Abstract
To better understand the components of an effective immune response to human immunodeficiency virus (HIV), the CD8+ T-cell responses to HIV, hepatitis C virus (HCV), and cytomegalovirus (CMV) were compared with regard to frequency, immunodominance, phenotype, and interleukin-2 (IL-2) responsiveness. Responses were examined in rare patients exhibiting durable immune-mediated control over HIV, termed long-term nonprogressors (LTNP) or elite controllers, and patients with progressive HIV infection (progressors). The magnitude of the virus-specific CD8+ T-cell response targeting HIV, CMV, and HCV was not significantly different between LTNP and progressors, even though their capacity to proliferate to HIV antigens was preserved only in LTNP. In contrast to HIV-specific CD8+ T-cell responses of LTNP, HLA B5701-restricted responses within CMV pp65 were rare and did not dominate the total CMV-specific response. Virus-specific CD8+ T cells were predominantly CD27+45RO+ for HIV and CD27−45RA+ for CMV; however, these phenotypes were highly variable and heavily influenced by the degree of viremia. Although IL-2 induced significant expansions of CMV-specific CD8+ T cells in LTNP and progressors by increasing both the numbers of cells entering the proliferating pool and the number of divisions, the proliferative capacity of a significant proportion of HIV-specific CD8+ T cells was not restored with exogenous IL-2. These results suggest that immunodominance by HLA B5701-restricted cells is specific to HIV infection in LTNP and is not a feature of responses to other chronic viral infections. They also suggest that poor responsiveness to IL-2 is a property of HIV-specific CD8+ T cells of progressors that is not shared with responses to other viruses over which immunologic control is maintained.
Gaining a better understanding of the immunologic control of human immunodeficiency virus type 1 (HIV-1) is among the most critical goals for the rational design of HIV vaccines and immunotherapies. Although most HIV-infected patients develop high-level viremia, CD4+ T-cell depletion, and progressive disease, a rare subgroup of patients variably termed long-term nonprogressors (LTNP) or elite controllers restrict HIV replication to below 50 copies of HIV RNA/ml plasma and remain disease free for up to 25 years without antiretroviral therapy (ART). Measurements of HIV-specific immune responses in these patients, in comparison with progressors, are providing insights into mechanisms that mediate immunologic control or loss of control in humans. Although the mechanisms of restriction of HIV replication remain incompletely understood, a number of lines of evidence suggest that it is mediated by HIV-specific CD8+ T cells (reviewed in reference 51). High frequencies of HIV-specific CD8+ T cells specific for the autologous virus are observed in both LTNP and untreated progressors, suggesting that differences in immunologic control are mediated not by quantitative but more likely by qualitative features of the immune response.
A number of qualitative features of the HIV-specific CD8+ T-cell response of LTNP or progressors have recently been proposed as the cause of immunologic control or loss of control, respectively. HLA B*5701 is highly overrepresented in LTNP, and the HIV-specific CD8+ T-cell response is highly focused on B5701-restricted peptides in B*5701+ LTNP but not in B*5701+ progressors (19, 50). In addition, there is a difference in surface markers between HIV- and cytomegalovirus (CMV)-specific CD8+ T cells thought to represent differences in maturation of the T-cell response (8). The CD8+ T cells of progressors are diminished in proliferative capacity and perforin upregulation in response to autologous HIV-infected CD4+ T cells (49). Recently, it has been proposed that this diminished proliferative capacity is due to a lack of paracrine or autocrine interleukin-2 (IL-2) production by HIV-specific CD4+ T cells or CD8+ T cells (41, 42, 75). Interpretation of proliferation studies is complicated by the fact that the effects of IL-2 were measured on the basis of 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) dye dilution of major histocompatibility complex (MHC) tetramer-positive cells. Because cell division over 6 days is an exponential function, IL-2 may induce small increases in the percentage of cells dividing or in the number of cell divisions that can result in large changes in the percent CFSElo cells, and yet the majority of antigen-specific cells may not proceed through the cell cycle. In addition, there are very limited data regarding whether the features of immunodominance, surface phenotype, and IL-2 responsiveness of HIV-specific CD8+ T cells extend to other chronic virus infections.
In the present study, we examined these qualitative features within the response to HIV, CMV, or hepatitis C virus (HCV) across patient groups. We observed that the high degree of focus upon B5701-restricted peptides found in LTNP does not extend to the HCV- or CMV-specific responses. The phenotype of HIV- or CMV-specific CD8+ T cells was highly variable and heavily influenced by the degree of viremia. In addition, when both the number of divisions and the percentage of cells dividing were analyzed, proliferation of HIV-specific CD8+ T cells was refractory to IL-2 stimulation, unlike that of CMV-specific cells. These results offer important insights into qualitative features of the HIV-specific CD8+ T-cell response, whether they extend to responses to other viruses, and whether they are associated with the presence or absence of immunologic control.
MATERIALS AND METHODS
Study population.
All patients in this study were previously recruited as part of a National Institute of Allergy and Infectious Diseases (NIAID) protocol and signed investigational review board-approved informed consent documents. LTNP patients had negative histories for opportunistic diseases, stable T-cell counts, set point HIV-1 RNA levels of <50 copies/ml (bDNA-based Versant HIV-1 RNA assay version 3.0; Bayer Diagnostics, Tarrytown, NY), and no ongoing ART. Progressors, defined as patients with a progressive decline in CD4+ T-cell counts and/or current or previously documented poor restriction of virus replication (HIV-1 RNA levels of >1,000 copies/ml) when not receiving ART, were divided into subgroups based on duration of infection, HIV-1 RNA set points, and treatment status. Untreated progressors either were ART naïve or had been off ART for at least 6 months prior to leukapheresis. Treated progressors received continuous ART. In addition, paired peripheral blood mononuclear cell (PBMC) samples reflecting “off therapy” and “on therapy” time points were obtained from each of nine HIV-seropositive patients undergoing an interruption of ART (66). Single-time-point or longitudinal PBMC samples from nine HIV-seronegative lung transplant recipients diagnosed with CMV pneumonitis were also obtained. HIV-1 infection in study participants was documented by HIV-1/2 immunoassay. CMV infection was confirmed using an enzyme-linked immunosorbent assay to detect CMV-specific immunoglobulin G. HCV infection was documented by HCV immunoassay, and HCV viral load was quantified by PCR. HLA class I/II typing was done by sequence-specific hybridization as described previously (6).
Q-PCR detection of CMV.
Patient and healthy donor blood samples were collected into EDTA-treated tubes. One-milliliter aliquots of plasma samples were prepared from the blood specimens. The MagNA Pure LC total nucleic acid (NA) isolation kit (large volume; Roche Applied Science, Indianapolis, IN) was used on the MagNA Pure LC instrument (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions to extract total NA from the 1-ml plasma aliquots and to elute the NA in 50 μl elution buffer. To verify that impurities that may contribute to quantitative real-time PCR (Q-PCR) inhibition were removed during the extraction, an internal control (linearized pBR322 plasmid) was added to the lysis/binding buffer from the MagNA Pure LC total NA isolation kit (large volume) at a final concentration of 200 copies/μl prior to the NA extraction step and then coextracted. A dilution series of a plasmid that contained CMV target sequence that was generated as described previously (9) was used to produce a quantitative reference standard curve for the quantification of positive samples in each Q-PCR experiment.
CMV DNA was detected and quantified with the RotorGene3000 system (Corbett Research/Robotics, San Francisco, CA) Primers, fluorescence resonance energy transfer detection probes, and the PCR conditions were essentially as previously described (9). Q-PCR was carried out with the QuantiTect Probe Mastermix (Qiagen, Valencia, CA) using 5 μl of extracted NA with a final reaction volume of 20 μl. The concentration of CMV target DNA was calculated by plotting the crossing point of each sample on the standard curve by using the RotorGene3000 software. The internal control was detected in a separate Q-PCR on the RotorGene3000 system with a second aliquot of extracted NA (5 μl) from the clinical sample.
Stimulation of CD8+ T cells for detection of gamma interferon (IFN-γ).
PBMCs, which had been obtained as described previously (49) and stored at −140°C, were thawed and rested overnight in 10% human AB (GemCell, Woodland, CA) medium prior to stimulation for 6 h with HCV peptides, CMV peptides, dendritic cell (DC) targets (CMV TB40E infected, HIVSF162 infected, or uninfected), phorbol myristate acetate (6.5 nM; Calbiochem, Darmstadt, Germany), and ionomycin (0.2 μM; Sigma-Aldrich, St. Louis, MO) or medium alone. For peptide stimulation experiments, autologous Epstein-Barr virus (EBV)-transformed B cells were pulsed with the relevant peptide/pool at a concentration of 0.5% PBMCs (200:1 effector/target cell [E:T] ratio). For HCV experiments, 441 14- to 18-mer peptides overlapping by 11 amino acids comprising the entire HCV 1a proteome were pooled according to gene product: core, E1, E2, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, Rockville, MD). The final peptide concentration was 4 μg/ml. For experiments using HIV Gag (NIH AIDS Research and Reference Reagent Program) or CMV pp65 (Multiple Peptide Systems, La Jolla, CA) peptide 123 or 138, respectively, 15-mers overlapping by 10 amino acids were used at a 2-μg/ml final concentration of each peptide. A peptide matrix was used for pp65 epitope mapping, whereby each peptide was present in two pools, as previously described (31, 32). Recognition of individual 15-mer peptides was then confirmed, and where applicable, 9-mers were constructed to identify optimal epitopes (see Table 2). To determine HLA restriction, heterologous B-cell targets matched at a single or limited number of HLA class I loci pulsed with the appropriate peptide were used as antigen-presenting cells (APC) at an E:T ratio of 10:1. To determine the total frequency of CMV- and HIV-specific CD8+ T cells, CMV TB40E-infected and HIVSF162-infected autologous and heterologous DC targets were used as APC at an E:T ratio of 1:1, which gave optimal responses with low background and minimal bystander activation and was relatively insensitive to variations in the level of infection.
TABLE 2.
Summary of cytotoxic T-lymphocyte epitopes derived from CMV pp65
| Peptide group and namea | Amino acid sequence | Position | Reference(s) |
|---|---|---|---|
| Newly identified epitopes and HLA class I restriction elements | |||
| B57 VF9 | VAFTSHEHF | 294-302 | |
| C6 TI9 | TRATKMQVI | 211-219 | |
| Previously reported epitopes and HLA class I restriction elements | |||
| A1 YY11 | YSEHPTFTSQY | 363-373 | 44, 57 |
| A2 NV9 | NLVPMVATV | 495-503 | 11, 72 |
| B35 IY9 | IPSINVHHY | 123-131 | 17 |
| B40 HL9 | HERNGFTVL | 267-275 | 35 |
| B44 EY10 | EFFWDANDIY | 512-521 | 71, 72 |
| B44 SY10 | SEHPTFTSQY | 364-373 | 13, 35 |
| B52 QV9 | QMWQARLTV | 155-163 | 31 |
| 15-mer peptides recognized in peptide matrix mapping experiments | |||
| 15 | PCHRGDNQLQVQHTY | 67-81 | |
| 33 | HRHLPVADAVIHASG | 139-153 | |
| 44 | TSAFVFPTKDVALRH | 183-197 | |
| 49 | ELVCSMENTRATKMQ | 203-217 | |
| 54 | YVKVYLESFCEDVPS | 223-237 | |
| 56 | FCEDVPSGKLFMHVT | 231-245 | |
| 64 | FMRPHERNGFTVLCP | 263-277 | |
| 94 | HTWDRHDEGAAQGDD | 383-397 | |
| 109 | ATACTAGVMTRGRLK | 443-457 |
Abbreviated as HLA class I restriction element followed by CMV peptide sequence identified as first and last amino acid symbols followed by sequence length.
PBMCs were aliquoted at 4 × 106 per stimulation tube (Starstedt Inc., Newton, NC) and stimulated with peptides or cell targets for 6 h prior to intracellular staining as previously described (50). In all peptide stimulation experiments, anti-CD28 and anti-CD49d were added for costimulation (1 μg/ml; BD Biosciences, San Jose, CA).
CMV preparation.
CMV strain TB40E, a kind gift from L. Hertel and E. Mocarski of Stanford University, was propagated in confluent human foreskin fibroblasts that were generously provided by J. Meier of the University of Iowa (23). Human foreskin fibroblasts were infected at a low multiplicity of infection, and infection was allowed to proceed until the fibroblasts exhibited nearly 100% cytopathic effect. Cell-free supernatant (75 ml) was then collected, centrifuged once at 1,300 rpm to remove debris, and then concentrated by ultracentrifugation at 80,000 × g for 70 min. The pelleted virus was then resuspended in 500 μl and stored in 35-μl aliquots at −80°C until used for DC infection (58).
Generation of CMV- and HIV-infected DCs.
DCs were generated as previously described (59). Briefly, cryopreserved PBMCs were thawed, washed twice in 10% fetal bovine serum (FBS; Gemini Bio-Products, West Sacramento, CA) medium, and incubated at 37°C in T75 flasks at a concentration of 2 × 106 cells/ml in 10% FBS medium. After 2 hours, nonadherent cells and medium were discarded. Adherent monocytes were washed twice with room-temperature phosphate-buffered saline (PBS) and incubated with 10% FBS medium supplemented with 6.5 ng/ml IL-4 (R&D, Minneapolis, MN) and 100 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech, Rocky Hill, NJ) for 7 days. Medium was replaced on days 2, 4, and 6. On day 7, DCs were pooled and counted. Residual CD8+ T cells at day 7 were depleted by automated magnetic cell sorting according to the manufacturer's protocol (AutoMACS; Miltenyi Biotec, Germany). Prior to infection, these cells were predominantly CD1Ahi and CD14lo.
Between 2 × 106 and 6 × 106 DCs were infected with 107 50% tissue culture infective doses of CMV TB40E (30 μl) or 5,000 50% tissue culture infective doses of HIVSF162 (50 μl). Cells and virus were resuspended in IL-4- and GM-CSF-containing medium and centrifuged at 2,400 rpm for 1.5 h at room temperature. After spinoculation, supernatant was aspirated, and cells were resuspended in IL-4- and GM-CSF-containing medium and plated in 24-well plates at a concentration of 106 cells/ml. In preliminary time course experiments, peak infection, determined by staining for the CMV immediate-early 1 (IE-1) or the HIV p24 gene product, occurred at 24 h and 4 days, respectively, of incubation. Subsequently, infection (CMV, 30 to 60%; HIV, 5 to 13%) and cell purity (>98%) were confirmed in all cases by flow cytometry. In experiments comparing HIV-infected DCs with HIV-infected primary CD4+ T-cell targets, autologous CD4+ T cells were positively selected from cryopreserved PBMCs by magnetic automated cell sorting and polyclonally stimulated prior to infection as previously described (49). Infection (21 to 47%) and cell purity (>98%) were confirmed in all cases by flow cytometry.
CFSE proliferation assays.
For proliferation experiments, PBMCs were labeled with CFSE (Molecular Probes, Eugene, OR) as previously described (49). CFSE-labeled PBMCs were incubated in 96-well, deep-well culture plates (PGC Scientifics, Frederick, MD) at a density of 106 PBMCs/well/ml with HCV or CMV peptides, anti-CD3 (Orthoclone OKT3, 1 μg/ml; Ortho Biotech, Bridgewater, NJ) and anti-CD28 (1 μg/ml) antibodies, or medium with or without IL-2 (20 IU/ml) for 6 days.
Flow cytometry.
Multiparameter flow cytometry was performed according to standard protocols (26). Surface and intracellular staining of DCs was done using the following antibodies: fluorescein isothiocyanate (FITC)-conjugated anti-CMV IE-1 (Chemicon International, Inc., Temecula, CA), FITC-conjugated anti-HIV p24 (Kc57-FITC; Beckman Coulter, Inc., Fullerton, CA), peridinin chlorophyll protein-conjugated anti-CD14 (BD PharMingen, San Diego, CA), and allophycocyanin-conjugated anti-CD1A (BD PharMingen). In order to stain infected and uninfected DCs for intracellular expression of the CMV IE-1 gene product, 2 × 105 DCs were pelleted and then placed on ice for 15 min prior to fixation and permeabilization with cold (stored at −70°C before use) absolute methanol (Fisher BioTech Grade, Fair Lawn, NJ) for 10 min at 4°C. The cells were washed with PBS and stained with 100 μl of FITC-conjugated anti-IE-1 at 37°C. After 1 h, the cells were washed three times with an 0.5% PBS-Tween 20 solution (Chemicon International, Inc.) and resuspended in 400 μl of PBS containing 1% formaldehyde. For intracellular staining of the HIV p24 gene product, DC or CD4+ T-cell targets were fixed and permeabilized with Cytofix/Cytoperm (BD PharMingen), washed, and then stained with the FITC-conjugated Kc57 antibody at 4°C for 30 min.
Surface and/or intracellular staining of PBMCs was performed using the following antibodies, which were purchased from BD Biosciences unless otherwise specified: FITC-conjugated anti-CD3 and anti-CD8; phycoerythrin (PE)-conjugated anti-CD3, anti-CD8, anti-CD69, and anti-IL-2; peridinin chlorophyll protein-conjugated anti-CD3, anti-CD8, and anti-CD45RA; allophycocyanin-conjugated anti-CD4, anti-IFN-γ, and anti-CD45RO; PE-Texas Red-conjugated anti-CD4 (Caltag Laboratories, Inc., Burlingame, CA); PE Alexa 700-conjugated anti-CD8 (Caltag Laboratories, Inc.); PE-Cy7-conjugated anti-tumor necrosis factor alpha; Alexa 700-conjugated anti-CD3; allophycocyanin-Cy7-conjugated anti-CD27 (eBiosciences, San Diego, CA); and Pacific blue-conjugated anti-CD3. CMV and HIV HLA class I tetramers conjugated to allophycocyanin and/or PE (Beckman Coulter, Inc.) were used to label epitope-specific CD8+ T cells (see Table 3). HLA-Cw0602-CMV pp65211-219 TRATKMQVI (TI9) allophycocyanin tetramer was conjugated at the NIH tetramer facility (Atlanta, GA). Cw*0602 cDNA was generously provided by H. Reyburn of Cambridge University (46). CCR7 staining was performed in two steps: murine anti-human CCR7 (BD PharMingen) was added at 37°C for 30 min, followed by Alexa 488-conjugated goat anti-mouse secondary antibody (Invitrogen Molecular Probes) at 4°C for 30 min. All other staining was performed at 4°C for 30 min.
TABLE 3.
Human leukocyte antigen class I tetramers
| Tetramera | Protein | Amino acid sequence | Position |
|---|---|---|---|
| CMV | |||
| A1 VY9 | pp50 | VTEHDTLLY | 245-253 |
| A2 NV9 | pp65 | NLVPMVATV | 495-503 |
| B7 TM10 | pp65 | TPRVTGGGAM | 417-426 |
| B8 EM9 | IE-1 | ELRRKMMYM | 199-207 |
| B35 IY9 | pp65 | IPSINVHHY | 123-131 |
| B57 VF9 | pp65 | VAFTSHEHF | 294-302 |
| C6 TI9 | pp65 | TRATKMQVI | 211-219 |
| HIV | |||
| A1 GY9 | Gag p17 | GSEELRSLY | 71-79 |
| A2 SL9 | Gag p17 | SLYNTVATL | 77-85 |
| A3 RK9 | Gag p17 | RLRPGGKKK | 20-28 |
| A24 RF10 | Nef | RYPLTFGWCF | 134-143 |
| B8 EI8 | Gag p24 | EIYKRWII | 260-267 |
| B8 FL8 | Nef | FLKEKGGL | 90-97 |
| B27 IK9 | Gag p17 | IRLRPGGKK | 19-27 |
| B27 KK10 | Gag p24 | KRWIILGLNK | 263-272 |
| B35 RY11 | Nef | RPQVPLRPMTY | 71-81 |
| B57 IW9 | Gag p24 | ISPRTLNAW | 147-155 |
| B57 KF11 | Gag p24 | KAFSPEVIPMF | 162-172 |
| B57 QW9 | Gag p24 | QASQEVKNW | 308-316 |
Abbreviated as HLA class I restriction element followed by virus peptide sequence identified as first and last amino acid symbols followed by sequence length.
Flow cytometry profiles were gated on CD3+ CD8+ lymphocytes, and 50,000 to 200,000 events were collected. Samples were analyzed on a FACSAria three-laser cytometer (Becton Dickinson) with FACSDiva software. Color compensations were performed for each patient's PBMCs using samples single stained for each of the fluorochromes used. Data were analyzed using FlowJo software (Tree Star, San Carlos, CA). In experiments with CFSE-labeled cells, the FlowJo Proliferation Platform provided detailed information about the division characteristics of gated tetramer+ CD8+ T cells. For each sample, a model fitting the cell population of interest was used to separate it into individual generations. When possible, a standard model was calculated from anti-CD3/anti-CD28-stimulated cells to define the nonproliferating and proliferating subsets and the individual divisions. This was then applied to subsequent samples. This type of analysis produced information about the fraction of cells of the original cell population that divided (percent divided) and the average number of divisions that these proliferating cells had undergone (proliferation index). To examine the effect of IL-2 on proliferation, these parameters for tetramer+ CD8+ T cells were generated in samples that had been stimulated with peptides plus IL-2 and compared to the values obtained from samples stimulated with peptides alone by calculating the delta percent divided (percent divided with IL-2 minus percent divided without IL-2) and the delta proliferation index (proliferation index with IL-2 minus proliferation index without IL-2) (FlowJo; Tree Star).
Statistical methods.
Independent groups were compared using the Wilcoxon two-sample test. The Wilcoxon signed-rank test was used to compare paired data. Correlation was determined by the Spearman rank method. Paired proportions were compared by the McNemar test. The Bonferroni method was used to adjust P values for multiple testing. Medians are reported.
RESULTS
Patient characteristics.
The clinical characteristics of the study patients are shown in Table 1. LTNP are included in a cohort who have been infected for a median of 17 years and yet typically maintain normal CD4+ T-cell counts (median, 834 [range, 456 to 1,830] cells/ml) and <50 copies of HIV RNA/ml plasma without ART. Additional detailed characteristics of this cohort are provided in previous studies (48-50). HLA B*5701 is highly overrepresented in LTNP but occurs in progressors at a frequency of 10%, similar to the U.S. Caucasian population (50). For this reason, additional patients with HLAB*57 were added to groups B, C, and D to permit comparisons of CD8+ T cells specific for HIV, HCV, or CMV that are restricted by these alleles.
TABLE 1.
Characteristics of coinfected patients
| Patient group and no. | HLA A | HLA Ba | HLA C | Yr of diagnosis | CD4 cell count | HIV VLb | HCV serostatus/VLc | CMV serostatus/VL |
|---|---|---|---|---|---|---|---|---|
| LTNP | ||||||||
| 4 | 1, 31 | 8, 57 | 6, 7 | 1985 | 1,190 | <50 | − | +/<50 |
| 6 | 11, 30 | 52, 57 | 7, 12 | 1986 | 606 | <50 | 6,000,000 | +/<50 |
| 8 | 11, 23 | 44, 57 | 4, 6 | 1985 | 473 | <50 | − | +/<50 |
| 11 | 32 | 14, 51 | 8, 14 | 1989 | 1,830 | <50 | − | +/<50 |
| 12 | 3, 11 | 7, 57 | 6, 7 | 1986 | 456 | <50 | <615 | +/<50 |
| 17 | 2, 26 | 38, 27 | 1, 12 | 1985 | 950 | <50 | − | +/<50 |
| 33 | 2, 30 | 13, 57 | 6 | 1995 | 972 | <50 | − | +/<50 |
| 34 | 1, 2 | 8, 57 | 7, 6 | 1989 | 958 | <50 | 2,375,950 | +/<50 |
| 37 | 30 | 42, 57 | 17, 18 | 1998 | 1,616 | <50 | − | +/<50 |
| 38 | 2, 24 | 44, 57 | 5, 6 | 1990 | 1,645 | <50 | − | +/<50 |
| 47 | 1, 74 | 57, 81 | 7, 18 | 1994 | 538 | <50 | − | +/<50 |
| 48 | 1, 33 | 8, 53 | 1, 4 | 1989 | 834 | <50 | − | +/<50 |
| 53 | 11, 80 | 27, 35 | 2, 4 | 1992 | 825 | <50 | − | +/<50 |
| 58 | 30, 74 | 15, 57 | 3, 8 | 1989 | 596 | <50 | − | +/<50 |
| 59 | 23, 30 | 7, 57 | 7, 15 | 1986 | 833 | <50 | − | +/<50 |
| Slow progressors | ||||||||
| 20 | 1 | 52, 57 | 6, 12 | 1985 | 1,307 | 28,954 | − | +/<50 |
| 21 | 1, 2 | 8, 27 | 1, 7 | 1984 | 564 | 7,148 | − | +/<50 |
| 35 | 2, 3 | 57 | 6, 7 | 2000 | 520 | 1,800 | 245,234 | +/<50 |
| 36 | 2 | 13, 44 | 5, 8 | 1996 | 614 | 1,770 | − | +/<50 |
| 44 | 1, 24 | 27, 37 | 2, 6 | 1986 | 789 | 1,370 | − | +/<50 |
| 45 | 2 | 45, 57 | 6 | 1990 | 791 | 9,607 | − | +/<50 |
| 46 | 2, 23 | 53, 57 | 6, 18 | 1984 | 461 | 1,543 | 2,383,710 | +/<50 |
| 54 | 2, 11 | 15, 37 | 4, 6 | 1989 | 649 | 1,289 | − | +/<50 |
| 55 | 1, 23 | 8, 57 | 7 | 1992 | 394 | 2,062 | <615 | +/<50 |
| 63 | 30 | 57, 81 | 18 | 1986 | 803 | 1,607 | + | +/<50 |
| Viremic progressors | ||||||||
| 103 | 2, 11 | 55, 57 | 3, 6 | 1991 | 545 | 1,406 | − | +/<50 |
| 105 | 2, 80 | 8, 57 | 7, 12 | 1990 | 720 | 1,220 | − | +/<50 |
| 107 | 3 | 40, 57 | 3, 7 | 1987 | 405 | 90,130 | − | +/<50 |
| 131 | 2, 11 | 35, 57 | 4, 6 | 1989 | 421 | 76,038 | − | +/<50 |
| 133 | 33, 68 | 15, 35 | 3, 16 | 2000 | 594 | 82,000 | 974,450 | +/<50 |
| 134 | 68 | 15, 27 | 3 | 2003 | 336 | 20,421 | <615 | +/<50 |
| 137 | 34, 68 | 53, 58 | 3, 4 | 2003 | 784 | 213,759 | − | +/<50 |
| 138 | 1, 68 | 15, 57 | 1, 7 | 1996 | 444 | 160,954 | − | +/<50 |
| 139 | 2, 32 | 27, 35 | 1, 4 | 1993 | 453 | 78,984 | − | +/<50 |
| 203 | 30, 32 | 18, 57 | 5, 6 | 1989 | 824 | 6,584 | − | +/<50 |
| Treated progressors (<50 copies) | ||||||||
| 101 | 1, 31 | 51, 57 | 6, 15 | 1986 | 602 | <50 | − | +/<50 |
| 113 | 1, 2 | 45, 57 | 7, 16 | 1991 | 574 | <50 | − | +/<50 |
| 114 | 1, 24 | 8, 57 | 6, 7 | 1989 | 292 | <50 | − | +/<50 |
| 115 | 1, 3 | 14, 57 | 6, 8 | 1995 | 567 | <50 | − | +/<50 |
| 118 | 2, 2 | 51, 57 | 6, 14 | 1990 | 668 | <50 | − | +/<50 |
| 124 | 11 | 35, 57 | 4, 6 | 1986 | 852 | <50 | − | +/<50 |
| 126 | 1, 68 | 8, 57 | 5, 8 | 1991 | 829 | <50 | 3,440,410 | +/<50 |
| 129 | 2, 32 | 15, 27 | 1, 2 | 1988 | 366 | <50 | − | +/<50 |
| 130 | 25, 68 | 18, 57 | 7, 12 | 1997 | 472 | <50 | <521 | +/<50 |
| 132 | 30, 66 | 40, 57 | 3, 7 | 1995 | 1,409 | <50 | − | +/<50 |
| 135 | 2, 3 | 42, 81 | 17, 18 | 1985 | 666 | <50 | 9,504,730 | +/<50 |
| 136 | 23, 68 | 14, 58 | 6, 8 | 1986 | 1,172 | <50 | − | +/<50 |
| 142 | 2, 68 | 27, 40 | 1, 3 | 1994 | 759 | <50 | − | +/<50 |
| 144 | 3, 26 | 7, 57 | 6, 7 | 1986 | 729 | <50 | − | +/<50 |
| 145 | 1, 68 | 15 | 2, 3 | 1994 | 1,914 | <50 | − | +/<50 |
HLA B57 is highlighted by underlining.
VL, viral load, expressed as number of copies/ml of plasma.
HCV viral load, expressed as International Units (IU)/ml of plasma.
Twelve patients in this cohort are seropositive for HCV; three are LTNP and nine are patients with evidence of HIV-1 disease progression. Of the three HIV/HCV-coinfected LTNP, patient 12 had a sustained virologic response following treatment with pegylated IFN and ribavirin, but the other two untreated patients had had HCV RNA levels persistently greater than 2 × 106 IU/ml plasma. Of the progressors, patients 55 and 134 spontaneously cleared their infection, and patient 130 was cured with antiviral therapy. Patient 46, initially diagnosed with HCV genotype 1a, was reinfected with genotype 1b following undetectable HCV RNA levels for 11 months posttreatment. In summary, the few HCV/HIV-coinfected patients in our cohort with immune-mediated control over HIV were not more likely to have cleared HCV infection, consistent with one prior report (38). All of the patients studied were CMV seropositive, but all had CMV DNA levels below 50 copies/ml plasma at the time of study.
Frequency, breadth, and MHC restriction of virus-specific responses in coinfected patients.
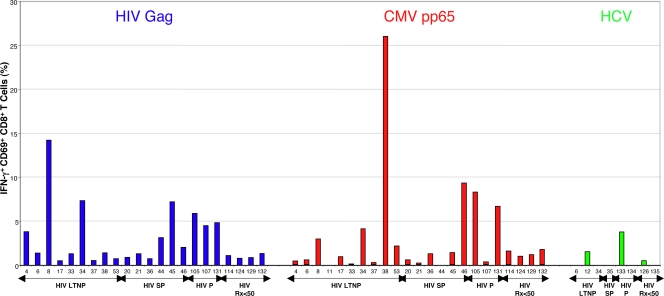
The relationship between levels of antigen and frequency and breadth of virus-specific CD8+ T cells was examined. Because the length of gene products can affect the number of epitopes detected and the frequency of responding CD8+ T cells (63), the response to CMV pp65 peptides was compared to the response to HIV Gag peptides. The amino acid lengths of these two gene products are roughly similar (499 and 561 for Gag and pp65, respectively). Compared with the cumulative frequency of IFN-γ-producing Gag-specific CD8+ T-cell responses, the total frequency of CMV pp65-specific responses was of similar magnitude (medians, 1.2% [range, 0 to 26%] versus 1.38% [range, 0.5 to 14.2%], respectively; P > 0.5) (Fig. 1). As reported previously, using various techniques to enumerate antigen-specific cells in HCV-monoinfected or HCV/HIV-coinfected patients, the frequencies of CD8+ T-cell responses to any HCV gene product in our cohort were very low compared to those to CMV or HIV (29, 38-40, 70). Using peptides spanning the entire HCV proteome, HCV-specific CD8+ T cells were detected in the peripheral blood of only three/eight patients and were of very low magnitude (Fig. 1). Although the small sample size did not permit meaningful statistical analysis, HCV-specific CD8+ T-cell frequencies did not appear to correlate with either HIV or HCV viral loads or CD4 counts (see Fig. S1 in the supplemental material). Furthermore, these cells from five coinfected patients did not proliferate to HCV peptides, even in three LTNP with preserved HIV-specific CD8+ T-cell proliferation (see Fig. S1 in the supplemental material). Given the low frequencies, the HCV-specific responses were not examined in further detail.
FIG. 1.
Comparison of CD8+ T-cell responses to HIV Gag, CMV pp65, and HCV. The percentages of CD8+ T cells that are CD69+ and IFN-γ+ in response to EBV-transformed B cells pulsed with HIV Gag (blue), CMV pp65 (red), or HCV (green) peptide pools are shown. Background activity against B cells not pulsed with peptides has been subtracted. Patient numbers are listed below each bar. Patients are subgrouped according to HIV disease status as LTNP (HIV LTNP), slow progressors (HIV SP), progressors (HIV P), and patients with antiretroviral-induced suppression of HIV RNA levels to <50 copies/ml (HIV Rx<50).
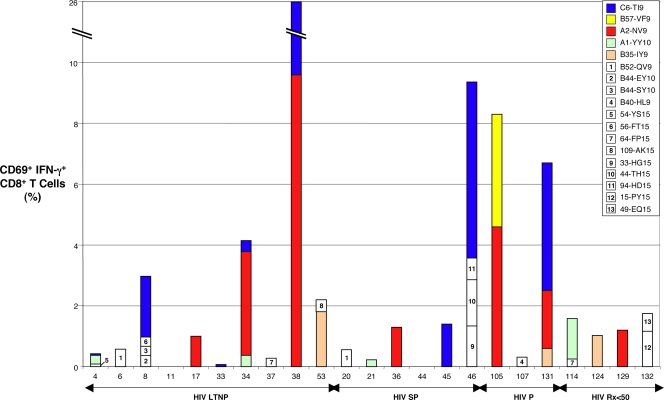
We then analyzed the frequency and breadth of immunodominant CMV-specific CD8+ T-cell responses and their MHC restriction elements in a subset of 23 CMV/HIV-coinfected patients using a peptide matrix in intracellular IFN-γ detection assays as previously described (31, 32). Two responses were frequently detected. In the first case, a response to the immunodominant HLA A0201-restricted NLVPMVATV (NV9) epitope (72) contained within peptides 121 and 122 was found in 6 of 11 (55%) HLA A*02+ patients (Table 2 and Fig. 2; see also Fig. S2 in the supplemental material). In the second case, a high-frequency response targeting the TRATKMQVI (TI9) epitope contained within peptides 50 and 51 was observed in eight patients who shared HLA B*57 and Cw*06 (see Fig. S2 in the supplemental material; also data not shown). Although CD8+ T-cell recognition of TI9 was previously reported to be HLA B13 restricted (32, 34, 53), Cw0602 was confirmed to be the major HLA class I restriction element in our cohort using heterologous B-cell lines matched at a limited number of class I alleles as APC (see Fig. S2 in the supplemental material). Of 12 Cw*0602+ patients, eight (67%) recognized the TI9 epitope, a prevalence exceeding the 55% of A*02+ patients who targeted the immunodominant HLA A02-restricted NV9 epitope (Fig. 2 and data not shown).
FIG. 2.
Summary of CMV pp65-specific CD8+ T-cell response mapping. The percentages of CD8+ T cells that are CD69+ and IFN-γ+ in response to CMV pp65 peptides following 6-hour stimulation are shown. Individual peptide-specific responses are color coded by specificity and HLA restriction if present in three or more patients. Two novel responses, seven previously reported CMV pp65 optimal epitope-specific CD8+ T-cell responses, and low-frequency responses targeting nine 15-mers in which the optimal epitopes and HLA restriction were not determined are shown (Table 2). Patient numbers are listed below each bar. Patients are subgrouped according to HIV disease status as LTNP (HIV LTNP), slow progressors (HIV SP), progressors (HIV P), and patients with antiretroviral-induced suppression of HIV RNA levels to <50 copies/ml (HIV Rx<50).
Identification of a B57-restricted CMV epitope was of interest to permit comparisons between B57-restricted CD8+ T-cell responses targeting HIV and CMV. The optimal epitope VAFTSHEHF (VF9) contained within peptide 71 had a planar motif compatible with B57 restriction and was recognized by the cells of the A*02+/B*57+ patient 105 (see Fig. S2 in the supplemental material). Although Cw12 was the restriction element in a previous report (35), B57 was confirmed to restrict CD8+ T-cell recognition of VF9 in the present case (Table 2 and Fig. 2; see also Fig. S2 in the supplemental material). Of 16 HLA B*57+ patients, only CD8+ T cells from patient 105 recognized this epitope. In addition, no other frequently targeted HLA B57-restricted epitope was identified. This is a striking contrast to HIV-specific CD8+ T-cell responses, where cells derived from all B*5701+ patients recognize some B5701-restricted HIV epitopes, and in LTNP, the majority of HIV-specific CD8+ T-cell responses are B5701 restricted (19, 50). The prevalence of recognition of the TI9 epitope by CD8+ T cells from 11 patients who were both HLA B*5701+ and Cw*06+ was significantly greater than recognition of the VF9 epitope (73% versus 0%, P = 0.005).
To examine characteristics of these CMV-specific CD8+ T cells with HIV-specific cells in the same patients, MHC class I tetramers were made for the HLA Cw0602-restricted CMV pp65211-219 TI9 and the HLA B5701-restricted CMV pp65294-302 VF9 epitopes (see Fig. S2 in the supplemental material). Cells from 12 of 14 (86%) HLA Cw*0602+ patients stained positively with the C6 TI9 tetramer, confirming the highly dominant nature of this response specificity. In contrast, cells from only 1 of 13 (7%) HLA B*5701+ patients stained positively with the B57 VF9 tetramer (data not shown). Similar to the results of the IFN-γ assays, the prevalence of positive staining with the C6 TI9 tetramer was significantly greater than that with the B57 VF9 tetramer in eight HLA B*5701+/Cw*0602+ patients (88 versus 0%, P = 0.02).
For most of the other dominant CD8+ T-cell responses detected in these CMV mapping experiments, HLA restriction had been previously described and was confirmed in a subset of patients (Table 2 and Fig. 2; also data not shown). Twenty-one of 23 patients had responses to CMV pp65, ranging in magnitude from 0.1 to 26% CD69+ IFN-γ+ CD8+ T cells. The median total CMV pp65-specific CD8+ T-cell frequency was not significantly different between 10 LTNP and 13 HIV+ progressors (0.8% [range, 0 to 26%] versus 1.3% [range, 0 to 9.4%], respectively; P > 0.5). The breadths or median numbers of pp65 peptides targeted were also similar between LTNP and progressors (1.5 [range, 0 to 4] versus 1 [range, 0 to 4], respectively; P > 0.5) (Fig. 2). The total frequencies and breadth of CMV pp65-specific CD8+ T-cell responses did not correlate with HIV RNA levels (P > 0.5).
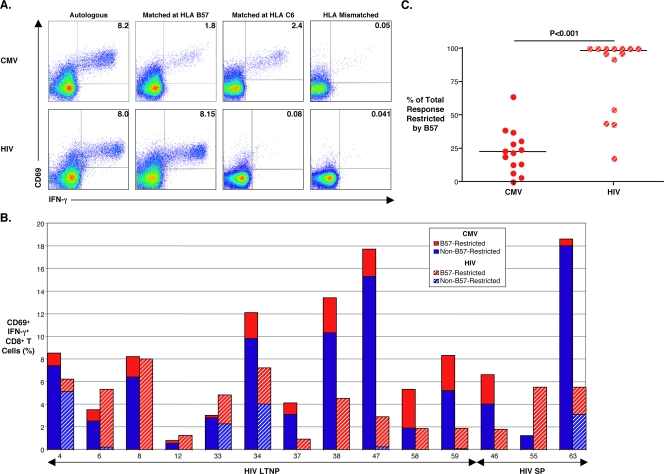
Given that CMV pp65 represents only a small portion of the CMV genome and that CMV gene products other than pp65 have been reported to elicit large CD8+ T-cell responses, we next examined the total CMV-specific CD8+ T-cell response in HIV/CMV-coinfected patients and determined the overall contribution of HLA B57 restriction (13, 32, 33, 63). Recently, it has been shown that certain endothelial strains of CMV (TB40E and VHLE) can productively infect monocyte-derived DCs, which can be used to stimulate CD8+ T cells (5, 18, 21, 23, 52, 58). In order to examine the total CD8+ T-cell response, CMV TB40E-infected autologous and heterologous DCs were used as APC. Similar to other reports, 30 to 60% of DCs expressed the IE-1 gene product at 24 h of infection (data not shown) (23, 52). Figure 3A (top row) shows an example where 8.1% of this representative patient's CD8+ T cells recognized autologous CMV TB40E-infected DCs. When stimulated with heterologous DC targets matched only at HLA B*57, 1.8% of CD8+ T cells responded, compared with a response of 2.4% using heterologous DCs matched only at HLA C*6. Minimal responses above background (<0.05%) were detected when effectors and targets were completely HLA mismatched or if effectors were derived from a CMV-seronegative donor (Fig. 3A and data not shown). In six randomly selected patients, the total CMV-specific CD8+ T-cell frequencies measured in this assay were greater than the frequencies measured against a pp65 peptide pool, suggesting that a significant proportion of CMV-specific CD8+ T cells recognize gene products other than pp65 (data not shown).
FIG. 3.
Determination of total CMV- and HIV-specific CD8+ T-cell responses and the contribution of HLA B57 restriction using virus-infected monocyte-derived DCs as APC. (A) Pseudocolor density plots are gated on CD8+ T cells from patient 8 showing CD69+ and IFN-γ+ expression in response to 6-hour stimulation with CMV TB40-infected (top row) or HIVSF162-infected (bottom row) DC targets at a 1:1 E:T ratio. DCs were autologous (first column), heterologous with a match only at HLA B*57 (second column), heterologous with a match only at Cw*06 (third column), or heterologous with complete mismatches at all HLA class I loci (fourth column). Numbers on plots reflect the percentages of the gated population expressing CD69 and IFN-γ after subtracting background values, which were measured against uninfected targets (not shown). (B) Total frequencies of CMV- (solid bars) and HIV-specific (hatched bars) CD8+ T cells measured by CD69+ and IFN-γ+ expression in response to virus-infected DC targets are shown for 11 B*5701+ LTNP and three B*5701+ slow progressors. The red bars signify the fractions of the total virus-specific CD8+ T-cell responses that are HLA B57 restricted. Patient numbers are listed below each bar. (C) Data in panel B expressed simply as the percentage of the total response restricted by B57. Solid black lines represent median values. Paired data were compared with the Wilcoxon signed-rank test.
The fraction of the total HIV-specific CD8+ T-cell response restricted by HLA B57 was also examined using a similar monocyte-derived DC system. Although we have previously examined the frequency of HIV-specific CD8+ T cells in response to autologous HIV-infected CD4+ T-cell targets, we have not examined the response to HIV-infected DCs (49). Therefore, potential differences in restriction patterns between CMV- and HIV-specific CD8+ T cells could be caused by differences in APC types, not differences in the responses to CMV in comparison to HIV. Consistent with previous reports, peak HIVSF162 infection on day 4 resulted in a median value of 6.1% (range, 5 to 13%) of DCs expressing the HIV p24 gene product (data not shown) (25, 45). In Fig. 3A, the CD8+ T-cell response to HIVSF162-infected autologous DCs is similar to the total CMV-specific response in this LTNP; however, the HIV-specific CD8+ T-cell response is completely restricted by HLA B57. To ensure the utility of HIVSF162-infected DCs as APC, CD8+ T-cell responses using these cell targets were compared with those of HIVSF162-infected CD4+ lymphoblast targets in a subset of eight B*57+ LTNP (50). Although the percentages of p24+ autologous DCs were significantly lower than the percentages of p24+ lymphoblasts (medians, 6.1% [range, 5 to 13%] versus 31.7% [range, 21 to 47%], respectively; P < 0.001), the HIV-specific CD8+ T-cell responses were of a similar magnitude (medians, 5.4% [range, 0.9 to 8%] versus 6.4% [range, 0.7 to 13.6%], respectively; P > 0.5). In addition, the proportions of the responses to infected DCs or lymphoblasts that were restricted by HLA B57 were similar (medians, 100% [range, 17.7 to 100%] versus 94.5% [range, 11 to 100%], respectively; P = 0.25; data not shown).
A summary of the total CMV- and HIV-specific CD8+ T-cell responses in 11 B*57+ LTNP and three B*57+ slow progressors is shown in Fig. 3B. With the use of monocyte-derived DC targets infected with CMV TB40E, the median total CMV-specific CD8+ T-cell response was 7.4% (range, 0.8 to 18.6%) with only a median of 22.6% (range, 0 to 64.2%) of the total response restricted by HLA B57 as determined by stimulation with heterologous (matched only at B*57) CMV-infected DCs (Fig. 3C). High-frequency, broadly directed CMV-specific CD8+ T-cell responses have also been reported in CMV-seropositive, HIV-seronegative subjects (13, 63). With the use of HIVSF162-infected DCs as APC, the median total HIV-specific CD8+ T-cell response was 4.7% (range, 0.9 to 8%) and was not significantly different from the total CMV-specific response (P = 0.07; Fig. 3B). However, unlike the CMV-specific responses, the HIV-specific CD8+ T-cell responses of B*57+ patients with low levels of HIV RNA were predominantly focused on B57-restricted epitopes, with a median of 98.2% (range, 17.7 to 100%) of the total response restricted by HLA B57 (P < 0.001; Fig. 3C). Thus, consistent with the pp65 peptide mapping data, HLA B57 appears to restrict a relatively small percentage of the total CMV-specific response. This observation was made in LTNP whose CD8+ T cells preferentially target B57-restricted HIV epitopes. These results suggest that the immunodominance of B57 in the response to HIV in LTNP does not extend to the response to CMV. Immunodominance of B57-restricted cells is therefore unlikely to be simply intrinsic to the B57 molecule itself or a response modifier but rather is influenced by other factors that are unique to HIV infection of LTNP.
Differences in surface phenotype of HIV- and CMV-specific CD8+ T cells are a consequence of viral load.
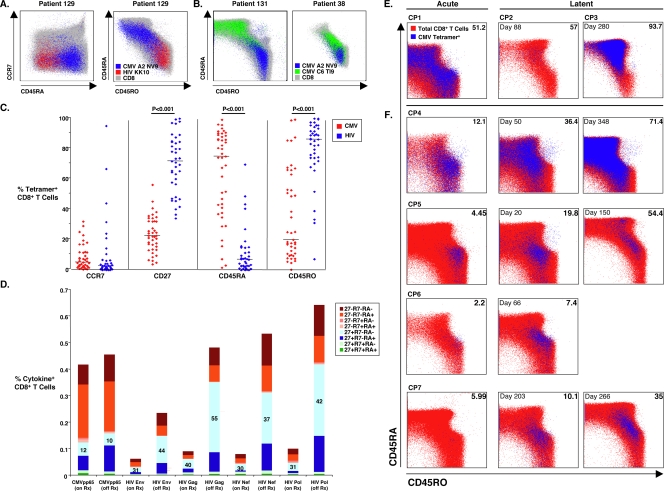
A comparison of the phenotype of HIV- and CMV-specific CD8+ T cells was also explored in these cohorts (Fig. 4). It has been suggested that the impaired functionality of HIV-specific CD8+ T cells is associated with a skewed surface phenotype in comparison to CMV-specific cells (8). HIV-specific CD8+ T cells of untreated progressors express a predominantly CD45RA−/CCR7− surface phenotype, which has been termed “pre-terminally differentiated,” whereas most CMV-specific cells have a CD45RA+/CCR7− phenotype, thought to be consistent with “terminally differentiated” effector cells. Based upon these designations, it was suggested that HIV-specific cells are not fully mature effectors in the context of viremia and that this inability to completely differentiate may be the basis of poor immunologic control of HIV replication (8, 14). We sought to confirm whether there are phenotypic differences based on cell surface markers between HIV- and CMV-specific CD8+ T cells and whether these differences are influenced by the level of HIV or CMV antigen.
FIG. 4.
Virus-specific CD8+ T-cell phenotype. (A) CCR7/CD45RA (left panel) and CD45RA/RO (right panel) expression of total CD8+ T cells (gray) overlaid with CMV (blue) and HIV (red) tetramer+ cells in PBMCs is shown for patient 129, an ART recipient with HIV RNA levels of <50 copies/ml. (B) Distinct CD45RA/RO phenotype of two CMV specificities, HLA A2 CMV NLVPMVATV (NV9) and Cw06 CMV TRATKMQVI (TI9) tetramer+ cells, overlaid on total CD8+ T cells (gray), is shown for a viremic progressor (patient 131, left) and an LTNP (patient 38, right). (C) The percentages of CMV- (43 tetramer stainings, red) and HIV-specific (39 tetramer stainings, blue) CD8+ T cells expressing CCR7, CD27, CD45RA, and CD45RO are depicted. Solid black lines represent median values. Comparisons were made using the Wilcoxon two-sample test. (D) Using Boolean gating analysis, the frequencies of different surface phenotypic subsets of antigen-specific CD8+ T cells, which had been enumerated by intracellular cytokine detection following 6-hour stimulation with 15-mer peptide pools spanning CMV pp65 or the major HIV gene products, are shown for “on therapy” (on Rx) and “off therapy” (off Rx) time points in nine patients prior to and during an interruption of ART. Numbers on the stacked bars reflect the percent expression of CCR7−/CD27+/CD45RA− CD8+ T cells, the HIV-specific phenotypic subset with the greatest expansion during treatment interruption. (E) CD45RA/RO surface phenotype of CMV tetramer+ CD8+ T cells was examined in PBMC samples obtained from a single time point during acute primary CMV infection in one HIV-seronegative lung transplant recipient (CP1) and during latent infection in two additional patients (CP2 and CP3). (F) Samples from four additional lung transplant recipients (CP4 to CP7) were evaluated during the acute primary infection and then longitudinally during latency. Numbers in left corners of plots denote days after documented acute infection, and numbers in right corners are percentages of CD45RA+ CMV tetramer+ CD8+ T cells.
Figure 4A shows phenotypic data for treated patient 129. The HIV Gag263-272 KRWIILGLNK (KK10)-specific cells in this patient are CCR7−/CD45RA−RO+ whereas the CMV pp65495-503 NLVPMVATV (NV9)-specific cells are CCR7−/CD45RA+RO−, consistent with prior reports (8, 14). However, upon examination of multiple specificities in additional patients, considerable heterogeneity in surface phenotype within a given patient and between patients was observed. The phenotypic data for two patients, patient 131, a viremic progressor, and patient 38, an LTNP, who both recognized the A2-restricted CMV pp65 NV9 epitope (blue) and the C6-restricted CMV pp65 TI9 epitope (green), are shown in Fig. 4B. Although CD8+ T cells specific for the C6-restricted CMV TI9 epitope are predominantly CD45RA+RO− for patient 131, these cells from patient 38 are more heterogenous in their CD45RA/RO expression. Furthermore, although the A2-restricted CMV NV9 epitope and C6-restricted CMV TI9 epitope are both within CMV pp65, the CD8+ T cells specific for these epitopes vary in their expression of CD45RA/RO for both of these patients. This heterogeneity in the CD45RA/RO phenotype of CMV-specific CD8+ T cells is consistent with findings reported for healthy CMV carriers who are HIV seronegative (73, 74).
For a group of LTNP (nine patients, 15 tetramer stainings), viremic progressors (10 patients, 14 tetramer stainings), and progressors with HIV RNA levels suppressed to <50 copies/ml by ART (Rx<50; 12 patients, 14 tetramer stainings), summary data of the surface phenotype of CD8+ T cells stained with a panel of CMV tetramers (Table 3) are shown in Fig. 4C. Although there was variability between patients and between specificities from a given individual, the CMV-specific CD8+ T cells of all groups were predominantly CCR7−/CD27−/CD45RA+RO−, consistent with previous findings (8, 14). In comparisons made between the three patient groups, no significant differences were observed in the surface phenotype of CMV tetramer+ cells (data not shown) (54). Consistent with previous reports, HIV-specific CD8+ T cells were predominantly CCR7−/CD27+/CD45RA−RO+ in 12 LTNP (15 tetramer stainings), 13 viremic progressors (15 tetramer stainings), and nine Rx<50 (nine tetramer stainings). No significant differences were noted in the surface phenotype of HIV tetramer+ cells between the three patient groups (data not shown). Comparing CMV and HIV tetramer+ CD8+ T cells, overall differences in CD27 and CD45RA/RO expression were significant and similar to results obtained in comparisons between CMV and HIV tetramer+ cells within each patient group (Fig. 4C and data not shown).
We next explored the potential influence of differences in the level of viremia on surface phenotype. We have recently observed that the level of viremia has a dramatic effect on the expression of CCR7 on HIV-specific CD4+ T cells (66). Longitudinal changes in CD45RA/RO expression have been observed during acute and convalescent EBV and CMV infections (7, 16, 73, 74). Thus, it was possible that differences in CD45RA/RO expression between CMV- and HIV-specific cells were a consequence of the marked differences in the levels of viremia between these two infections. We therefore examined the surface phenotype of virus-specific CD8+ T cells in two longitudinal cohorts: HIV-infected patients during an interruption of therapy (Fig. 4D) and CMV-infected patients during acute CMV viremia following lung transplantation (Fig. 4F). In the first group, the phenotypes of the CD8+ T cells targeting the major gene products of HIV that expanded following an interruption of ART were predominantly CD45RA−/RO+, suggesting that HIV viremia skewed the surface phenotype to CD45RO+. In contrast, the frequencies and phenotype of CMV tetramer+ cells in these same patients were relatively stable. The surface phenotype of CMV pp65-specific CD8+ T cells was also studied cross-sectionally in three HIV-seronegative lung transplant recipients (CP1 to CP3, Fig. 4E) and longitudinally in four additional patients (CP4 to CP7, Fig. 4F). In these patients, there was a trend for most CMV-specific cells to be CD45RO+ during the acute primary infection and to subsequently revert to a CD45RA+ phenotype during latency. Taken together, these results are consistent with prior work in EBV and CMV in HIV-seronegative patients (7, 16, 73, 74) and suggest that differences in surface phenotype of HIV- and CMV-specific cells may be due not to differences in maturation and function but rather to levels of antigen. They also suggest that the designation of terminally differentiated effectors as CD45RA+ may not be correct, given that patients who ultimately control CMV viremia have a skewing toward a CD45RO+ phenotype during peak viremia.
Proliferation of a significant proportion of HIV-specific CD8+ T cells from progressors could not be restored with IL-2.
We then examined the proliferative capacity and IL-2 responsiveness of CMV-specific CD8+ T cells in comparison to HIV-specific cells. CFSE-labeled PBMCs derived from 10 LTNP and 18 progressors (11 viremic and seven Rx<50) were stimulated with the peptide pools spanning HIV Gag or CMV pp65 for 6 days and assessed for CFSE dilution. As shown previously, HIV Gag-specific CD8+ T-cell proliferation was significantly greater in LTNP than in progressors (median percent CFSElo cells, 46.4 [range, 2 to 64.3] versus 4.6 [0.5 to 41.4], respectively; P < 0.001; data not shown) (5, 27, 49). In contrast, CMV pp65-specific CD8+ T-cell proliferation values were similar between LTNP and progressors (median percent CFSElo cells, 17.4 [range, 0.2 to 75] versus 6.8 [range, 0.4 to 49.5], respectively; P = 0.5; data not shown). In untreated patients, pp65-specific CD8+ T-cell proliferation was not correlated with HIV RNA levels (R = −0.3, P = 0.5); however, Gag-specific CD8+ T-cell proliferation was inversely correlated with HIV RNA levels (R = −0.6, P = 0.01), as reported previously (5, 27, 49).
The role of IL-2 in the rescue of CMV- and HIV-specific CD8+ T-cell proliferation was then assessed. It has previously been shown that the percentage of CMV-specific CFSElo CD8+ T cells is greatly increased upon the addition of IL-2 in immunocompetent CMV-seropositive donors (69, 74). Recently, it has also been suggested that IL-2 reverses the proliferative defect seen in HIV-specific CD8+ T cells of progressors (41). However, this observation was made on the basis of increases in the percentages of CFSElo tetramer+ cells. Because cell division causes exponential increases in CFSElo cells, small changes in the number of cell divisions or the fraction of cells entering the cell cycle can produce very large changes in the percentages of CFSElo cells. To compare the effects of IL-2 on CMV- and HIV-specific CD8+ T cells, PBMCs were stimulated with HIV and/or CMV optimal epitopes in the presence or absence of IL-2 and stained with HIV and/or CMV tetramers on day 6. In preliminary experiments, virus-specific CD8+ T cells proliferated in response to optimal epitopes without significant bystander proliferation of nonspecific cells (see Fig. S3 in the supplemental material). To assess proliferation of CFSE-labeled tetramer+ cells in greater detail, proliferation analysis software was used to determine the changes in the average number of cell divisions that dividing cells had undergone (proliferation index) and the proportion of the original cell population entering cycle and dividing at least one time (percent divided; Fig. 5).
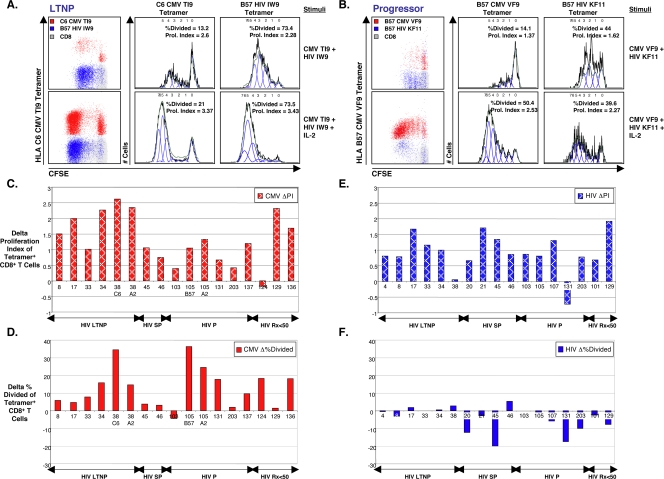
FIG. 5.
Effects of IL-2 on CMV- versus HIV-specific CD8+ T-cell proliferation. (A and B) Representative proliferation analysis of virus-specific CD8+ T cells is shown for one LTNP (A) (patient 33) and one progressor (B) (patient 105). Stimulation conditions are shown on the far right. With gating on CD8+ T cells (gray), dot plots (left columns) depict the proliferation of CMV (red) and HIV (blue) tetramer+ CD8+ T cells in response to the corresponding CMV and HIV optimal epitopes with (bottom rows) and without (top rows) IL-2 at 6 days for each patient. Proliferation analyses of gated CMV (middle columns) and HIV (right columns) tetramer+ CD8+ T cells are shown. Percent divided is defined as the percentage of cells of the original sample that divided. Proliferation index is the average number of divisions that divided cells had undergone. (C to F) Proliferation analyses of gated CMV (C and D) (red bars, n = 15 patients) and HIV (E and F) (blue bars, n = 17 patients) tetramer+ CD8+ T cells are shown. Delta proliferation index of CMV (C) or HIV (E) tetramer+ CD8+ T cells is the proliferation index when cells were stimulated with peptide and IL-2 minus the proliferation index when cells were stimulated with peptide alone. Delta percent divided of CMV (D) or HIV (F) tetramer+ CD8+ T cells is the percent divided when cells were stimulated with peptide and IL-2 minus the percent divided when cells were stimulated with peptide alone. Patient numbers are listed below each bar. Patients are subgrouped according to HIV disease status as LTNP (HIV LTNP), slow progressors (HIV SP), progressors (HIV P), and patients with antiretroviral-induced suppression of HIV RNA levels to <50 copies/ml (HIV Rx<50). The Wilcoxon signed-rank test was used to compare paired data.
Consistent with previous findings, IL-2 was able to induce large expansions of CMV-specific CD8+ T cells from both LTNP and progressors in a 6-day assay. In two representative patients, the percentage of CMV tetramer+ cells that entered the proliferating pool increased from 13.2% to 21% in the LTNP (Fig. 5A, middle column) and from 14.1% to 50.4% (Fig. 5B, middle column) in the progressor when stimulation conditions included IL-2. The average number of divisions of proliferating CMV tetramer+ cells increased from 2.6 to 3.37 in the LTNP (Fig. 5A, middle column) and from 1.37 to 2.53 in the progressor (Fig. 5B, middle column) with peptides plus IL-2 compared with peptides alone. Changes in these parameters were expressed as the delta proliferation index (proliferation index with IL-2 minus proliferation index without IL-2) and the delta percent divided (percent divided with IL-2 minus percent divided without IL-2). Summary data for 15 patients (17 CMV tetramer stainings) are shown in Fig. 5C and D. IL-2 induced significant increases in both the proliferation index (P < 0.001) and percent divided (P < 0.001) of CMV-specific CD8+ T cells, with a median delta proliferation of 1.2 divisions (range, −0.2 to 2.6 divisions; Fig. 5C) and the median delta percent divided of 9.5% (range, −4.4 to 36.6%; Fig. 5D).
The effects of IL-2 on HIV-specific CD8+ T-cell proliferation, measured with the same parameters, were also examined for the two representative patients (Fig. 5A and B, left and right columns) and summarized for 17 patients (17 tetramer stainings) in Fig. 5E and F. Consistent with previous reports, even in the absence of IL-2, most HIV-specific CD8+ T cells from HIV-infected LTNP were CFSElo at 6 days (Fig. 5A, left and right panels of top row). Similar to the effect of IL-2 on CMV-specific cells, the addition of IL-2 produced significant increases in the proliferation index of HIV-specific CD8+ T cells in the 17 patients (P < 0.001), with a median delta proliferation index of 0.86 divisions (range, −0.7 to 1.9 divisions; Fig. 5E). The positive net effect of IL-2 on the proliferation index of CMV-specific CD8+ T cells was similar to the positive net effect of IL-2 on the proliferation index of HIV-specific CD8+ T cells (medians, 1.2 versus 0.86, respectively, P = 0.2; Fig. 5C and E). However, unlike the case with CMV-specific CD8+ T cells, there was a striking lack of an effect of IL-2 on the percentage of HIV-specific cells recruited into the proliferating pool (median delta percent divided, −2.8 [range, −20.1 to 5.4]; P = 0.12; Fig. 5F). The minimal effect of IL-2 observed in the percent divided of HIV-specific CD8+ T cells of LTNP was not unexpected, considering that nearly all cells divided even in the absence of IL-2 (Fig. 5A and data not shown). The positive net effect of IL-2 on the percent divided of CMV-specific CD8+ T cells was significantly different from the absence of an effect of IL-2 on the percent divided of HIV-specific CD8+ T cells (medians, 9.5% versus −2.8%, respectively; P < 0.001; Fig. 5D and F).
In order to control for autocrine or paracrine production of IL-2 or other factors affecting proliferation, a similar analysis was performed including only data from 12 patients in which CMV- and HIV-specific CD8+ T-cell proliferation were studied in the same tube using CMV and HIV tetramers labeled with different fluorochromes (similar to results shown in Fig. 5A and B). Under these conditions, the net positive effect of IL-2 on the proliferation indices of both CMV- and HIV-specific cells was similar (medians, 1.1 [0.4 to 2.6] versus 0.9 [−0.7 to 1.9] divisions, respectively; P = 0.35; data not shown). In contrast, the effect of IL-2 on the percent divided was vastly different between CMV-specific and HIV-specific cells, with an extraordinary lack of an increase in the percentage of HIV-specific cells that divided (medians for percent divided, 5.2 [range, −4.4 to 36.3] versus −1.5 [range, −20.1 to 5.4], respectively; P < 0.01; data not shown), consistent with the results in Fig. 5D and F. In summary, although IL-2 induced significant increases in both the number of CMV-specific CD8+ T cells recruited into the cell cycle and the number of cell divisions that they underwent, the increases in CFSElo HIV-specific cells induced by exogenous IL-2 were completely attributable to increases in the number of divisions of cells already in the cell cycle. IL-2 did not recruit more HIV-specific CD8+ T cells to enter the cell cycle and the majority of HIV-specific cells of progressors did not divide despite 6 days of stimulation with peptide and IL-2.
DISCUSSION
The results of the present study provide insights regarding the HIV-specific CD8+ T-cell response through comparisons with responses to other chronic virus infections in coinfected individuals. Marked differences in immunodominance, surface phenotype, and IL-2 responsiveness were observed between HIV- and CMV- or HCV-specific CD8+ T cells. Although the vast majority of HIV-specific CD8+ T-cell responses of HLA B*57+ LTNP are B5701 restricted, this pattern was not observed in the CMV-specific responses of the same patients. Although some differences in surface phenotype were observed between HIV- and CMV-specific CD8+ T cells, these differences were not intrinsic to the response to these viruses but rather appeared to be secondary to the level of viremia. Lastly, HIV-specific CD8+ T cells were only poorly responsive to exogenous IL-2. Taken together, these results suggest that the extraordinary immunodominance of HLA B5701-restricted cells in LTNP and poor responsiveness to IL-2 in progressors are unique features of the HIV-specific CD8+ T-cell response.
The differences in immunodominance patterns between HIV- and CMV-specific cells may provide some insights regarding the immunologic control of HIV in B*5701+ LTNP. HLA B5701-restricted cells preferentially proliferate in vitro, expand in vivo during acute infection and vaccination, and persist in the blood during chronic infection (19, 27, 30, 49, 62). It is believed that the extraordinary immunodominance of B5701-restricted cells in LTNP is a functional association that suggests direct involvement of the B5701 molecule in immunologic control and not simply linkage disequilibrium between B*5701 and another gene responsible for immune protection. Because the percentage of HIV-infected patients who are B*5701+ progressors is similar to the frequency of B*5701 in the U.S. population, it was thought that immunologic control in LTNP may be mediated through a combination of B*5701 with another gene acting as a response modifier. Such a mechanism has been suggested in some work where a subset of Bw4 alleles (80I) in combination with KIR3DL1 is associated with slower progression to AIDS (47). However, these associations were made in cohorts that were predominantly progressors. We have been unable to confirm such an association in our LTNP in comparison with progressor cohorts when controlled for MHC (L. Shupert, personal communication). If a KIR or other response modifier operated in conjunction with B5701, we would expect to observe some immunodominant B5701-restricted CMV-specific responses in LTNP, given that the genome of CMV is approximately 20-fold larger than that of HIV-1. Similarly, one would expect to observe B5701-restricted CMV responses if induction of immunodominant responses was intrinsic to the B5701 molecule. However, within all of pp65, we observed only one response, and this response was found in only a single patient. A broader examination of total CMV-specific responses did not reveal immunodominance of B*5701. These results suggest that the immunodominance of B5701-restricted HIV-specific responses is less likely due to a response modifier or to properties intrinsic to the B5701 molecule and rather is specific to the interaction of HIV with B5701.
The results of the present study also demonstrate phenotypic differences between HIV- and CMV-specific cells and the effects of viremia on these phenotypes observed in the peripheral blood. We observed considerable heterogeneity in the surface phenotypic analyses of HIV- and CMV-specific CD8+ T cells. The surface phenotype for HIV- or CMV-specific cells was predominantly CCR7−. HIV-specific cells were predominantly CD27+/CD45RO+, and CMV-specific cells were predominantly CD27−/CD45RA+, consistent with prior results (4, 8, 14). However, these differences were not observed to be intrinsic to HIV- or CMV-specific cells but rather were a consequence of the level of viremia in patients undergoing a treatment interruption or patients with CMV viremia. These results are very consistent with prior studies of EBV and CMV infections where MHC tetramer+ cells were observed to be CD45RO+ during peak viremia at a time when they also exhibited maximal cytotoxicity and CD45RA+ during convalescence (7, 16, 73, 74). Similarly, proliferating HIV-specific cells are almost entirely CD45RO+ after 6 days of reexposure to antigen (49, 60, 74). It should be noted that designation of CD45RA+ cells as effectors was made using redirected cytotoxic T-lymphocyte assays in which most of the CD8+ T cells had not been recently exposed to their cognate antigen and were more likely to acquire this phenotype. It is likely that surface phenotypes would be more heterogeneous if they were determined using assays that measure the antigen-specific response during viremia. We, and others, have observed that surface phenotypes of CD4+ or CD8+ T cells are not reliably associated with a particular effector function (4, 14, 24, 55, 66, 67, 73, 74). The matter is further complicated in some studies in humans where the timing of exposure to antigen and the trafficking pattern of antigen-specific cells are uncertain.
We also observed dramatic differences in the abilities of HIV-and CMV-specific CD8+ T cells to proliferate in response to IL-2. In one prior report, an increase in CFSElo HIV-specific cells was observed following incubation with either IL-2 or autologous HIV-specific CD4+ T cells (41). Based upon this observation, it was suggested that diminished proliferative capacity of HIV-specific CD8+ T cells is due to the effects of diminished T-cell “help.” However, it should be noted that changes in the percentage of CFSElo cells in such studies are likely a result of the exponential expansion of extremely small numbers of dividing cells. If tetramer-positive cells are gated, only 0.5% dividing cells are required to produce 32% CFSElo cells in six twofold divisions. In this case, greater than 99.5% of tetramer+ cells have not divided. These numbers are consistent with the relationship between the frequency and proliferation of HIV-specific CD4+ T cells (28, 66). In addition, prior studies have not determined whether the effects of IL-2 on HIV-specific CD8+ T cells were to increase the number of cells dividing or to simply increase the number of divisions that cells in the cell cycle underwent. The results of the present study are clearly consistent with the latter. HIV-specific CD8+ T cells were particularly refractory to increases in the percent divided in response to IL-2. This result is consistent with our prior work, where conditions that cause large increases in secreted IL-2 in vitro, such as addition of autologous CD4+ T-cell blasts or stimulating cells with anti-CD3/CD28, did not restore the proliferative capacity of HIV-specific CD8+ T cells of progressors to the level observed in LTNP (49). In addition, these results are also consistent with prior work in which administration of IL-2 in vivo to progressor patients neither restores in vitro CD8+ T-cell proliferation nor reduces viral load (1, 10). We previously found no significant differences in the percent expression of the IL-2 receptor α chain CD25 on unstimulated CMV or HIV tetramer+ CD8+ T cells between LTNP and progressors but did demonstrate that proliferating CFSElo CMV and HIV tetramer+ CD8+ T cells were CD25hi in both patient groups (S. A. Migueles, unpublished observations). How these and other factors might account for these differences in virus-specific CD8+ T-cell proliferation and IL-2 responsiveness are presently unknown and are the focus of intensive investigation.
It should be noted that some of the differences in frequency, surface phenotype, proliferative capacity, or IL-2 responsiveness among cells specific for HIV, HCV, or CMV observed in the present study may be attributable to differences in virus biology. In individuals with chronic HCV infection, HCV-specific CD4+ and CD8+ T-cell responses are barely detectable in the peripheral blood, even though vigorous and broadly directed responses have been associated with spontaneous recovery from acute infection and have been implicated in clearance after interferon and ribavirin therapy for chronic infection (12, 37, 40, 64, 65, 70). More recently, T-cell responses were confirmed to mediate protective immunity against HCV by in vivo depletion studies in the chimpanzee model (20, 61). In our small cohort of HCV/HIV-coinfected patients, the HCV-specific CD8+ T-cell responses were infrequently detected and of very low magnitude regardless of the level of immunologic control of HCV or HIV, which is consistent with one previous study (38), but not others (3, 68), of coinfected patients. This discordance in measurable HCV- and HIV-specific responses in the peripheral blood of coinfected patients is somewhat surprising, since high levels of virus replication characterize both of these chronic infections. Diminished frequencies of HCV-specific CD8+ T cells have been ascribed to deficient priming by antigen-presenting liver sinusoidal endothelial cells (43), hepatic compartmentalization (22), suppression by HCV proteins or regulatory T cells (2, 36), impaired proliferation (70), or exhaustion (38). In addition to low frequencies, impaired effector function of HCV-specific CD8+ T cells has been observed (38, 70). Whether these features simply reflect a unique pattern of virus-host interaction or also account for the immune dysfunction responsible for HCV persistence remains to be determined.
Because HCV-specific T-cell frequencies were too low to permit a meaningful evaluation, we relied on comparisons with CMV-specific responses as other investigators have done, realizing that infections with CMV or HIV are also distinct for a number of reasons. Studies in humans and animal models have demonstrated a central role for CD4+ and CD8+ T cells in controlling viral replication and preventing clinically significant disease (56). Although the levels of viral replication are considerably lower in chronic CMV infection than in chronic HIV infection, comparable frequencies of CMV-specific CD8+ T cells are readily detectable in the periphery. Unique target cell tropisms, which may impact virus replication kinetics and the efficiency of antigen presentation, might predispose to the accumulation of immune responses of increased magnitude and breadth in the peripheral blood disproportionate to the level of viremia. Furthermore, heightened immune responses in the context of an intermittently replicating virus might relate to the overall size and tremendous complexity of CMV, which possesses a double-strand DNA genome encoding approximately 200 open reading frames for proteins greater than 80 amino acids in length (63). Other inherent features potentially distinguishing CMV from HIV infection may include timing of antigen encounter, cytokine milieu during the priming event, anatomic sites of virus turnover, activation state of effectors and APC, and recirculation pathways of virus-specific cells (4, 14, 15). Where possible, we made an effort to control for some of these differences in our assays by standardizing antigen-presenting systems, using comparable peptides and target cell types, and analyzing the results in the context of the size of viral gene products and the level of viremia. Regardless, these factors might lead to distinct phenotypic and functional profiles of virus-specific CD8+ T cells without impacting their capacity to control virus replication.
The results of the present study demonstrate several phenotypic and functional features of the HIV-specific CD8+ T-cell response that are not shared with responses to other viruses. A better understanding of the factors that direct the immunodominance of HLA B5701 in LTNP and signals that might induce proliferation of HIV-specific CD8+ T cells in progressors may have important implications for prophylactic and therapeutic vaccines for HIV.
Supplementary Material
Acknowledgments
This research was supported (in part) by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 7 January 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrams, D. I., J. D. Bebchuk, E. T. Denning, R. T. Davey, L. Fox, H. C. Lane, J. Sampson, R. Verheggen, D. Zeh, and N. P. Markowitz. 2002. Randomized, open-label study of the impact of two doses of subcutaneous recombinant interleukin-2 on viral burden in patients with HIV-1 infection and CD4+ cell counts of > or = 300/mm3: CPCRA 059. J. Acquir. Immune Defic. Syndr. 29221-231. [DOI] [PubMed] [Google Scholar]
- 2.Accapezzato, D., V. Francavilla, M. Paroli, M. Casciaro, L. V. Chircu, A. Cividini, S. Abrignani, M. U. Mondelli, and V. Barnaba. 2004. Hepatic expansion of a virus-specific regulatory CD8(+) T cell population in chronic hepatitis C virus infection. J. Clin. Investig. 113963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alatrakchi, N., V. Di Martino, V. Thibault, and B. Autran. 2002. Strong CD4 Th1 responses to HIV and hepatitis C virus in HIV-infected long-term non-progressors co-infected with hepatitis C virus. AIDS 16713-717. [DOI] [PubMed] [Google Scholar]
- 4.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8379-385. [DOI] [PubMed] [Google Scholar]
- 5.Arrode, G., J. S. Finke, H. Zebroski, F. P. Siegal, and R. M. Steinman. 2005. CD8+ T cells from most HIV-1-infected patients, even when challenged with mature dendritic cells, lack functional recall memory to HIV gag but not other viruses. Eur. J. Immunol. 35159-170. [DOI] [PubMed] [Google Scholar]
- 6.Bunce, M., C. M. O'Neill, M. C. Barnardo, P. Krausa, M. J. Browning, P. J. Morris, and K. I. Welsh. 1995. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 46355-367. [DOI] [PubMed] [Google Scholar]
- 7.Callan, M. F., L. Tan, N. Annels, G. S. Ogg, J. D. Wilson, C. A. O'Callaghan, N. Steven, A. J. McMichael, and A. B. Rickinson. 1998. Direct visualization of antigen-specific CD8+ T cells during the primary immune response to Epstein-Barr virus In vivo. J. Exp. Med. 1871395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champagne, P., G. S. Ogg, A. S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G. P. Rizzardi, S. Fleury, M. Lipp, R. Forster, S. Rowland-Jones, R. P. Sekaly, A. J. McMichael, and G. Pantaleo. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410106-111. [DOI] [PubMed] [Google Scholar]
- 9.Cortez, K. J., S. H. Fischer, G. A. Fahle, L. B. Calhoun, R. W. Childs, A. J. Barrett, and J. E. Bennett. 2003. Clinical trial of quantitative real-time polymerase chain reaction for detection of cytomegalovirus in peripheral blood of allogeneic hematopoietic stem-cell transplant recipients. J. Infect. Dis. 188967-972. [DOI] [PubMed] [Google Scholar]
- 10.Davey, R. T., Jr., R. L. Murphy, F. M. Graziano, S. L. Boswell, A. T. Pavia, M. Cancio, J. P. Nadler, D. G. Chaitt, R. L. Dewar, D. K. Sahner, A. M. Duliege, W. B. Capra, W. P. Leong, M. A. Giedlin, H. C. Lane, and J. O. Kahn. 2000. Immunologic and virologic effects of subcutaneous interleukin 2 in combination with antiretroviral therapy: a randomized controlled trial. JAMA 284183-189. [DOI] [PubMed] [Google Scholar]
- 11.Diamond, D. J., J. York, J. Y. Sun, C. L. Wright, and S. J. Forman. 1997. Development of a candidate HLA A*0201 restricted peptide-based vaccine against human cytomegalovirus infection. Blood 901751-1767. [PubMed] [Google Scholar]
- 12.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, E. A. Wierenga, T. Santantonio, M. C. Jung, D. Eichenlaub, and G. R. Pape. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 3461006-1007. [DOI] [PubMed] [Google Scholar]
- 13.Elkington, R., S. Walker, T. Crough, M. Menzies, J. Tellam, M. Bharadwaj, and R. Khanna. 2003. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 775226-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellefsen, K., A. Harari, P. Champagne, P. A. Bart, R. P. Sekaly, and G. Pantaleo. 2002. Distribution and functional analysis of memory antiviral CD8 T cell responses in HIV-1 and cytomegalovirus infections. Eur. J. Immunol. 323756-3764. [DOI] [PubMed] [Google Scholar]
- 15.Emery, V. C. 2001. Investigation of CMV disease in immunocompromised patients. J. Clin. Pathol. 5484-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faint, J. M., N. E. Annels, S. J. Curnow, P. Shields, D. Pilling, A. D. Hislop, L. Wu, A. N. Akbar, C. D. Buckley, P. A. Moss, D. H. Adams, A. B. Rickinson, and M. Salmon. 2001. Memory T cells constitute a subset of the human CD8+CD45RA+ pool with distinct phenotypic and migratory characteristics. J. Immunol. 167212-220. [DOI] [PubMed] [Google Scholar]
- 17.Gavin, M. A., M. J. Gilbert, S. R. Riddell, P. D. Greenberg, and M. J. Bevan. 1993. Alkali hydrolysis of recombinant proteins allows for the rapid identification of class I MHC-restricted CTL epitopes. J. Immunol. 1513971-3980. [PubMed] [Google Scholar]
- 18.Gerna, G., E. Percivalle, D. Lilleri, L. Lozza, C. Fornara, G. Hahn, F. Baldanti, and M. G. Revello. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86275-284. [DOI] [PubMed] [Google Scholar]
- 19.Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retrovir. 121691-1698. [DOI] [PubMed] [Google Scholar]
- 20.Grakoui, A., N. H. Shoukry, D. J. Woollard, J. H. Han, H. L. Hanson, J. Ghrayeb, K. K. Murthy, C. M. Rice, and C. M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302659-662. [DOI] [PubMed] [Google Scholar]
- 21.Grigoleit, U., S. Riegler, H. Einsele, K. Laib Sampaio, G. Jahn, H. Hebart, P. Brossart, F. Frank, and C. Sinzger. 2002. Human cytomegalovirus induces a direct inhibitory effect on antigen presentation by monocyte-derived immature dendritic cells. Br. J. Haematol. 119189-198. [DOI] [PubMed] [Google Scholar]
- 22.He, X. S., B. Rehermann, F. X. Lopez-Labrador, J. Boisvert, R. Cheung, J. Mumm, H. Wedemeyer, M. Berenguer, T. L. Wright, M. M. Davis, and H. B. Greenberg. 1999. Quantitative analysis of hepatitis C virus-specific CD8(+) T cells in peripheral blood and liver using peptide-MHC tetramers. Proc. Natl. Acad. Sci. USA 965692-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 777563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hislop, A. D., N. H. Gudgeon, M. F. Callan, C. Fazou, H. Hasegawa, M. Salmon, and A. B. Rickinson. 2001. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. J. Immunol. 1672019-2029. [DOI] [PubMed] [Google Scholar]
- 25.Holl, V., M. Peressin, S. Schmidt, T. Decoville, S. Zolla-Pazner, A. M. Aubertin, and C. Moog. 2006. Efficient inhibition of HIV-1 replication in human immature monocyte-derived dendritic cells by purified anti-HIV-1 IgG without induction of maturation. Blood 1074466-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, K., B. Fowlkes, I. Schmid, and J. Giorgi. 1995. Preparation of cells and reagents for flow cytometry, p. 5.3.1-5.3.23. In J. Coligan, A. Kruisbeek, D. Margulies, E. Sheevac, and W. Strober (ed.), Current protocols in immunology, vol. 1. Green Publishing Associates, New York, NY. [DOI] [PubMed] [Google Scholar]
- 27.Horton, H., I. Frank, R. Baydo, E. Jalbert, J. Penn, S. Wilson, J. P. McNevin, M. D. McSweyn, D. Lee, Y. Huang, S. C. De Rosa, and M. J. McElrath. 2006. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J. Immunol. 1777406-7415. [DOI] [PubMed] [Google Scholar]
- 28.Iyasere, C., J. C. Tilton, A. J. Johnson, S. Younes, B. Yassine-Diab, R. P. Sekaly, W. W. Kwok, S. A. Migueles, A. C. Laborico, W. L. Shupert, C. W. Hallahan, R. T. Davey, Jr., M. Dybul, S. Vogel, J. Metcalf, and M. Connors. 2003. Diminished proliferation of human immunodeficiency virus-specific CD4+ T cells is associated with diminished interleukin-2 (IL-2) production and is recovered by exogenous IL-2. J. Virol. 7710900-10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlsson, A. C., J. N. Martin, S. R. Younger, B. M. Bredt, L. Epling, R. Ronquillo, A. Varma, S. G. Deeks, J. M. McCune, D. F. Nixon, and E. Sinclair. 2003. Comparison of the ELISPOT and cytokine flow cytometry assays for the enumeration of antigen-specific T cells. J. Immunol. Methods 283141-153. [DOI] [PubMed] [Google Scholar]
- 30.Kaslow, R. A., C. Rivers, J. Tang, T. J. Bender, P. A. Goepfert, R. El Habib, K. Weinhold, and M. J. Mulligan. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 758681-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 1851709-1716. [DOI] [PubMed] [Google Scholar]
- 32.Kern, F., I. P. Surel, N. Faulhaber, C. Frommel, J. Schneider-Mergener, C. Schonemann, P. Reinke, and H. D. Volk. 1999. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J. Virol. 738179-8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan, N., M. Cobbold, R. Keenan, and P. A. Moss. 2002. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J. Infect. Dis. 1851025-1034. [DOI] [PubMed] [Google Scholar]
- 34.Kiecker, F., M. Streitz, B. Ay, G. Cherepnev, H. D. Volk, R. Volkmer-Engert, and F. Kern. 2004. Analysis of antigen-specific T-cell responses with synthetic peptides—what kind of peptide for which purpose? Hum. Immunol. 65523-536. [DOI] [PubMed] [Google Scholar]
- 35.Kondo, E., Y. Akatsuka, K. Kuzushima, K. Tsujimura, S. Asakura, K. Tajima, Y. Kagami, Y. Kodera, M. Tanimoto, Y. Morishima, and T. Takahashi. 2004. Identification of novel CTL epitopes of CMV-pp65 presented by a variety of HLA alleles. Blood 103630-638. [DOI] [PubMed] [Google Scholar]
- 36.Large, M. K., D. J. Kittlesen, and Y. S. Hahn. 1999. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J. Immunol. 162931-938. [PubMed] [Google Scholar]
- 37.Lasarte, J. J., M. Garcia-Granero, A. Lopez, N. Casares, N. Garcia, M. P. Civeira, F. Borras-Cuesta, and J. Prieto. 1998. Cellular immunity to hepatitis C virus core protein and the response to interferon in patients with chronic hepatitis C. Hepatology 28815-822. [DOI] [PubMed] [Google Scholar]
- 38.Lauer, G. M., T. N. Nguyen, C. L. Day, G. K. Robbins, T. Flynn, K. McGowan, E. S. Rosenberg, M. Lucas, P. Klenerman, R. T. Chung, and B. D. Walker. 2002. Human immunodeficiency virus type 1-hepatitis C virus coinfection: intraindividual comparison of cellular immune responses against two persistent viruses. J. Virol. 762817-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lauer, G. M., K. Ouchi, R. T. Chung, T. N. Nguyen, C. L. Day, D. R. Purkis, M. Reiser, A. Y. Kim, M. Lucas, P. Klenerman, and B. D. Walker. 2002. Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J. Virol. 766104-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lechner, F., D. K. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 1911499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lichterfeld, M., D. E. Kaufmann, X. G. Yu, S. K. Mui, M. M. Addo, M. N. Johnston, D. Cohen, G. K. Robbins, E. Pae, G. Alter, A. Wurcel, D. Stone, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lichterfeld, M., G. Pantaleo, and M. Altfeld. 2005. Loss of HIV-1-specific T cell proliferation in chronic HIV-1 infection: cause or consequence of viral replication? AIDS 191225-1227. [DOI] [PubMed] [Google Scholar]
- 43.Limmer, A., J. Ohl, C. Kurts, H. G. Ljunggren, Y. Reiss, M. Groettrup, F. Momburg, B. Arnold, and P. A. Knolle. 2000. Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat. Med. 61348-1354. [DOI] [PubMed] [Google Scholar]
- 44.Longmate, J., J. York, C. La Rosa, R. Krishnan, M. Zhang, D. Senitzer, and D. J. Diamond. 2001. Population coverage by HLA class-I restricted cytotoxic T-lymphocyte epitopes. Immunogenetics 52165-173. [DOI] [PubMed] [Google Scholar]
- 45.Lore, K., A. Smed-Sorensen, J. Vasudevan, J. R. Mascola, and R. A. Koup. 2005. Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J. Exp. Med. 2012023-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandelboim, O., H. T. Reyburn, M. Vales-Gomez, L. Pazmany, M. Colonna, G. Borsellino, and J. L. Strominger. 1996. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 184913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31429-434. [DOI] [PubMed] [Google Scholar]
- 48.Migueles, S. A., and M. Connors. 2001. Frequency and function of HIV-specific CD8(+) T cells. Immunol. Lett. 79141-150. [DOI] [PubMed] [Google Scholar]
- 49.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 31061-1068. [DOI] [PubMed] [Google Scholar]
- 50.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Migueles, S. A., J. C. Tilton, and M. Connors. 2004. Advances in understanding immunologic control of HIV infection. Curr. HIV/AIDS Rep. 112-17. [DOI] [PubMed] [Google Scholar]
- 52.Moutaftsi, M., A. M. Mehl, L. K. Borysiewicz, and Z. Tabi. 2002. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 992913-2921. [DOI] [PubMed] [Google Scholar]
- 53.Nastke, M. D., L. Herrgen, S. Walter, D. Wernet, H. G. Rammensee, and S. Stevanovic. 2005. Major contribution of codominant CD8 and CD4 T cell epitopes to the human cytomegalovirus-specific T cell repertoire. Cell. Mol. Life Sci. 6277-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papagno, L., V. Appay, J. Sutton, T. Rostron, G. M. Gillespie, G. S. Ogg, A. King, A. T. Makadzanhge, A. Waters, C. Balotta, A. Vyakarnam, P. J. Easterbrook, and S. L. Rowland-Jones. 2002. Comparison between HIV- and CMV-specific T cell responses in long-term HIV infected donors. Clin. Exp. Immunol. 130509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ravkov, E. V., C. M. Myrick, and J. D. Altman. 2003. Immediate early effector functions of virus-specific CD8+CCR7+ memory cells in humans defined by HLA and CC chemokine ligand 19 tetramers. J. Immunol. 1702461-2468. [DOI] [PubMed] [Google Scholar]
- 56.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2831-844. [DOI] [PubMed] [Google Scholar]
- 57.Retiere, C., V. Prod'homme, B. M. Imbert-Marcille, M. Bonneville, H. Vie, and M. M. Hallet. 2000. Generation of cytomegalovirus-specific human T-lymphocyte clones by using autologous B-lymphoblastoid cells with stable expression of pp65 or IE1 proteins: a tool to study the fine specificity of the antiviral response. J. Virol. 743948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riegler, S., H. Hebart, H. Einsele, P. Brossart, G. Jahn, and C. Sinzger. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81393-399. [DOI] [PubMed] [Google Scholar]
- 59.Sallusto, F., and A. Lanzavecchia. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1791109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandberg, J. K., N. M. Fast, and D. F. Nixon. 2001. Functional heterogeneity of cytokines and cytolytic effector molecules in human CD8+ T lymphocytes. J. Immunol. 167181-187. [DOI] [PubMed] [Google Scholar]
- 61.Shoukry, N. H., A. Grakoui, M. Houghton, D. Y. Chien, J. Ghrayeb, K. A. Reimann, and C. M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 1971645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Streeck, H., M. Lichterfeld, G. Alter, A. Meier, N. Teigen, B. Yassine-Diab, H. K. Sidhu, S. Little, A. Kelleher, J. P. Routy, E. S. Rosenberg, R. P. Sekaly, B. D. Walker, and M. Altfeld. 2007. Recognition of a defined region within p24 Gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 817725-7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sylwester, A. W., B. L. Mitchell, J. B. Edgar, C. Taormina, C. Pelte, F. Ruchti, P. R. Sleath, K. H. Grabstein, N. A. Hosken, F. Kern, J. A. Nelson, and L. J. Picker. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6578-582. [DOI] [PubMed] [Google Scholar]
- 65.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 1941395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tilton, J. C., M. R. Luskin, A. J. Johnson, M. Manion, C. W. Hallahan, J. A. Metcalf, M. McLaughlin, R. T. Davey, Jr., and M. Connors. 2007. Changes in paracrine interleukin-2 requirement, CCR7 expression, frequency, and cytokine secretion of human immunodeficiency virus-specific CD4+ T cells are a consequence of antigen load. J. Virol. 812713-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unsoeld, H., S. Krautwald, D. Voehringer, U. Kunzendorf, and H. Pircher. 2002. Cutting edge: CCR7+ and CCR7- memory T cells do not differ in immediate effector cell function. J. Immunol. 169638-641. [DOI] [PubMed] [Google Scholar]
- 68.Valdez, H., N. L. Carlson, A. B. Post, R. Asaad, P. S. Heeger, M. M. Lederman, P. V. Lehmann, and D. D. Anthony. 2002. HIV long-term non-progressors maintain brisk CD8 T cell responses to other viral antigens. AIDS 161113-1118. [DOI] [PubMed] [Google Scholar]
- 69.van Leeuwen, E. M., L. E. Gamadia, P. A. Baars, E. B. Remmerswaal, I. J. ten Berge, and R. A. van Lier. 2002. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. J. Immunol. 1695838-5843. [DOI] [PubMed] [Google Scholar]
- 70.Wedemeyer, H., X. S. He, M. Nascimbeni, A. R. Davis, H. B. Greenberg, J. H. Hoofnagle, T. J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 1693447-3458. [DOI] [PubMed] [Google Scholar]
- 71.Weekes, M. P., M. R. Wills, K. Mynard, A. J. Carmichael, and J. G. Sissons. 1999. The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J. Virol. 732099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wills, M. R., A. J. Carmichael, K. Mynard, X. Jin, M. P. Weekes, B. Plachter, and J. G. Sissons. 1996. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J. Virol. 707569-7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wills, M. R., A. J. Carmichael, M. P. Weekes, K. Mynard, G. Okecha, R. Hicks, and J. G. Sissons. 1999. Human virus-specific CD8+ CTL clones revert from CD45ROhigh to CD45RAhigh in vivo: CD45RAhighCD8+ T cells comprise both naive and memory cells. J. Immunol. 1627080-7087. [PubMed] [Google Scholar]
- 74.Wills, M. R., G. Okecha, M. P. Weekes, M. K. Gandhi, P. J. Sissons, and A. J. Carmichael. 2002. Identification of naive or antigen-experienced human CD8(+) T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8(+) T cell response. J. Immunol. 1685455-5464. [DOI] [PubMed] [Google Scholar]
- 75.Zimmerli, S. C., A. Harari, C. Cellerai, F. Vallelian, P. A. Bart, and G. Pantaleo. 2005. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support C D4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. USA 1027239-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.