a result of accumulation of most of the M protein at the plasma membrane. No further increase in the accumulation of M protein in the plasma membrane was detected with increasing times after ReAsH labeling (data not shown). Overall, these results indicate that the M protein reaches the plasma membrane less than 30 min after synthesis, confirming previous observations (2) that the plasma membrane association of the M protein is rapid and occurs less than 5 min after synthesis. Additionally, our data show that the M protein continues to accumulate at the plasma membrane for 2 1/2 h after synthesis. The specificities of the dyes to bind to the M proteins were further confirmed by examining the immunostaining pattern of M with anti-M MAb (Fig. 6D, H, and L), which closely overlapped with the red and green staining. A similar dynamic distribution of the Mtc protein was also noted in cells ectopically expressing the protein (data not shown), indicating that the membrane localization properties and intracellular distribution of the M protein were not influenced by the presence of the viral NCs or other viral proteins.
FIG. 6.
Dynamic imaging of existing and newly synthesized pools of M protein by dual biarsenical labeling. BHK-21 cells infected with VSV-ΔM-Mtc were treated with ReAsH for 30 min at 4 hpi (A, E, and I), washed, and then labeled immediately (A, B, C, and D) or 1 h (E, F, G, and H) or 2 h (I, J, K, and L) later with FlAsH for 30 min (B, F, and J). Following biarsenical dye labeling, the cells were washed, fixed, immunostained with anti-M MAb and Alexa 350-conjugated secondary antibody (D, H, and L), and examined by confocal fluorescence microscopy as described in Materials and Methods. Merged images of cells stained with ReAsH and FlAsH are shown in panels C, G, and K.
Examination of virus uncoating in infected cells.
We next wanted to determine if attachment of the dually fluorescent virus particles to the cell surface and subsequent entry and uncoating of the particles in the cytoplasm can be visualized. BHK-21 cells grown on a glass-bottomed culture dish were infected at 4°C for 45 min with dually fluorescent virus particles to synchronize virus infection. Following washing of the infected cells to remove the unadsorbed virus particles, multiple dually fluorescent (yellow) particles were seen attached to the cellular plasma membrane when observed under a fluorescence microscope (data not shown). To determine the average time it takes for virus particles to enter the cells, fuse with the endosomes, and release the viral NCs into the cytoplasm, we performed a time course experiment. We reasoned that the intact virus particles in the cytoplasm would appear as yellow dots when excited with both lasers simultaneously, while the PeGFP-containing virus NCs released into the cytoplasm would appear as green dots due to the loss of the ReAsH-labeled Mtc-containing viral envelope. We also reasoned that tracking of the input particles and the NCs would be unequivocal under conditions in which new NCs were not synthesized.
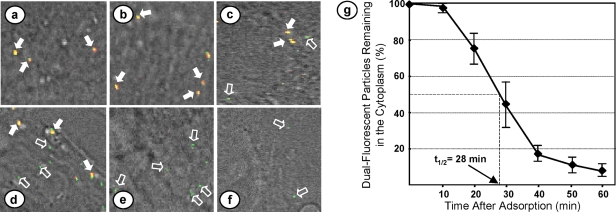
To carry out the study, BHK-21 cells on glass coverslips were first treated with cycloheximide for 1 h to inhibit viral protein synthesis and therefore block new NC synthesis. The cells were then infected with the dually fluorescent particles at a multiplicity of infection (MOI) of 10. Virus adsorption was allowed for 45 min at 4°C. Following adsorption, the cells were extensively washed with cold phosphate-buffered saline and incubated in medium at 37°C in the presence of cycloheximide. Coverslips were collected immediately or at 10-min intervals after incubation, and the cells were fixed and examined under a confocal fluorescence microscope for detection of yellow dots (intact virus particles) as well as green dots (released NCs) in the cytoplasm. As shown in Fig. 7a, only intact virus particles (yellow) were detected on the surfaces of the infected cells immediately following virus adsorption. Ten min after adsorption, the number of yellow dots detected remained largely unchanged. However, as the time of incubation increased, smaller numbers of yellow dots could be detected in the cytoplasm, with concomitant increases in the number of green dots (Fig. 7b to f). Counting about 20 to 25 infected cells and approximately 8 to 12 yellow and green dots per cell, we found that the majority of the virus particles (yellow dots) released the NCs (green dots) into the cytoplasm between 20 min and 40 min after adsorption. We performed the experiment three times and counted the number of virus particles remaining intact at various times after adsorption. Our results (Fig. 7g) showed that it takes approximately 28 min for 50% of virus particles to uncoat their NCs and release them into the cytoplasm. Furthermore, we found that even after 60 min of incubation, nearly 10% of the particles still remained intact in the cytoplasm.
FIG. 7.
Uncoating of dually fluorescent virus particles in infected cells. BHK-21 cells on glass coverslips were treated with cycloheximide for 1 h and then infected with ReAsH-labeled VSV-PeGFP-ΔM-Mtc at 4°C. Following adsorption, the cells were washed and incubated in cycloheximide-containing medium at 37°C. Coverslips were collected immediately or at various times after incubation, fixed with paraformaldehyde, and examined under a confocal fluorescence microscope. (a to f) Representative images of areas of cells immediately after or 10, 20, 30, 40, and 50 min after incubation, showing the intact virions (yellow dots), some of which are identified by filled arrows, and the released NCs (green dots), some of which are identified by open arrows. (g) Numbers of intact virions remaining in the cytoplasm at various times postadsorption. Error bars show standard deviations from three independent experiments. The time taken for 50% of virions to have released their NCs (t1/2 of uncoating) is shown.
Interestingly, we consistently observed that the number of green dots gradually decreased 40 min after incubation and that very few green dots could be detected after 60 min. The reasons for this are unclear, but it is possible that the polymerase molecules (L and PeGFP complexes) associated with the released NCs were coming off the N-RNA templates following transcription.
DISCUSSION
In the present study, we described a strategy for generating a dually fluorescent recombinant VSV. A VSV genome encoding PeGFP in place of wt P and Mtc in place of wt M allowed recovery of infectious virus. Cells infected with this virus and treated with the biarsenical red dye ReAsH produced progeny virions that appeared green under a fluorescence microscope due to incorporation of PeGFP and appeared red due to incorporation of ReAsH-labeled Mtc. This work describes for the first time the recovery of infectious rhabdoviruses encoding and incorporating a tagged M protein. Using VSV encoding Mtc, we show by sequential labeling with the two biarsenical dyes that newly synthesized M protein reaches the plasma membrane within 30 min after synthesis. We further show that the transport and plasma membrane localization of M protein are not dependent on microtubules, which are generally used for transport of macromolecules inside the cell. Additionally, using the dually fluorescent virus, we show that following virus adsorption, it takes approximately 28 min for one-half of the virus particles to enter a cell and release the NCs into the cytoplasm in a synchronized infection model.
In the past few years, the use of fluorescent proteins as tags has been critical in understanding the dynamic events of virus infection in living cells (for reviews, see references 8, 22, and 23). Using viruses genetically tagged with fluorescent proteins or with envelopes labeled with lipophilic dyes, several aspects of virus biology and virus-cell interactions have been examined. Fluorescent viruses have been used to study intracellular transport of viral capsids, capsid assembly, genome recombination, virus entry and egress, gene expression, tissue tropism and pathogenesis, and RNP transport and also for live tracking of single particles in infected cells (7, 29, 33, 34, 40, 43, 50, 55). For an enveloped virus such as VSV, which enters cells by endocytosis, infection proceeds by fusion of the viral envelope with the endosomal membrane, and subsequently, the NC is released into the cytoplasm. To study the early events during virus entry and the mechanisms of NC uncoating in the cytoplasm, dual-fluorescently labeled virus particles in which the NC core is labeled with one fluorescent color and the viral envelope is labeled with another fluorescent color would be highly desirable. Therefore, we sought to generate a dually fluorescent VSV where eGFP is fused in frame with the P protein (component of the viral NC core) and mRFP is fused in frame with the M protein (a component of the viral envelope). Multiple attempts to rescue such a virus failed, indicating that MmRFP protein with mRFP fused at the carboxy terminus of the M protein is not competent to support production of infectious VSV. Recently, similar attempts to recover rabies virus encoding a fluorescently tagged M protein were also unsuccessful (27). Likewise, Ebola virus VP40 (the counterpart of matrix protein) fused with GFP also did not support virus-like particle assembly (45). However, we were successful in generating a recombinant VSV encoding MmRFP as an extra cistron in the G-L intergenic junction (Fig. 1C). It should be noted that the fusion of mRFP to the M protein at either the carboxy terminus or the amino terminus retained the properties of M protein-like plasma membrane association and transcription inhibition of VSV minireplicons (S. C. Das, D. Panda, and A. K. Pattnaik, unpublished data), yet the M fusion proteins were not incorporated into the progeny virions. On the other hand, the Mtc protein with a small tag at the carboxy terminus of M faithfully recapitulated the properties of M protein and supported the assembly of infectious virus. It is possible that the large size of the MmRFP fusion protein interferes with the virus assembly functions of the M protein. Since the viruses encoding MmRFP in an extra cistron grew to reasonable titers (Fig. 1D), it appears that the MmRFP fusion protein does not impart a trans-dominant-negative phenotype on the assembly functions of the wt M protein.
The method of tc tagging and biarsenical dye labeling of proteins in vivo was developed recently and holds promise to overcome the size constraints of fluorescent protein tagging (24, 38, 39). One significant advantage of the system is that the tc hairpin can bind specifically and irreversibly to the biarsenical dyes immediately after synthesis, so the proteins can be visualized by fluorescence microscopy. The biarsenical labeling reagents can also be applied sequentially to label existing and nascently synthesized pools of proteins differentially and thus can allow for visualization of protein dynamics in living cells (20, 38, 46, 48). Therefore, tc tagging of M protein and sequential labeling of M with both dyes offer significant advantages over conventional detection of M protein with anti-M antibody staining. Using the two biarsenical dyes (ReAsH and FlAsH) and a sequential labeling approach, we have found that M reaches the plasma membrane within 30 min of synthesis and continues to accumulate there for 2 1/2 hours after synthesis (Fig. 6). Previous biochemical and membrane fractionation studies have determined that plasma membrane association of the M protein is rapid and occurs less than 5 min after synthesis (2). Although our data presented here are not as precise as those in the previous study (2) due to the limitation of the biarsenical dye labeling method, which requires incubation with the dye for at least 30 min, our approach has been very useful in observing the protein dynamics of the existing and newly synthesized pools of M protein.
Previous studies with VSV (11, 12, 26, 37), rabies virus (41), parainfluenza virus (13), and measles virus (49) have suggested that the matrix protein binds to NCs. Moreover, the M proteins of measles virus, parainfluenza virus, and Ebola virus drive budding of progeny virions by specific interaction with the viral NCs (13, 45, 49). In cells infected with VSV-PeGFP-ΔM-Mtc and labeled with ReAsH, we were unable to observe interaction of M protein with the viral NCs either inside the cytoplasm or at the plasma membrane, consistent with previous reports (18, 54). It is possible that during virus assembly, initial interaction of a limited number of M molecules (beyond the limits of detection of fluorescence microscopy) with the NCs at the plasma membrane renders the NCs assembly competent, which then recruits more M protein to the assembly site. Thus, it appears that the mode of assembly of VSV might be different from that of other enveloped viruses at the plasma membrane.
The M protein of VSV is synthesized as a soluble fraction inside the cytoplasm and is transported toward the plasma membrane (2, 5, 12, 18). Early biochemical studies indicated that the M protein associates with the plasma membrane independent of VSV G protein (5). However, the mode of transport of the M protein toward the plasma membrane is unclear. Since the M protein has been shown to physically associate with tubulin (42), we reasoned that the M protein could be transported to the plasma membrane in a microtubule-dependent mechanism. However, our results (Fig. 5) suggest a microtubule-independent mechanism for the transport of the M protein to the plasma membrane. Additionally, disruption of actin filaments, another cytoskeletal structure, also did not affect M protein transport (data not shown). Since we had previously shown that the viral NCs are transported toward the cell periphery in a microtubule-dependent manner (14), our data presented here clearly support the idea that the M protein and the NCs are transported toward the plasma membrane independent of each other. Further work will be necessary to determine the mode of transport of the M protein to the plasma membrane.
During VSV infection, the M protein dissociates from other viral components and is distributed throughout the cytoplasm (47). However, the step at which the M protein dissociates from the viral NCs is unclear. It has been demonstrated that fusion of the viral envelope with the endosomal membrane and the release of the NC into the cytoplasm are two independent but successive steps in the endocytic pathway of VSV infection (31). Release of viral NCs into the lumen of the endosomal vesicle occurs by the fusion of the viral envelope with the membranes of the endosomes. However, the NC release into the cytoplasm may require a back-fusion event in which the internal vesicles fuse with the membranes of the late endosome (31). These studies have shown that fusion of the endosomal membrane with the viral envelope occurs approximately 20 to 25 min after virus entry (31). Using the dually fluorescent virus, our observation that NCs are released from virus particles with a half-life of uncoating of approximately 28 min suggests that the back-fusion event which is necessary for the release of the NCs into the cytoplasm following fusion of the viral envelope with the membranes of the endosomes must be a fast process requiring less than 10 min.
The potential use of the tc-tagged virus particles reported here for studies on virus entry and uncoating as well as virus assembly and egress in infected cells is exciting. The tc motif can be processed after ReAsH labeling for both live-cell imaging and photoconversion of diaminobenzidine to allow direct correlation of live-cell images with high-resolution electron microscopic data (20, 21). Thus, VSV-PeGFP-ΔM-Mtc and VSV-ΔM-Mtc viruses will be valuable tools in studies to understand the mechanisms of VSV assembly and the role of the M protein in the process. The precise location of the M protein inside the virion is unclear. Some studies suggest that the M protein forms a bridge between the G protein and the viral NC (11, 26, 36, 44), while other studies suggest that the M protein forms a cigar-shaped scaffold around which the viral NC is wrapped (3, 4). Since the ReAsH dye can be used for photoconversion of diaminobenzidine, which can allow direct correlation of live-cell images with electron microscopy (20, 21), the ReAsH-labeled VSV-ΔM-Mtc viruses could be used to reexamine the internal organization of the M protein in purified VSV.
Acknowledgments
We thank You Zhou and Terry Fangman of the microscopy core facility, Center for Biotechnology, UNL, for help with fluorescence microscopic studies and members of the Pattnaik laboratory for helpful comments on the manuscript. We also thank D. Lyles for anti-M Mab and R. Tsien for the plasmid encoding mRFP. We appreciate the support of the UNL Statistical Helpdesk for statistical analysis.
This investigation was supported by Public Health Service grant AI-34956 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 19 January 2009.
REFERENCES
- 1.Adams, S. R., R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis, and R. Y. Tsien. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 1246063-6076. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, P. H., S. A. Moyer, and D. F. Summers. 1976. Assembly of vesicular stomatitis virus glycoprotein and matrix protein into HeLa cell plasma membranes. J. Mol. Biol. 102613-631. [DOI] [PubMed] [Google Scholar]
- 3.Barge, A., J. Gagnon, A. Chaffotte, P. Timmins, J. Langowski, R. W. Ruigrok, and Y. Gaudin. 1996. Rod-like shape of vesicular stomatitis virus matrix protein. Virology 219465-470. [DOI] [PubMed] [Google Scholar]
- 4.Barge, A., Y. Gaudin, P. Coulon, and R. W. Ruigrok. 1993. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J. Virol. 677246-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, J. E., and P. J. Fusco. 1988. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J. Cell Biol. 1071707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blondel, D., G. G. Harmison, and M. Schubert. 1990. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J. Virol. 641716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandenburg, B., L. Y. Lee, M. Lakadamyali, M. J. Rust, X. Zhuang, and J. M. Hogle. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, E. M., and T. J. Hope. 2005. Gene therapy progress and prospects: viral trafficking during infection. Gene Ther. 121353-1359. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, E. M., O. Perez, M. Melar, and T. J. Hope. 2007. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology 360286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 997877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong, L. D., and J. K. Rose. 1994. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J. Virol. 68441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong, L. D., and J. K. Rose. 1993. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J. Virol. 67407-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 751117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, S. C., D. Nayak, Y. Zhou, and A. K. Pattnaik. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 806368-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, S. C., and A. K. Pattnaik. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 786420-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, S. C., and A. K. Pattnaik. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 798101-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finke, S., K. Brzozka, and K. K. Conzelmann. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 7812333-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flood, E. A., and D. S. Lyles. 1999. Assembly of nucleocapsids with cytosolic and membrane-derived matrix proteins of vesicular stomatitis virus. Virology 261295-308. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaietta, G., T. J. Deerinck, S. R. Adams, J. Bouwer, O. Tour, D. W. Laird, G. E. Sosinsky, R. Y. Tsien, and M. H. Ellisman. 2002. Multicolor and electron microscopic imaging of connexin trafficking. Science 296503-507. [DOI] [PubMed] [Google Scholar]
- 21.Gaietta, G. M., B. N. Giepmans, T. J. Deerinck, W. B. Smith, L. Ngan, J. Llopis, S. R. Adams, R. Y. Tsien, and M. H. Ellisman. 2006. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc. Natl. Acad. Sci. USA 10317777-17782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giepmans, B. N., S. R. Adams, M. H. Ellisman, and R. Y. Tsien. 2006. The fluorescent toolbox for assessing protein location and function. Science 312217-224. [DOI] [PubMed] [Google Scholar]
- 23.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124741-754. [DOI] [PubMed] [Google Scholar]
- 24.Griffin, B. A., S. R. Adams, and R. Y. Tsien. 1998. Specific covalent labeling of recombinant protein molecules inside live cells. Science 281269-272. [DOI] [PubMed] [Google Scholar]
- 25.Hwang, L. N., N. Englund, and A. K. Pattnaik. 1998. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J. Virol. 721805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaptur, P. E., R. B. Rhodes, and D. S. Lyles. 1991. Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J. Virol. 651057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klingen, Y., K. K. Conzelmann, and S. Finke. 2008. Double-labeled rabies virus: live tracking of enveloped virus transport. J. Virol. 82237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koser, M. L., J. P. McGettigan, G. S. Tan, M. E. Smith, H. Koprowski, B. Dietzschold, and M. J. Schnell. 2004. Rabies virus nucleoprotein as a carrier for foreign antigens. Proc. Natl. Acad. Sci. USA 1019405-9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 1009280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lampe, M., J. A. Briggs, T. Endress, B. Glass, S. Riegelsberger, H. G. Krausslich, D. C. Lamb, C. Brauchle, and B. Muller. 2007. Double-labelled HIV-1 particles for study of virus-cell interaction. Virology 36092-104. [DOI] [PubMed] [Google Scholar]
- 31.Le Blanc, I., P. P. Luyet, V. Pons, C. Ferguson, N. Emans, A. Petiot, N. Mayran, N. Demaurex, J. Faure, R. Sadoul, R. G. Parton, and J. Gruenberg. 2005. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 7653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121168-174. [DOI] [PubMed] [Google Scholar]
- 33.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 1014204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 1025832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyles, D. S., and C. E. Rupprecht. 2007. Rhabdoviridae, p. 1363-1408. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 36.Lyles, D. S., M. McKenzie, and J. W. Parce. 1992. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J. Virol. 66349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyles, D. S., M. O. McKenzie, and R. R. Hantgan. 1996. Stopped-flow, classical, and dynamic light scattering analysis of matrix protein binding to nucleocapsids of vesicular stomatitis virus. Biochemistry 356508-6518. [DOI] [PubMed] [Google Scholar]
- 38.Machleidt, T., M. Robers, and G. T. Hanson. 2007. Protein labeling with FlAsH and ReAsH. Methods Mol. Biol. 356209-220. [DOI] [PubMed] [Google Scholar]
- 39.Martin, B. R., B. N. Giepmans, S. R. Adams, and R. Y. Tsien. 2005. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 231308-1314. [DOI] [PubMed] [Google Scholar]
- 40.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mebatsion, T., F. Weiland, and K. K. Conzelmann. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melki, R., Y. Gaudin, and D. Blondel. 1994. Interaction between tubulin and the viral matrix protein of vesicular stomatitis virus: possible implications in the viral cytopathic effect. Virology 202339-347. [DOI] [PubMed] [Google Scholar]
- 43.Muller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 7810803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newcomb, W. W., and J. C. Brown. 1981. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J. Virol. 39295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panchal, R. G., G. Ruthel, T. A. Kenny, G. H. Kallstrom, D. Lane, S. S. Badie, L. Li, S. Bavari, and M. J. Aman. 2003. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. USA 10015936-15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perlman, M., and M. D. Resh. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic 7731-745. [DOI] [PubMed] [Google Scholar]
- 47.Rigaut, K. D., D. E. Birk, and J. Lenard. 1991. Intracellular distribution of input vesicular stomatitis virus proteins after uncoating. J. Virol. 652622-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudner, L., S. Nydegger, L. V. Coren, K. Nagashima, M. Thali, and D. E. Ott. 2005. Dynamic fluorescent imaging of human immunodeficiency virus type 1 Gag in live cells by biarsenical labeling. J. Virol. 794055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runkler, N., C. Pohl, S. Schneider-Schaulies, H. D. Klenk, and A. Maisner. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell. Microbiol. 91203-1214. [DOI] [PubMed] [Google Scholar]
- 50.Rust, M. J., M. Lakadamyali, F. Zhang, and X. Zhuang. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 792754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8404-407. [PubMed] [Google Scholar]
- 53.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 10116034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swinteck, B. D., and D. S. Lyles. 2008. Plasma membrane microdomains containing vesicular stomatitis virus M protein are separate from microdomains containing G protein and nucleocapsids. J. Virol. 825536-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Messling, V., D. Milosevic, and R. Cattaneo. 2004. Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc. Natl. Acad. Sci. USA 10114216-14221. [DOI] [PMC free article] [PubMed] [Google Scholar]