Abstract
Mammalian genomes harbor a large number of retroviral elements acquired as germ line insertions during evolution. Although many of the endogenous retroviruses are defective, several contain one or more intact viral genes that are expressed under certain physiological or pathological conditions. This is true of the endogenous polytropic retroviruses that generate recombinant polytropic murine leukemia viruses (MuLVs). In these recombinants the env gene sequences of exogenous ecotropic MuLVs are replaced with env gene sequences from an endogenous polytropic retrovirus. Although replication-competent endogenous polytropic retroviruses have not been observed, the recombinant polytropic viruses are capable of replicating in numerous species. Recombination occurs during reverse transcription of a virion RNA heterodimer comprised of an RNA transcript from an endogenous polytropic virus and an RNA transcript from an exogenous ecotropic MuLV RNA. It is possible that homodimers corresponding to two full-length endogenous RNA genomes are also packaged. Thus, infection by an exogenous virus may result not only in recombination with endogenous sequences, but also in the mobilization of complete endogenous retrovirus genomes via pseudotyping within exogenous retroviral virions. We report that the infection of mice with an ecotropic virus results in pseudotyping of intact endogenous viruses that have not undergone recombination. The endogenous retroviruses infect and are integrated into target cell genomes and subsequently replicate and spread as pseudotyped viruses. The mobilization of endogenous retroviruses upon infection with an exogenous retrovirus may represent a major interaction of exogenous retroviruses with endogenous retroviruses and may have profound effects on the pathogenicity of retroviral infections.
Although the existence of endogenous retroviruses in the genomes of vertebrate species, including mice and humans, has been known for many years, it was not until the sequencing of the human and mouse genomes that the extent to which we harbor endogenous retroviral sequences was fully appreciated. It has been estimated that our germ line has incurred about 40,000 infections by retroviruses over the course of evolution and that retroviral sequences account for 8 to 10% of our total DNA (20, 38). Most of these retrovirus elements are defective, and many consist only of solo long terminal repeats (LTRs) generated by recombination between the two LTRs generated during the integration process. Others that retain coding sequences are often transcriptionally silent; however, some of the sequences are expressed in a very controlled manner throughout the lifetime of the host and appear to be modulated under various physiological and pathological circumstances (4, 5, 23, 26, 29). There are an increasing number of studies indicating that many of these elements have been utilized for a number of physiological processes. These include processes at the genomic level, such as transcriptional control of several genes (3, 7, 22, 25), at the transcript level, by interaction of viral RNAs with proteins (35), and at the protein level, such as the role of endogenous retrovirus envelope proteins in the fusion of placental trophoblasts (6, 13, 33). The observations that endogenous retrovirus sequences are expressed during the lifetimes of animals may reflect requirements for the transcription and translation of some of the endogenous retroviruses, even though the expression of these viruses may have deleterious consequences. Thus, some restriction factors likely reflect mechanisms evolved by the host to control endogenous viruses that are obliged to be transcribed due to a physiological role.
Among the most extensively studied groups of endogenous retroviruses are the endogenous polytropic retroviruses of mice (11, 12, 15, 17, 19, 36). These viruses, like some human retroviruses, are expressed in a tightly controlled fashion during the lifetime of the host and have not been found to produce infectious viruses, even though some of them possess intact env genes and are transcribed (1, 9, 28, 31, 36). They do, however, interact with exogenous viruses that have infected the mouse. Upon infection, the exogenous viruses recombine with the env gene sequences of the endogenous polytropic MuLVs to generate host range variants that utilize a different cell surface receptor for infectious entry (11, 15, 16, 17, 32, 37). The generation of such recombinant viruses is instrumental in the induction of disease by a number of exogenous retroviruses.
Recombination between the exogenous and endogenous retrovirus genomes requires transcription of a complete endogenous provirus to an RNA strand that is copackaged with an exogenous murine leukemia virus (MuLV) transcript as a heterodimeric virion RNA. Upon subsequent infection, the heterodimer can undergo recombination during reverse transcription (RT) (16, 37). Although the endogenous polytropic proviruses are transcribed, replication of the endogenous polytropic viruses in the absence of recombination has not been observed. This may, in many cases, reflect defects, such as point mutations or deletions, in the endogenous viral genome but may also be influenced by the activities of various restriction factors (14, 27). The fact that exogenous MuLVs are capable of replicating in mice indicates that they have evolved mechanisms to circumvent the activities of at least some of the restriction factors. Thus, exogenous retroviruses might facilitate, through complementation, active replication of endogenous retroviruses.
In this report, we present evidence that infection of mice by an exogenous virus results in the mobilization of complete endogenous retroviruses. This includes proviruses that are severely defective and possess large deletions, as well as proviruses that are full length. Furthermore, the transferred sequences are transcribed and packaged into virions released from the newly infected cells.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T3 cells were used for the propagation of viruses and assays of MuLVs. Mus dunni (21) and Fischer rat embryo (FRE) cells and mink lung fibroblasts (ATTC CCL64), all of which are devoid of endogenous polytropic proviruses, were used as targets to assess the transfer of endogenous retroviruses. All cells were maintained in tissue culture media supplemented with glutamine (2 mM), penicillin (100 units/ml), streptomycin (25 μg/ml), gentamicin (50 μg/ml), and amphotericin B (2 μg/ml). NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated bovine calf serum. M. dunni and FRE cells were maintained in RPMI supplemented with 10% heat-inactivated fetal calf serum, and mink cells were maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum. The ecotropic MuLV F-MuLV 57 (30) was obtained as a virus stock after transfection of 3T3 cells with a plasmid carrying the provirus.
Mice.
The mice utilized in this study were of the NFS/N strain maintained as an inbred colony at Rocky Mountain Laboratories. The mice were infected by intraperitoneal injection within 24 h of birth and sacrificed at various times after inoculation. Spleens and thymuses were removed and dissociated by mincing the tissue, suspending the minced tissue in medium (DMEM containing 10% bovine calf serum), and passing it through successively smaller syringe needles (from 18 to 23 gauge). Cells from the tissues were then cocultivated for 12 h with target cell lines seeded at 105 cells per 60-mm tissue culture dish. The target cells were carried for several (10 to 30) passages before the isolation of cellular genomic DNA and harvesting of released viruses from the tissue culture media. All animal procedures were done in accordance with the guidelines of the National Institutes of Health and Rocky Mountain Laboratories animal care and use committees.
Generation of a genomic DNA lambda library of infected FRE cells and detection of lambda clones containing polytropic MuLV LTRs.
DNA was isolated from a clonal cell line derived from FRE cells coinfected with splenocytes from an F-MuLV-infected NFS/N mouse 34 days after infection. Approximately 1.5 × 107 cells were suspended in 3 ml of lysing buffer from a DNA isolation kit (Gentra Systems, Inc.; D5000A), and the DNA was thereafter purified according to the manufacturer's instructions. The purified DNA was partially digested with MboI and sedimented on a 10 to 30% linear glycerol gradient in 100 mM NaCl, 10 mM Tris-HCl, pH 7.4, and 1 mM EDTA. The gradient was fractionated, and a sample of each fraction was analyzed on a 0.4% agarose gel (Seakem GTG). Fractions containing fragments of approximately 15 to 25 kb were pooled; precipitated with ethanol; resuspended in 10 mM Tris, 1 mM EDTA; and cloned into the Lambda Fix vector (Stratagene) according to the manufacturer's instructions. The library was screened using a probe to the endogenous polytropic MuLV LTR that was generated and labeled by chemiluminescence using a PCR DIG Probe Synthesis Kit (Roche; catalog no. 11 636 090 910). The PCR was accomplished using a primer set consisting of the forward primer BL8313 (AAGCTAGCTGCAGTAACGCCATTTTGC) and the reverse primer BL8564RC (GAGGTGCACAGTGCTCTGG). BL8313 corresponds to bases 2024 to 2050 of the prototypic endogenous PT (polytropic structural subclass) provirus MX33 and primes near the 5′ end of the U3 region of the LTR (36). BL8564RC corresponds to the reverse complement sequence of bases 2301 to 2319 of MX33 and primes within the unique 190-bp insert of the U3 region of the LTR (18). The template was a plasmid containing an endogenous intermediate polytropic (iPT) provirus designated NB1, isolated from a lambda library of the NFS/N mouse genome (9). Development of the filters to identify plaques homologous to the probe was done according to the manufacturer's instructions. Lambda DNA preparations were performed as previously detailed (9).
PCR analyses and DNA sequencing.
PCR amplification of LTR U3 sequences of endogenous polytropic proviruses or virion RNA was accomplished using the primer set described above (BL8313 and BL8564RC) with the puReTaq Ready-To-Go PCR system (Amersham Pharmacia Biotech Inc.; no. 27-9558-01) or Pfu Turbo DNA polymerase (Stratagene; no. 600153) according to the manufacturer's instructions. RT-PCR amplification was accomplished using SuperScript III reverse transcriptase (Invitrogen; no.18080-044), followed by PCR as described above. The generation of RT-PCR products was dependent upon reverse transcriptase and was unaffected by treatment with DNase, indicating that the template corresponded to virion RNA rather than a DNA contaminant. PCR amplification and nucleotide sequencing of proviruses in genomic DNA was accomplished using a set of 39 reverse primers and 40 forward primers approximately evenly distributed across a consensus polytropic provirus genome. This consensus sequence was constructed from alignments of polytropic proviruses detected in the Mus musculus genome (38). Similarly, lambda DNA containing polytropic MuLV sequences was sequenced using this battery of primers.
Nucleotide sequence accession numbers.
The sequences of the mobilized polytropic viruses characterized from FRE clonal cell lines 5, 15, and 51 (see Fig. 3 and 4) have been deposited in GenBank under accession no. FJ544576, FJ544577, and FJ544578, respectively.
FIG. 3.
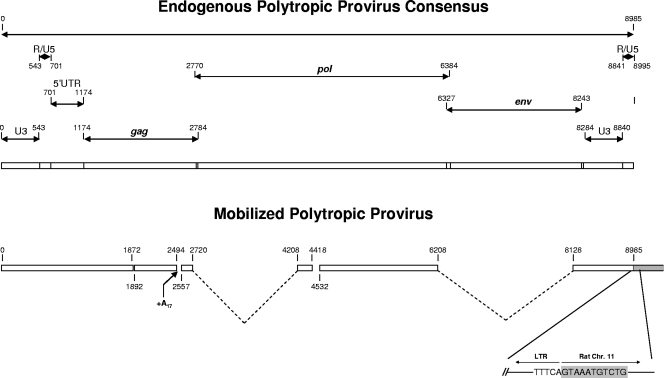
Structure of an NFS/N polytropic provirus transferred to FRE cells and its integration junction with rat genomic sequences. A provirus was isolated from a lambda library derived from a clonal cell line of FRE cells (clone 5) that had been cocultivated with splenocytes from an NFS/N mouse 34 days after infection with F-MuLV. The structure was derived from an alignment of the nucleotide sequence of the transferred provirus with an endogenous polytropic provirus consensus sequence. An apparent insertion of 17 A residues occurred at the position of a small deletion from 2494 to 2557. Large deletions are indicated by dashed lines extending down from the bar diagram. The proviral DNA 3′ terminus and flanking sequence present in a FRE clonal cell are indicated in the expanded region. FRE rat sequences are shaded. Identical sequences were obtained from three individual lambda clones from a genomic library of the clonal cell line.
FIG. 4.
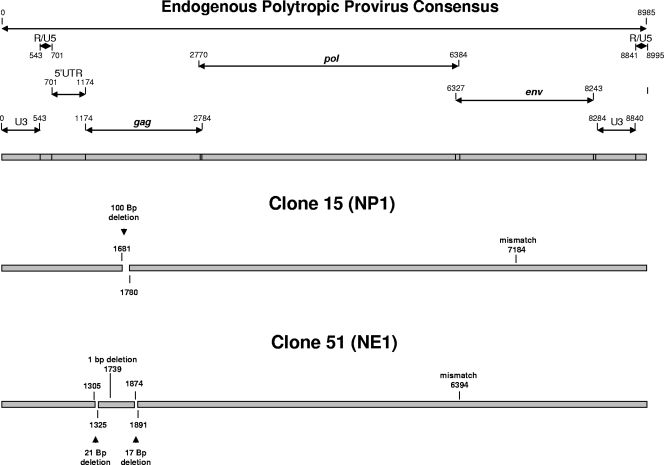
Structures of two NFS/N polytropic proviruses transferred to FRE cells. The proviruses were sequenced from PCRs of genomic DNAs from clonal FRE cell lines each containing a single NFS/N polytropic provirus. The structures were derived from alignments of the nucleotide sequences of the transferred proviruses with an endogenous polytropic provirus consensus sequence. It is nearly certain that the LTR sequences were derived from both the 3′ and 5′ LTRs, although the presence of both complete LTRs could not be formally determined by these analyses. With that qualification, we have depicted the proviruses as complete structures possessing both LTRs. The provirus found in clone 15 contains a deletion of 100 bp in the Gag region. It differs from the NP1 proviral env gene found in the NFS/N genome by only 1 base at position 7184 in the consensus sequence. The provirus found in clone 51 contains three small deletions in the Gag region and differs from the NE1 proviral env gene found in the NFS/N genome by a single mismatch at residue 6394 in the consensus sequence.
RESULTS
Detection of NFS/N endogenous polytropic LTR sequences in the genomic DNA of rat cell lines cocultivated with F-MuLV-infected mouse tissues and in virion RNA released from the cells.
Nearly all polytropic proviruses of NFS/N mice contain a unique insert of 190 to 212 bp in the U3 region of their LTRs that is not found in any of the exogenous MuLVs (18). In this regard, although recombinant polytropic MuLVs that contain sequences derived from endogenous polytropic gag, pol, and env genes have been described (8), no recombinant MuLVs have been identified that contain an endogenous polytropic LTR. We utilized this feature to assess the transfer of mouse endogenous LTR sequences to target cell lines. The transfer of such sequences might signal the transfer of intact polytropic proviruses. Newborn mice were infected with F-MuLV, and after approximately 7 weeks, the splenocytes or thymocytes of the infected mice were cocultivated overnight with FRE cells, which are devoid of endogenous polytropic proviruses. After cocultivation, the FRE cells were transferred through multiple (10 to 30) passes to eliminate residual tissue cells from the mouse. Cellular genomic DNA was then prepared from the cocultivated cells and assayed for the presence of the endogenous NFS/N polytropic U3 region. This was accomplished by utilizing a PCR with a forward primer (BL8313) near the beginning of the U3 region and a reverse primer (BL8564RC) within the unique insert of endogenous polytropic proviral LTRs.
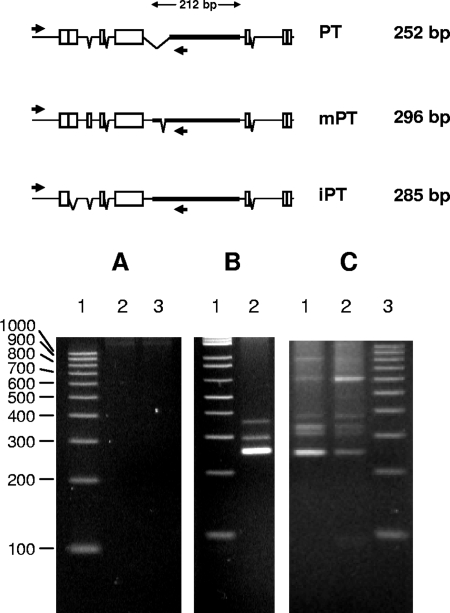
Three structural subclasses of polytropic proviruses termed the PT, the modified polytropic (mPT), and the iPT proviruses have been identified in NFS/N mice (9, 36). Each subclass is distinguishable by characteristic features of the U3 region of its LTR. As a result, three sizes of amplicons from the PCR analyses might be expected, depending on the structural subclass of the polytropic provirus (Fig. 1). No PCR products were detected using genomic DNA of FRE cells or of FRE cells that had been cocultivated with splenocytes from an uninfected NFS/N mouse (Fig. 1A). In contrast, PCR products were readily detected in FRE cells that had been cocultivated with splenocytes from NFS/N mice infected with F-MuLV (Fig. 1B). The products included one major product of approximately 250 bp and two less prominent bands corresponding to products of approximately 300 and 350 bp. Sequence analysis of the prominent 250-bp product revealed that it corresponded to the expected product of the LTR of the PT subclass of endogenous NFS/N proviruses, while analyses of the minor bands were inconclusive. Very similar results indicating the transfer of polytropic LTRs from infected mouse splenocytes were observed from several mice of the same age or older using M. dunni or mink lung fibroblasts as target cells; both of which are devoid of endogenous polytropic proviral sequences. The transfer of LTR sequences was not observed in cocultivation experiments using thymocytes from F-MuLV-infected NFS/N mice (data not shown). The spleen, rather than the thymus, is the major target tissue for F-MuLV.
FIG. 1.
PCR amplification of the LTR U3 region of endogenous polytropic proviruses detected in FRE cells cocultivated with splenocytes from F-MuLV-infected NFS/N mice and RT-PCR amplification of virion RNA released from the cocultivated FRE cells. The schematics depict the U3 regions of the LTRs of the respective endogenous polytropic provirus subclasses. Adjacent blocks indicate direct repeats, whereas isolated blocks indicate regions that are repeated in the U3 regions of other retroviruses. Deletions relative to the U3 regions of other viruses are depicted by the breaks in the lines connected by the downward extended Vs. The inserts ranging from 190 bp to 212 bp are indicated by the thick horizontal lines. The annealing sites of the forward and reverse primers are indicated by arrows, and the amplicon sizes are indicated to the right of the schematics. (A) Lane 1, 100-bp ladder; lane 2, PCR products from genomic DNA of uninfected FRE cells; lane 3, PCR products from genomic DNA of FRE cells cocultivated with splenocytes from an uninfected NFS/N mouse (77 days old). (B) Lane 1, 100-bp ladder; lane 2, PCR products from genomic DNA from FRE cells cocultivated with splenocytes from an NFS/N mouse infected with F-MuLV (34 days old). (C) Lanes 1 and 2, RT-PCR products from virion RNA released from FRE cells cocultivated with splenocytes from two different NFS/N mice infected with F-MuLV (34 days old); lane 3, 100-bp ladder.
It was of interest to determine if the newly acquired proviruses in the FRE cells were transcribed and packaged into virions released from those cells. Virions released from cells were harvested, and their RNAs were analyzed by RT-PCR for the presence of NFS/N polytropic LTR sequences. The RNA from virions released from the cocultivated cells did indeed contain LTR U3 sequences from NFS mice (Fig. 1C), which was confirmed by sequence analyses.
Analyses of genomic DNA from clonal lines of FRE cells cocultivated with F-MuLV-infected NFS/N splenocytes.
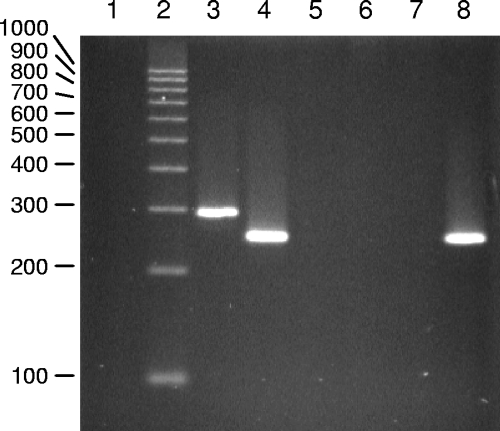
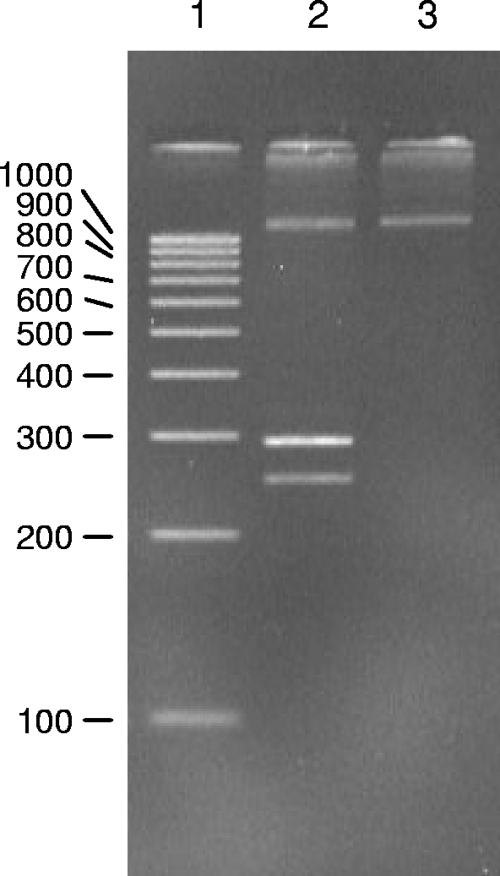
Numerous colonies of the FRE cells were isolated, and their DNAs were analyzed for the presence of NFS/N polytropic LTRs to determine the frequency of infected target cells. Analyses of 116 clones from FRE cells cocultivated with splenocytes from two different NFS/N mice identified eight target FRE single-cell colonies containing an endogenous LTR from the infected NFS/N mouse. Each clonal line positive for an NFS/N-derived LTR contained only a single type of polytropic LTR (Fig. 2). The transferred LTRs detected in the clonal cell lines corresponded to LTRs from the PT and mPT subclasses of polytropic proviruses, as was evident from the sizes of the amplicons (Fig. 1A; see Fig. 4), as well as by sequence analyses (data not shown). Six clonal cell lines contained PT LTRs, and two contained mPT LTRs. LTRs from the iPT class of proviruses, which comprise a small proportion of NFS/N endogenous proviruses (Fig. 1), were not detected. The observation that each clonal line contained a single type of polytropic LTR (Fig. 2), coupled with the finding that about 7% of the clonal lines tested positive for a polytropic LTR, suggested that most of the positive clones harbored a single polytropic provirus.
FIG. 2.
PCR amplification of the LTR U3 region of endogenous polytropic proviruses from clonal cell lines of FRE cells derived from cells cocultivated with F-MuLV-infected NFS/N splenocytes. Lane 2, 100-bp ladder; lanes 1 and 3 to 8, PCR products from the DNAs of seven clonal cell lines. The cell lines in lanes 3 and 4 tested positive for an mPT LTR and a PT LTR, respectively. The cell lines in lanes 5 to 7 tested negative for polytropic LTRs, and the cell line in lane 7 tested positive for a PT LTR.
LTRs detected in FRE cells cocultivated with F-MuLV-infected NFS/N splenocytes signal the transfer of intact endogenous mouse proviruses.
The detection of polytropic LTRs in the DNA of cocultivated FRE cells strongly suggested that endogenous proviruses from the mouse had been transferred to the rat cells. Alternatively, the detected proviruses could correspond to a new type of recombinant between the exogenous ecotropic retrovirus and the endogenous polytropic proviruses that retained the polytropic LTR. To distinguish between these possibilities, a lambda library was generated from the genomic DNA of one of the clonal cell lines that harbored a provirus containing a polytropic LTR, and the library was subsequently screened for lambda clones containing polytropic LTR sequences. Three lambda clones containing the polytropic LTR were isolated from the library and subjected to sequence analysis. One of the clones contained an entire provirus, while the provirus sequences of the other two clones had been truncated during the cloning process and retained only the 3′ region of the proviral genome. Sequence comparisons of the proviruses and their flanking sequences indicated that the three clones corresponded to clones of the same integrated provirus.
The provirus transferred to the rat cells did not contain discernible ecotropic F-MuLV sequences, indicating that it corresponded to a complete endogenous NFS/N provirus rather than a new type of recombinant virus. It exhibited several deletions that would render the virus replication defective, including a large deletion encompassing nearly all of the env gene sequences (Fig. 3).
The 3′-flanking sequences of all three lambda clones were identical and were located at the same position on chromosome 11 of the rat genome. This corroborated our conclusion from the sequence data that the clones corresponded to the same integrated provirus that was precisely integrated into the rat genome rather than being the result of the fusion of mouse and rat cells or of inadvertent transfection of the rat cells by mouse DNA. Furthermore, the provirus exhibited the conserved 3′ CA dinucleotide proviral terminus immediately adjacent to rat sequences, typical of integrants mediated by MuLV (Fig. 3).
PCR analyses of the genomic DNA from several additional FRE clonal cell lines that harbor a single polytropic LTR identified two proviruses closely corresponding to previously characterized NFS/N proviruses. These proviruses corresponded to NE1 and NP1, both of which are PT proviruses (Fig. 4). The transferred proviruses differed by only one or two env gene nucleotides, respectively, from the previously characterized NFS/N proviruses (9). These differences likely reflect point mutations incurred during retroviral replication subsequent to mobilization by F-MuLV. Both of these proviruses contain intact env genes, and one, NP1, is a frequent participant in recombination with ecotropic MuLVs (1).
The transfer of intact endogenous mouse proviruses occurs by pseudotyping within ecotropic MuLV virions.
Mice infected with F-MuLV express both ecotropic and polytropic Env proteins. The transfer of the endogenous polytropic retroviruses of mice to rat cells could occur through infection utilizing polytropic Env proteins or, alternatively, through pseudotyping of polytropic genomes within ecotropic F-MuLV virions (10, 24, 34). Pseudotyping of the NFS/N endogenous retrovirus genomes was assessed by comparing the transfer of proviruses to uninfected FRE cells to the transfer of proviruses to FRE cells chronically infected with F-MuLV. Viral interference renders F-MuLV-infected cells refractory to reinfection by ecotropic virions, including endogenous proviruses pseudotyped by F-MuLV. Thus, pseudotyping by the ecotropic virus would be reflected in a decrease in the transfer of endogenous genomes to F-MuLV-infected FRE cells compared to uninfected FRE cells. For these experiments, we utilized young mice, 10 days postinfection, to minimize the level of replication-competent recombinant polytropic MuLVs that also might be capable of pseudotyping endogenous retrovirus genomes. PCRs revealed that the proviruses were readily transferred to uninfected FRE cells. In contrast, the F-MuLV-infected cells were nearly impervious to the transfer of mobilized endogenous polytropic genomes, indicating that the polytropic genomes released from the mouse splenocytes were extensively pseudotyped (Fig. 5).
FIG. 5.
Block in the transfer of NFS/N polytropic virus to FRE cells by retroviral interference. FRE cells or FRE cells chronically infected with F-MuLV were cocultivated with splenocytes from NFS/N mice inoculated with F-MuLV or from uninfected NFS/N mice as controls. The U3 region of the LTR was amplified by PCR to assess the transfer of NFS proviruses to the FRE cells. Lane 1, 100-bp ladder; lane 2, FRE cells cocultured with 10-day-old F-MuLV-infected NFS mouse splenocytes; lane 3, F-MuLV-infected FRE cells cocultured with 10-day-old F-MuLV-infected NFS mouse splenocytes. PCRs performed with DNA derived from cocultivated FRE cells show robust amplification of both polytropic (∼250-bp) and modified polytropic (∼300-bp) LTRs (lane 2). The PCRs performed with DNA from cocultivated FRE cells that had been previously infected with F-MuLV exhibited a striking decrease in the products of both polytropic LTRs (lane 3).
DISCUSSION
Recombination between exogenous ecotropic MuLVs and endogenous polytropic MuLVs, a process that requires full-length transcription of the participating endogenous polytropic provirus, occurs quite frequently in infected mice. Recombination occurs during RT of the heterodimeric RNA; thus, the full-length copy of the endogenous provirus is capable of being packaged within virions. We surmised that if full-length transcripts of the endogenous polytropic proviruses are packaged as RNA heterodimers, it is likely that they are also packaged as RNA homodimers that might subsequently be transferred to different cells in the absence of recombination. The transfer of the endogenous proviruses was signaled in our analyses by detection of the endogenous polytropic U3 regions in target FRE cells. This region has never been detected in a recombinant polytropic virus and would likely be indicative of transfer of the entire endogenous genome. This was confirmed by analysis of the complete genomes of transferred proviruses. Furthermore, analyses of flanking genomic sequences in the target rat cells suggested that the transferred NFS/N provirus had inserted into the rat genome in a manner typical of MuLV-mediated integration.
We have recently characterized many of the endogenous retroviruses of NFS/N mice and noted some features of these proviruses that suggest that the proviruses had undergone active replication during recent periods of their evolution (9). In this regard, one of the endogenous polytropic MuLVs was clearly a recombinant between two distinct classes of the endogenous viruses. Since recombination occurs through the copackaging of the RNAs of each parent in a heterodimeric RNA, this observation strongly suggested that both types of viruses had simultaneously replicated in the same cell, recombined, and subsequently reintegrated into the germ line. A second observation suggesting active replication of the endogenous viruses was the presence of identical lethal mutations in different proviruses. This suggested that a replication-defective endogenous retroviruses was transcribed and pseudotyped within a replication-competent retrovirus and subsequently reentered the germ line. Periods of active replication of endogenous retroviruses may have been initiated through widespread infection of mouse populations by exogenous retroviruses capable of circumventing restriction factors present in the mouse.
Recombinant viruses released from mice infected with the ecotropic MuLVs are frequently pseudotyped within ecotropic virions (10, 24, 34). It seems likely that this might also be the case with the transfer of the endogenous proviruses, and certainly with those proviruses that exhibit deletions rendering them replication defective. Indeed, our observation that the transfer was blocked in target cells chronically infected with the ecotropic F-MuLV confirmed that the transfer of endogenous proviruses to target cells was facilitated by viral pseudotyping. Pseudotyping may provide an effective means to evade host replication restriction factors that have been circumvented by the ecotropic virus. Our studies suggest that the packaging and infectious transfer of endogenous retroviruses after infection of the host by exogenous retroviruses may be a frequent occurrence.
We have demonstrated that the proviruses transferred to the FRE target cells were transcribed and released into progeny virions, indicating the potential for further spread. Furthermore, the transfer of endogenous viruses after infection by exogenous viruses occurs early after infection, before recombinant viruses are detected, and may represent the major interaction of exogenous viruses with endogenous retroviruses. The finding that endogenous viruses are mobilized by infection of the host with exogenous viruses may have far-reaching implications in retrovirus evolution, as well as in retroviral pathogenesis. The spread of intact endogenous proviruses to cells of other species strongly suggests that a similar spread is occurring in the infected host, particularly early after infection, when inhibition by viral interference would be minimal. The expression of viral gene products has been implicated in pathological processes, such as the involvement of polytropic SU proteins in autoimmune murine lupus nephritis (2). The spread of endogenous proviruses in the host could potentially facilitate the inappropriate expression of viral proteins, which could in turn trigger a pathological response. Furthermore, retroviral spread can result in insertional mutagenesis, as well as transcriptional activation, in infected cells. Clearly, the mobilization of one virus by another is clinically relevant and could contribute to the pathogenicity of exogenous retroviruses, such as the MuLVs, human immunodeficiency virus, and human T-cell leukemia virus.
Acknowledgments
We thank A. Kolokithas, K. Peterson, and K. Hasenkrug for helpful discussions.
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 30 December 2008.
REFERENCES
- 1.Alamgir, A. S., N. Owens, M. Lavignon, F. Malik, and L. H. Evans. 2005. Precise identification of endogenous proviruses of NFS/N mice participating in recombination with Moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs. J. Virol. 794664-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudino, L., K. Yoshinobu, N. Morito, S. Kikuchi, L. Fossati-Jimack, B. J. Morley, T. J. Vyse, S. Hirose, T. N. Jorgensen, R. M. Tucker, C. L. Roark, B. L. Kotzin, L. H. Evans, and S. Izui. 2008. Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J. Immunol. 1812846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieche, I., A. Laurent, I. Laurendeau, L. Duret, Y. Giovangrandi, J. L. Frendo, M. Olivi, J. L. Fausser, D. Evain-Brion, and M. Vidaud. 2003. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol. Reprod. 681422-1429. [DOI] [PubMed] [Google Scholar]
- 4.Bock, M., and J. P. Stoye. 2000. Endogenous retroviruses and the human germline. Curr. Opin. Genet. Dev. 10651-655. [DOI] [PubMed] [Google Scholar]
- 5.de Parseval, N., and T. Heidmann. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 110318-332. [DOI] [PubMed] [Google Scholar]
- 6.Dunlap, K. A., M. Palmarini, M. Varela, R. C. Burghardt, K. Hayashi, J. L. Farmer, and T. E. Spencer. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl. Acad. Sci. USA 10314390-14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn, C. A., P. Medstrand, and D. L. Mager. 2003. An endogenous retroviral long terminal repeat is the dominant promoter for human β1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 10012841-12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, L. H., and M. W. Cloyd. 1984. Generation of mink cell focus-forming viruses by Friend murine leukemia virus: recombination with specific endogenous proviral sequences. J. Virol. 49772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, L. H., M. Lavignon, M. Taylor, and A. S. Alamgir. 2003. Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice. J. Virol. 7710327-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischinger, P. J., C. S. Blevins, and N. M. Dunlop. 1978. Genomic masking of nondefective recombinant murine leukemia virus in Moloney virus stocks. Science 201457-459. [DOI] [PubMed] [Google Scholar]
- 11.Fischinger, P. J., S. Nomura, and D. P. Bolognesi. 1975. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc. Natl. Acad. Sci. USA 725150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel, W. N., and J. M. Coffin. 1994. Endogenous nonecotropic proviruses mapped with oligonucleotide probes from the long terminal repeat region. Mamm. Genome 5275-281. [DOI] [PubMed] [Google Scholar]
- 13.Frendo, J. L., D. Olivier, V. Cheynet, J. L. Blond, O. Bouton, M. Vidaud, M. Rabreau, D. Evain-Brion, and F. Mallet. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 233566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 3861-85. [DOI] [PubMed] [Google Scholar]
- 15.Hartley, J. W., N. K. Wolford, L. J. Old, and W. P. Rowe. 1977. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. USA 74789-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 871556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishimoto, A., A. Adachi, K. Sakai, T. Yorifuji, and S. Tsuruta. 1981. Rapid emergence of mink cell focus-forming (MCF) virus in various mice infected with NB-tropic Friend virus. Virology 113644-655. [DOI] [PubMed] [Google Scholar]
- 18.Khan, A. S., and M. A. Martin. 1983. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc. Natl. Acad. Sci. USA 802699-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, A. S., W. P. Rowe, and M. A. Martin. 1982. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J. mice: identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J. Virol. 44625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander, E. S. et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 21.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landry, J. R., A. Rouhi, P. Medstrand, and D. L. Mager. 2002. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 191934-1942. [DOI] [PubMed] [Google Scholar]
- 23.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavignon, M., and L. Evans. 1996. A multistep process of leukemogenesis in Moloney murine leukemia virus-infected mice that is modulated by retroviral pseudotyping and interference. J. Virol. 703852-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medstrand, P., J. R. Landry, and D. L. Mager. 2001. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 2761896-1903. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, A., Y. Okazaki, J. Sugimoto, T. Oda, and Y. Jinno. 2003. Human endogenous retroviruses with transcriptional potential in the brain. J. Hum. Genet. 48575-581. [DOI] [PubMed] [Google Scholar]
- 27.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3799-808. [DOI] [PubMed] [Google Scholar]
- 28.Obata, Y., H. Ikeda, E. Stockert, and E. A. Boyse. 1975. Relation of GIX antigen of thymocytes to envelope glycoprotein of murine leukemia virus. J. Exp. Med. 141188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okahara, G., S. Matsubara, T. Oda, J. Sugimoto, Y. Jinno, and F. Kanaya. 2004. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84982-990. [DOI] [PubMed] [Google Scholar]
- 30.Oliff, A. I., G. L. Hager, E. H. Chang, E. M. Scolnick, H. W. Chan, and D. R. Lowy. 1980. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J. Virol. 33475-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver, P. L., and J. P. Stoye. 1999. Genetic analysis of Gv1, a gene controlling transcription of endogenous murine polytropic proviruses. J. Virol. 738227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rein, A. 1982. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology 120251-257. [DOI] [PubMed] [Google Scholar]
- 33.Rote, N. S., S. Chakrabarti, and B. P. Stetzer. 2004. The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta 25673-683. [DOI] [PubMed] [Google Scholar]
- 34.Sitbon, M., J. Nishio, K. Wehrly, and B. Chesebro. 1985. Pseudotyping of dual-tropic recombinant viruses generated by infection of mice with different ecotropic murine leukemia viruses. Virology 140144-151. [DOI] [PubMed] [Google Scholar]
- 35.Song, X., Y. Sun, and A. Garen. 2005. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl. Acad. Sci. USA 10212189-12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 612659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuhlmann, H., and P. Berg. 1992. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J. Virol. 662378-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterston, R. H. et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420520-562. [DOI] [PubMed] [Google Scholar]