Abstract
The HA2 glycopolypeptide (gp) is highly conserved in all influenza A virus strains, and it is known to play a major role in the fusion of the virus with the endosomal membrane in host cells during the course of viral infection. Vaccines and therapeutics targeting this HA2 gp could induce efficient broad-spectrum immunity against influenza A virus infections. So far, there have been no studies on the possible therapeutic effects of monoclonal antibodies (MAbs), specifically against the fusion peptide of hemagglutinin (HA), upon lethal infections with highly pathogenic avian influenza (HPAI) H5N1 virus. We have identified MAb 1C9, which binds to GLFGAIAGF, a part of the fusion peptide of the HA2 gp. We evaluated the efficacy of MAb 1C9 as a therapy for influenza A virus infections. This MAb, which inhibited cell fusion in vitro when administered passively, protected 100% of mice from challenge with five 50% mouse lethal doses of HPAI H5N1 influenza A viruses from two different clades. Furthermore, it caused earlier clearance of the virus from the lung. The influenza virus load was assessed in lung samples from mice challenged after pretreatment with MAb 1C9 (24 h prior to challenge) and from mice receiving early treatment (24 h after challenge). The study shows that MAb 1C9, which is specific to the antigenically conserved fusion peptide of HA2, can contribute to the cross-clade protection of mice infected with H5N1 virus and mediate more effective recovery from infection.
Highly pathogenic avian influenza (HPAI) virus H5N1 strains are currently causing major morbidity and mortality in poultry populations across Asia, Europe, and Africa and have caused 385 confirmed human infections, with a fatality rate of 63.11% (37, 39). Preventive and therapeutic measures against circulating H5N1 strains have received a lot of interest and effort globally to prevent another pandemic outbreak. Influenza A virus poses a challenge because it rapidly alters its appearance to the immune system by antigenic drift (mutating) and antigenic shift (exchanging its components) (5). The current strategies to combat influenza include vaccination and antiviral drug treatment, with vaccination being the preferred option. The annual influenza vaccine aims to stimulate the generation of anti-hemagglutinin (anti-HA) neutralizing antibodies, which confer protection against homologous strains. Current vaccines have met with various degrees of success (31). The facts that these strategies target the highly variable HA determinant and that predicting the major HA types that pose the next epidemic threat is difficult are significant limitations to the current antiviral strategy. In the absence of an effective vaccine, therapy is the mainstay of control of influenza virus infection.
Therefore, therapeutic measures against influenza will play a major role in case a pandemic arises due to H5N1 strains. Currently licensed antiviral drugs include the M2 ion-channel inhibitors (rimantidine and amantidine) and the neuraminidase inhibitors (oseltamivir and zanamivir). The H5N1 viruses are known to be resistant to the M2 ion-channel inhibitors (2, 3). Newer strains of H5N1 viruses are being isolated which are also resistant to the neuraminidase inhibitors (oseltamivir and zanamivir) (5, 17). The neuraminidase inhibitors also require high doses and prolonged treatment (5, 40), increasing the likelihood of unwanted side effects. Hence, alternative strategies for treatment of influenza are warranted.
Recently, passive immunotherapy using monoclonal antibodies (MAbs) has been viewed as a viable option for treatment (26). The HA gene is the most variable gene of the influenza virus and also the most promising target for generating antibodies. It is synthesized as a precursor polypeptide, HA0, which is posttranslationally cleaved to two polypeptides, HA1 and HA2, linked by a disulfide bond. MAbs against the HA1 glycopolypeptide (gp) are known to neutralize the infectivity of the virus and hence provide good protection against infection (12). However, they are less efficient against heterologous or mutant strains, which are continuously arising due to antigenic shift and, to an extent, drift. Recent strategies for alternative therapy explore the more conserved epitopes of the influenza virus antigens (18, 33), which not only have the potential to stimulate a protective immune response but are also conserved among different subtypes, so as to offer protection against a broader range of viruses.
The HA2 polypeptide represents a highly conserved region of HA across influenza A virus strains. The HA2 gp is responsible for the fusion of the virus and the host endosomal membrane during the entry of the virus into the cell (16). Previously, anti-HA MAbs that lacked HA inhibition activity were studied and were found to reduce the infectivity of non-H5 influenza virus subtypes by inhibition of fusion during viral replication (14). They are known to block fusion of the virus to the cell membrane at the postbinding and prefusion stage, thereby inhibiting viral replication. Furthermore, in vivo studies show that anti-HA2 MAbs that exhibit fusion inhibition activity contribute to protection and recovery from H3N2 influenza A virus infection (8). It is interesting that although the HA2 gp is generally conserved, the fusion peptide represents the most conserved region of the HA protein. So far, there have been no studies on the possible therapeutic effects of MAbs, specifically against the fusion peptide of HA, on lethal HPAI H5N1 infections.
Previous studies have suggested that HA2 could contain a potential epitope responsible for the induction of antibody-mediated protective immunity (9). In the present study, a panel of MAbs against HA2 gp was characterized for their respective epitopes by epitope mapping. The therapeutic and prophylactic efficacies of these MAbs were evaluated in mice challenged with HPAI H5N1 virus infection.
MATERIALS AND METHODS
Cells and viruses.
Chinese hamster ovary (CHO-K1) cells and Madin-Darby canine kidney (MDCK) cells were obtained from American Type Culture Collection. They were cultured in Ham's F12-K medium and Dulbecco's minimal essential medium, respectively, both supplemented with 10% fetal bovine serum, 100 mg/ml streptomycin, and 100 units penicillin, and were maintained at 37°C in 5% CO2. Avian influenza virus H5N1 (A/goose/Guangdong/97) was inactivated with betapropiolactone (11) and used for RNA extraction to amplify the HA0 gene. The human HPAI H5N1 virus A/Indonesia/TLL013/06 was obtained from the Ministry of Health, Indonesia. The clade 1.0 virus A/Vietnam/1203/2004 (H5N1) was rescued by reverse genetics (36). Briefly, the HA and NA genes were synthesized by GenScript, based on the sequences from the NCBI influenza virus database. The synthetic HA and NA genes were cloned into a dual-promoter plasmid for influenza A virus reverse genetics. Dual-promoter plasmids were obtained from the Centers for Disease Control and Prevention, Atlanta, GA. The reassortant virus was rescued by transfecting plasmids containing HA and NA together with the remaining six gene plasmids derived from A/Puerto Rico/8/34 (H1N1) into a coculture of 293T and MDCK cells, using Lipofectamine 2000 (Invitrogen Corp). At 72 h posttransfection, culture medium from the transfected cells was inoculated into 11-day-old embryonated chicken eggs or MDCK cells. The HA and NA genes of reassortant viruses were sequenced to confirm the presence of the introduced HA and NA genes and the absence of mutations.
The viruses were propagated in the allantoic cavities of 11-day-old chicken embryos, and the allantoic fluid was harvested from the eggs after 48 h. Virus titers were determined using standard hemagglutination assays (38). The virus was then clarified by centrifugation at 20,000 × g for 20 min at 4°C and stored at −80°C. All experiments with live viruses were performed in a biosafety level 3 containment laboratory, and all animal experiments were carried out in an animal biosafety level 3 containment laboratory in compliance with CDC/NIH and WHO recommendations (20, 35).
Production of recombinant HA proteins.
The total RNA was extracted from H5N1 (A/goose/Guangdong/97) by using a commercial guanidium-phenol solution (Trizol; Invitrogen). The HA gene and the HA2 gene were amplified from the cDNA and cloned into pQE-30 vector (Qiagen, Germany), using standard cloning techniques for expression in bacteria. The clones were transformed into Escherichia coli M15/pREP4 competent cells for protein expression. The transformed E. coli M15 cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.5 to 0.6 in Luria-Bertani (LB) medium containing ampicillin (100 μg/ml), and protein expression was induced by the addition of 1 mmol/liter IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h with shaking. The histidine-tagged protein was purified on a Ni-nitrilotriacetic acid column (Qiagen, Germany), using standard protocols.
MAb production.
BALB/c mice were immunized twice subcutaneously at regular intervals of 2 weeks with 25 μg of recombinant H5N1 HA0 antigen in 0.1 ml of phosphate-buffered saline (PBS), which was emulsified with an equal volume of adjuvant (SEPPIC, France). Thereafter, mice were boosted intravenously with 25 μg of recombinant antigen 3 days before the fusion of splenocytes with SP2/0 cells (41). Hybridoma culture supernatants were screened by immunofluorescence assays (IFA) as described below. Hybridomas that produced specific antibody were cloned by limiting dilution, expanded, and further subcultured. The hybridoma cultures were harvested, and cell debris was removed by centrifugation at 400 × g for 10 min. The supernatant was collected and stored at −20°C. Positive clones were checked for isotype by using a 1-minute isotyping kit (Amersham Bioscience, England) as described in the manufacturer's protocol. The MAbs were purified using protein A Sepharose beads (Millipore). The purity of the antibodies was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis. The MAbs were then tested by neutralization assay and standard HI assay as described below.
IFA.
Sf-9 and MDCK cells cultured in 96-well plates were infected with recombinant baculovirus harboring the truncated H5N1 HA1 and HA0 genes or with avian influenza virus H5N1 Indonesian strains, namely, H5N2 and H5N3 strains, respectively. At 36 h (for Sf-9 cells) and 24 to 48 h (for MDCK cells) postinfection, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature and washed thrice with PBS, pH 7.4. The fixed cells were incubated with hybridoma culture fluid at 37°C for 1 h. Cells were rinsed thrice with PBS and incubated with a 1:40 dilution of fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin (Dako, Denmark). Cells were rinsed again in PBS prior to scoring of results with an epifluorescence microscope (Olympus, Japan) with appropriate barrier and excitation filters for optimized fluorescein isothiocyanate visualization (32).
Western blotting.
MAbs were evaluated by immunoblotting assays. The recombinant H5N1 HA2 protein and whole purified Indonesian H5N1 virus were fractionated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions. The separated proteins were electrotransferred and immobilized on a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk in PBS containing 0.1% Tween 20 (PBST) at 37°C for 1 h. The membrane was subsequently incubated with hybridoma supernatant containing the MAb, rinsed in PBST, and incubated with horseradish peroxidase (HRP)-conjugated rabbit anti-mouse immunoglobulin followed by incubation with ECL reagents (Amersham Biosciences) to detect bound MAb (6).
Epitope mapping of MAbs.
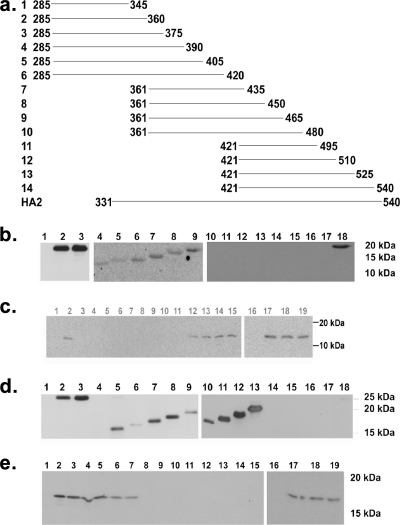
The MAbs were first confirmed to bind to the recombinant HA2 protein by Western blotting analysis. A series of 14 fragments of the HA2 polypeptide were designed so as to detect the exact region of the HA2 molecule (epitope) to which the MAbs bound. Fourteen overlapping fragments encoding the HA2 protein of the A/goose/Guangdong/1997 strain (Fig. 1a) were amplified by PCR and cloned into the pQE-30 expression vector. The fusion peptides were expressed in E. coli as described above and analyzed by Western blotting (6). After a 15- to 20-amino-acid stretch was identified as the epitope region, a panel of 15 mutants of the appropriate fragment was generated, with substitutions at amino acid residues 331 to 345 and 346 to 360. Each construct included an amino acid mutation to alanine. The point mutants were generated using a PCR-based site-directed mutagenesis protocol and verified by sequence analysis. These mutants were also expressed as His fusion peptides as described above.
FIG. 1.
(a) Plan for epitope mapping of anti-HA2 MAbs. The gene segments coding for fragments 1 to 14 above were PCR amplified and cloned into pQE-30 vector, and the peptides were expressed as histidine-tagged proteins. The cell lysates of bacteria expressing the 14 fragments were used for Western blotting analysis of the MAbs to map their respective epitopes (the numbers indicate the amino acid number on mature HA0). (b) Western blot analysis of the 14 fragments. MAb 1C9 was used as the primary antibody, followed by HRP-conjugated anti-mouse immunoglobulin, and the membrane was developed using ECL reagents. Lane 1, protein molecular weight marker; lanes 2 and 18, recombinant HA2 gp; lane 3, HA2 from whole virus; lanes 4 to 17, 14 fragments. Only fragments 1 to 6 show positive results, while fragments 7 to 14 show negative results. The samples were run in three different gels but processed identically. (c) Results of Western blot analysis of point mutants also expressed as histidine fusion peptides. The membrane was developed using ECL reagents. The analysis was carried out to deduce the amino acid sequence required for MAb 1C9 binding. Lanes 1 and 16, protein marker; lanes 2 and 17, wild-type fragment 1; lanes 3 to 15, 18, and 19, overexpressed point mutants. Positive results were seen only in lanes with wild-type fragment 1 and the I340A (lane 12), E341A (lane 13), G342A (lane 14), G343A (lane 15), W344A (lane 18), and Q345A (lane 19) point mutants, indicating the absence of the role of the respective amino acids in binding to the HA2 gp. Negative results were seen in lanes with the G331A (lane 3), L332A (lane 4), F333A (lane 5), G334A (lane 6), A335A (lane 7), I336A (lane 8), A337G (lane 9), G338A (lane 10), and F339A (lane 11) point mutants, indicating the role of these amino acids in forming the epitope of MAb 1C9. Lane numbers of the respective point mutants are indicated in brackets. The samples were run in two different gels but processed identically. (d) Western blot analysis of the 14 fragments. MAb 1B12 was used as the primary antibody, followed by HRP-conjugated anti-mouse immunoglobulin, and the membrane was developed using ECL reagents. Lane 1, protein molecular weight marker; lane 2, recombinant HA2 gp; lane 3, HA2 from whole virus; lanes 4 to 17, 14 fragments. Only fragments 2 to 10 show positive results, while fragments 1and 11 to 14 show negative results. (e) Results of Western blot analysis of point mutants also expressed as histidine fusion peptides. The membrane was developed using ECL reagents. The analysis was carried out to determine the amino acid sequence of the minimal epitope for MAb 1B12. Lane 1 and lane 16, protein marker; lanes 2 and 17, wild-type fragment 2; lanes 3 to 15, 18, and 19, overexpressed point mutants. Positive results were seen only in lanes with the G346A (lane 3), M347A (lane 4), V348A (lane 5), D349A (lane 6), G350A (lane 7), E359A (lane 18), and Q360A (lane 19) point mutants, indicating the absence of the role of the respective amino acids in binding to the MAb. Negative results were seen in lanes with the W351A (lane 8), Y352A (lane 9), G353A (lane 10), Y354A (lane 11), H355A (lane 12), H356A (lane 13), S357A (lane 14), and N358A (lane 15) point mutants, indicating the role of these amino acids in binding to MAb 1B12. Lane numbers of the respective point mutants are indicated in brackets. The samples were run in two different gels but processed identically.
Cell-cell fusion inhibition assay.
CHO-K1 cells expressing the HA0 precursor were used for the cell-cell fusion assay (10). Cells were grown to confluence in 24-well culture plates. When appropriate, cells were treated with tosylsulfonyl phenylalanyl chloromethyl ketone-trypsin (2.5 μg/ml) for 10 min at 37°C to cleave HA0 into HA1 and HA2. The cell monolayer was washed with 1× PBS, pH 7.0, and then incubated for 30 min at 37°C with the MAbs at the concentrations indicated. The pH was decreased to 5 for 1 min, and then the cells were incubated with complete medium until syncytium formation was observed in the control well. The cells were then fixed with 4% (wt/vol) paraformaldehyde and stained with 1% (wt/vol) toluidine blue in PBS.
Challenge studies against influenza H5N1 virus infection.
To determine the therapeutic efficacy of MAbs, inbred specific-pathogen-free BALB/c mice aged 4 to 6 weeks were used. Six mice (n = 6) per group were anesthetized with ketamine-xylazine and intranasally infected with five 50% mouse lethal doses (MLD50) of two different H5N1 strains (A/Vietnam/1203/2004 from clade 1 and A/Indonesia/TLL013/06 from clade 2.1). The MLD50 was determined by the method of Reed and Muench (24).
(i) Prophylactic efficacy.
To determine the prophylactic efficacy of the MAbs, groups of mice were pretreated intraperitoneally with 5 mg/kg of body weight, 10 mg/kg, or 0 mg/kg (PBS) of MAb prior to the viral challenge. After 24 h, mice were challenged with 5 MLD50 of the two different H5N1 strains. Mice were observed daily to monitor body weight and mortality. Monitoring continued until all animals died or until day 14 after challenge. Serum samples were collected on days 7, 10, and 14 from the mice surviving the viral challenge for evaluation of the humoral immune response to the viral infection by determining the HI titers.
(ii) Therapeutic efficacy.
To determine the therapeutic efficacy of the MAbs, each group of mice was treated via the intraperitoneal route with 5 mg/kg, 10 mg/kg, or 0 mg/kg (PBS) of each anti-HA2 MAb 1 or 3 days after viral challenge. Mice were observed daily to monitor body weight and mortality. Sampling was done as mentioned above.
Another set of mice was maintained (n = 10) for each experimental group with animals infected with clade 2.1 for quantitative PCR and histopathology experiments. On days 3, 6, and 9 postchallenge, mice were euthanized by a lethal dose of sodium pentobarbital. Viral infectivity titers were determined for the group of animals treated with MAb 1C9 1 day after challenge with clade 2.1 virus. To determine the viral titers following challenge, the lungs of the mice were homogenized in 2 ml of sterile PBS. The homogenized suspensions were titrated on monolayers of MDCK cells in quadruplicate (27). The viral titers were calculated by the Reed and Muench method (24) and expressed as log10 50% tissue culture infective doses (TCID50)/ml.
For histopathology, a lung sample was collected in 10% (wt/vol) buffered formalin solution, embedded in paraffin, and sectioned. Sections were stained with hematoxylin and eosin and were analyzed for pathology.
Determination of viral load by real-time PCR.
About 200 μl of lung homogenate was treated with 1 ml Trizol reagent to extract RNA. Total RNA (500 ng/sample) was used for cDNA synthesis, using 20 U of avian myeloblastosis virus reverse transcriptase (Roche). The cDNA suspension was used for amplification in a quantitative real-time PCR. A DyNAmo Capillary SYBR green qPCR kit (Finnzymes) was used for the PCR assay. Briefly, the cDNA was amplified in 20 μl containing 0.25 μmol of forward primer (5′-GAAATCAAACAGATTAGTCCTTGC-3′), 0.25 μmol of reverse primer (5′-CCTGCCATCCTCCCTCTATAAA-3′), and 1× DyNAmo master mix. Reactions were performed in a Light Cycler instrument (Roche) under the following conditions: 10 min at 95°C, followed by 50 cycles of 95°C for 10 s, 57°C for 5 s, and 72°C for 9 s. Fluorescence signals from these reactions were captured at the end of the extension step in each cycle. To determine the specificity of the assay, PCR products were subjected to melting curve analysis at the end of the assay (65 to 95°C at 0.1°C/s). The murine glyceraldehyde-3-phosphate dehydrogenase gene was used as an internal positive control to check for the presence of PCR-inhibitory substances. To determine the dynamic range of real-time quantitative PCR, serial dilutions of in vitro-transcribed RNA containing the target sequence were made, subjected to the real-time quantitative PCR assay, and used to construct a standard curve. The assay was able to distinguish 10-fold differences in concentration over a range of 1,000 to 109 copies, and no signal was observed in the water control. Relative quantification was obtained in triplicate for each experimental sample by using the standard curve method. The viral titers were expressed as a measure of the number of copies of viral RNA in the lungs of infected mice.
HI assay.
HI assays were performed as described previously (23). Briefly, purified hybridoma supernatant was treated with receptor-destroying enzyme (RDE) and subsequently serially diluted (twofold) in V-bottomed 96-well plates and mixed with H5N1 virus (A/Indonesia/TLL013/06). Plates were incubated for 30 min at room temperature, and 1% chicken red blood cells were added to each well. The HI end point was the highest serum dilution in which agglutination was not observed.
Neutralization assay.
Neutralization assays were performed as described previously (27). A dose of 100 TCID50 of viruses was used in each of the virus neutralization assays. Supernatants were tested at a starting dilution of 1:10. Briefly, twofold dilutions of hybridoma supernatants were mixed with virus suspension containing 100 TCID50 of purified H5N1 virus and incubated at 37°C for 1 h. The virus-antibody mix was then transferred onto MDCK cell monolayers in 96-well plates at 37°C for 1 h. One hundred microliters of serum-free Dulbecco's modified Eagle's medium was added to each well, and the cells were subsequently incubated at 37°C in a 5% CO2 incubator. Cytopathic effect was observed every 24 h for 48 to 72 h.
RESULTS
Characterization of MAbs.
A panel of nine hybridoma clones secreting MAbs to H5 HA0 antigen was screened by IFA against Indonesian H5N1 influenza virus strains and other non-H5 subtypes. The results showed that MAb 1C9 and MAb 1B12 strongly reacted with MDCK cells infected with H5 subtypes and yielded positive cytoplasmic immunofluorescence patterns (data not shown). IFA with SF9 cells infected with baculovirus expressing the HA0 gene on the surface yielded positive results. However, negative results were observed for IFA with SF9 cells infected with baculovirus harboring the HA1 gene (data not shown). By these results, we concluded that the epitope recognized by the antibodies was specific to the HA2 region. Immunoblotting with the whole virus lysates showed a positive signal at 27 kDa, which we believe to be HA2. This was also further confirmed by a positive Western blot using recombinant HA2 protein (Fig. 1b). With this result, we concluded that the MAbs were indeed against the HA2 region of the virus, and since they showed positive results on the Western blot, we concluded that the antibodies were against linear epitopes on the HA2 gp. The HI assay showed that the antibodies had no HI titers. The neutralization titers of these MAbs, recorded against 100 TCID50 of virus, were found to be <10. The isotype of both MAbs was determined to be immunoglobulin G1.
Epitope mapping of MAbs.
MAbs 1C9 and 1B12 recognized the HA2 domain of H5 in Western blot assays, suggesting that the epitope is linear or readily refolded through short-range interactions. This property was exploited to map the HA epitope targeted by MAbs 1C9 and 1B12. To this end, partially overlapping expressed HA fragments were probed by Western blot analysis (Fig. 1a). MAb 1C9 reacted with fragments 1 to 6 and did not react with the other fragments (Fig. 1b). MAb 1B12 was found to be positive with fragments 2 to 10 but negative with fragments 1 and 11 to 14 (Fig. 1c). These data indicated that the epitope of 1C9 comprised amino acids 331 to 345 of HA0 (amino acids 1 to 15 of HA2). Similarly, the epitope of MAb 1B12 comprised amino acids 346 to 360 of HA0 (amino acids 15 to 30 of HA2). Subsequent analysis of a panel of expression constructs with single amino acid changes by Western blotting indicated that MAb 1C9 bound to I340A, E341A, G342A, G343A, W344A, and Q345A mutants. In contrast, 1C9 failed to bind to G331A, L332A, F333A, G334A, A335G, I336A, A337G, G338A and F339A mutants (Fig. 1c). These data indicate that amino acids 331 to 339 (GLFGAIAGF) participate in formation of the antigenic site for MAb 1C9. Similar analysis of MAb 1B12 revealed the involvement of WYGYHHSN (amino acids 351 to 358) in the formation of its antigenic site (Fig. 1e).
Cell-cell fusion of CHO-K1 cells expressing HA0.
The HA0 precursor of influenza A virus HA was expressed on the surfaces of CHO-K1 cells. Upon treatment with trypsin and low pH, HA0 undergoes a conformational change from the native to the fusion-active form, resulting in polykaryon formation. We studied the influence of the anti-HA2 MAbs 1C9 and 1B12 on this fusion process. Normal polykaryon formation was observed in the control well (Fig. 2b). Polykaryon formation was completely inhibited by MAb 1C9 at a concentration of 100 μg/ml (Fig. 2e) and partially inhibited at a concentration of 50 μg/ml (Fig. 2d). MAb 1B12, however, did not show any fusion inhibition, even at high concentrations (data not shown). The irrelevant MAb 3H5 did not show any inhibition of polykaryon formation (Fig. 2f).
FIG. 2.
Inhibition of cell-cell fusion of CHO cells expressing the HA0 precursor. (a) Cells without trypsin treatment following pH 5 treatment. (b) Cells after trypsin treatment and following pH 5 treatment. Cells are also shown after trypsin treatment and following pH 5 treatment in the presence of 10 μg/ml of MAb 1C9 (c), in the presence of 50 μg/ml of MAb 1C9 (d), in the presence of 100 μg/ml of MAb 1C9 (e), and in the presence of 100 μg/ml of nonspecific MAb 3H5 (f).
Prophylactic efficacy of anti-HA2 MAbs against H5N1 infection.
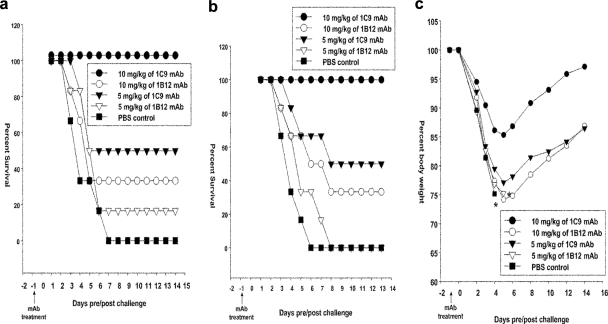
To evaluate the prophylactic efficacy of the MAbs, different groups of experimental mice (n = 6/group) were treated with the indicated dosages of MAbs 1 day prior to challenge with 5 MLD50 of HPAI H5N1 strains from clade 1.0 or clade 2.1. Untreated mice challenged with the H5N1 viruses (PBS control group) showed the most rapid decline in body weight, culminating in 100% mortality within 6 days after viral challenge. Pretreatment with 10 mg/kg of MAb 1C9 provided 100% protection against 5 MLD50 of two H5N1 strains, from clade 1.0 (Fig. 3a) and clade 2.1 (Fig. 3b), while pretreatment with 5 mg/kg provided 50% protection against 5 MLD50 of H5N1 viruses of both clades. The group of mice pretreated with 10 mg/kg of 1C9 showed only a 15% loss in body weight on day 5 and had regained their body weight by day 14 after lethal challenge (Fig. 3c). However, MAb 1B12 provided only 33% protection against the same dose of virus, even at 10 mg/kg. MAb 1B12 at 5 mg/kg provided inadequate protection (<16%) against both H5N1 strains (Fig. 3a and b). Mice in this group lost up to 25% of their body weight and did not regain it completely even at 14 days postchallenge.
FIG. 3.
Protection of mice from lethal H5N1 viral challenge after pretreatment with MAbs. Each group of mice was pretreated intraperitoneally with 10 mg/kg, 5 mg/kg, or 0 mg/kg (PBS) of MAb 1C9 or 1B12 1 day before challenge with mouse-adapted Indonesian HPAI H5N1 strains, either A/Vietnam/1203/04 clade 1.0 virus (a) or A/TLL013/06 clade 2.1 virus (b and c). Mice were monitored for survival (a and b) throughout a 14-day observation period. The results are expressed in terms of percent survival. (c) Mice were monitored for weight loss throughout the 14-day observation period. The results are expressed in terms of percent body weight (at the beginning of the trial).
Therapeutic efficacy of anti-HA2 MAbs against H5N1 infection.
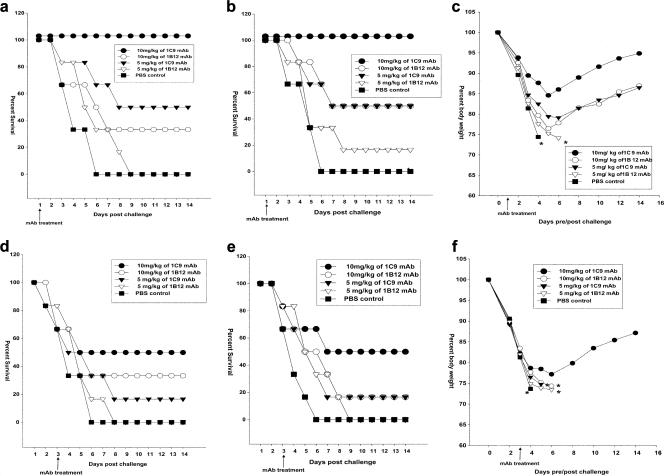
Different groups of experimental mice (n = 6/group) were treated with the indicated doses of MAbs 1 or 3 days after challenge with 5 MLD50 of HPAI H5N1 strains from clade 1.0 or clade 2.1. The progress of infection was indicated by various trends of decreases in body weight in the different groups. For mice challenged with the H5N1 viruses, untreated mice showed the most rapid decline in body weight, culminating in 100% mortality within 6 days after viral challenge. On day 3, the body weight of mice who succumbed from this group was below 75% of the original body weight (Fig. 4c). Upon treatment 1 day after infection with MAb 1C9, mice showed up to a 15% loss of body weight. However, after 5 days, they steadily regained their body weight (Fig. 4c). At the higher concentration of 10 mg/kg body weight, MAb 1C9 completely protected mice from disease upon challenge with clade 2.1 (Fig. 4b) and clade 1 viruses (Fig. 4a). The same concentrations of MAb 1B12, however, could provide only 50% and 33% protection against lethal infection with clade 2.1 and clade 1 viruses, respectively. The mice showed about a 23% loss of body weight, and from 5 days after challenge, the mice gradually regained only about 8 to 10% of the lost body weight (Fig. 4c). The groups of mice treated with 5 mg/kg of 1C9 showed a loss of body weight of up to 22%. The mortality studies showed that this concentration provided 50% protection against 5 MLD50 of both clades of virus. MAb 1B12 at 5 mg/kg provided inadequate protection (<20.0%) against both H5N1 strains (Fig. 4a and b). The groups of mice (n = 6/group) were also treated with the indicated doses of MAbs 3 days after challenge with the two H5N1 strains. Three days after the challenge (before treatment), 16.6 to 33.3% (one or two mice of six mice/group) of the mice in each group died, and the therapeutic efficacy of the MAbs was tested only in the surviving mice. The 10-mg/kg dose of MAb 1C9 provided 75% (three of four mice survived) protection against H5N1 strains (Fig. 4d and e). Five milligrams of 1C9 and 10 mg of 1B12 provided 50% (two of four mice survived) and 25% (one of four mice survived) protection, respectively, against 5 MLD50 of H5N1 strains (Fig. 4d and e).
FIG. 4.
Protection of mice from lethal H5N1 viral challenge. Each group of mice was treated intraperitoneally with 10 mg/kg, 5 mg/kg, or 0 mg/kg (PBS) of MAb 1C9 or 1B12 at 1 day post-viral challenge (a) and 3 days post-viral challenge (d) with clade 1 virus A/Vietnam/1203/04 or at 1 day post-viral challenge (b) and 3 days post-viral challenge (e) with clade 2 virus A/Indonesia/TLL013/06. Mice were monitored for survival throughout a 14-day observation period. The results are expressed in terms of percent survival. Mice were also monitored for weight loss at 1 day post-viral challenge (c) and 3 days post-viral challenge (f) with clade 2 virus A/Indonesia/TLL013/06 throughout a 14-day observation period. The results are expressed in terms of percent body weight (at the beginning of the trial).
Histopathology.
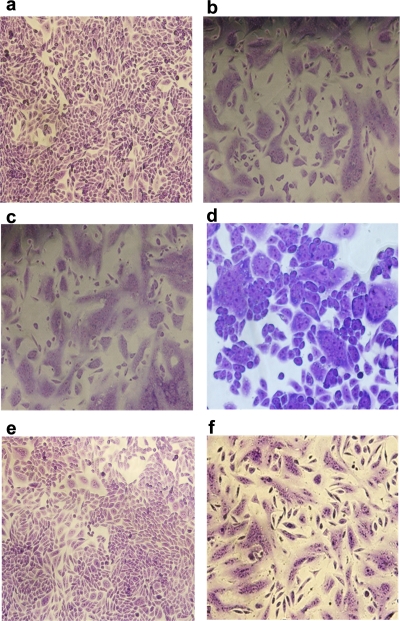
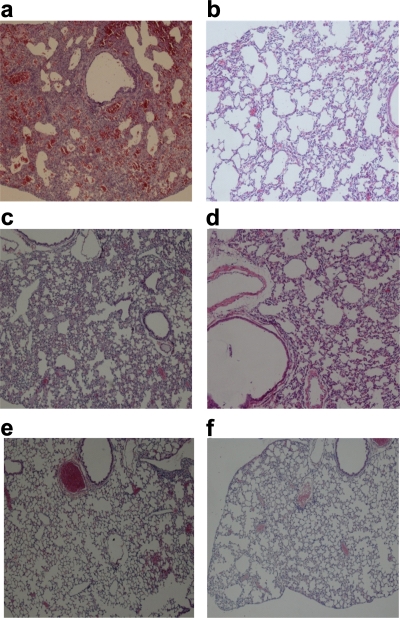
Histopathology studies were performed only for the lungs of mice treated with MAb 1C9 at 24 h pre- or post-viral challenge with clade 2.1 virus. On day 6 postinfection, lungs of untreated mice had pulmonary lesions consisting of moderate to severe necrotizing bronchitis and moderate to severe histiocytic alveolitis with associated pulmonary edema (Fig. 5b). The uninfected mice lacked lesions in the lungs (Fig. 5a). Mice treated (24 h pre- or postchallenge) with 5 mg/kg of 1C9 had minimal to moderate bronchitis (Fig. 5c and e), and mice treated (24 h pre- or postchallenge) with 10 mg/kg of 1C9 had minimal bronchitis.
FIG. 5.
Photomicrographs of hematoxylin- and eosin-stained lung sections of mice infected with clade 2.1 H5N1 virus at 6 days postchallenge. (a) Infected and untreated mice. Normal morphology was seen in uninfected mice (b), infected mice treated with 5 mg/kg of 1C9 at 24 h postchallenge (c), infected mice treated with 10 mg/kg of 1C9 at 24 h postchallenge (d), infected mice treated with 5 mg/kg of 1C9 24 h prior to viral challenge (e), and infected mice treated with 10 mg/kg of 1C9 24 h prior to viral challenge (f).
Determination of viral loads.
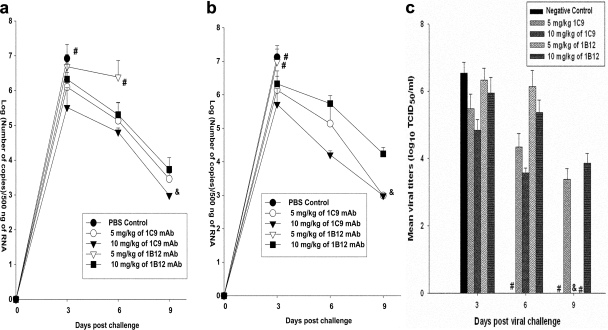
Real-time PCR was used to evaluate the kinetics of the influenza A virus load in the lung samples of the animals infected with clade 2.1 virus. The viral titers were expressed as a measure of the number of copies of viral RNA. Titers of viruses in the lungs of mice treated with MAbs 1C9 and 1B12 at 24 h pre- or postchallenge were compared with those for the untreated mice. Mice receiving 10 mg/kg of 1C9 before or after viral challenge showed significantly lower viral loads on all days, and viral titers were below the detection limit (<1,000 copies) on day 9 after viral challenge. Mice receiving 5 mg/kg of the same MAb showed lesser viral clearance than did mice receiving 10 mg/kg of 1C9, in a dose-dependent manner. However, mice receiving 5 mg/kg or 10 mg/kg of 1B12 showed insignificant viral clearance (Fig. 6a and b). Viral infectivity titers were measured only for the mice challenged with clade 2.1 virus and treated 1 day after challenge. Similar results were observed even when the viral infectivity titers were measured (Fig. 6c). Mice receiving 10 mg/kg of MAb 1C9 1 day after viral challenge showed significantly lower viral loads which were below the detection limit on day 9 after viral challenge.
FIG. 6.
Measurement of viral loads in lungs of treated mice. (a) Mice were pretreated with 10 mg/kg, 5 mg/kg, or 0 mg/kg (PBS) of MAb 1C9 or 1B12 1 day before challenge with mouse-adapted Indonesian HPAI H5N1 (A/TLL013/06 [clade 2.1]) virus. The viral loads were measured in the lungs of the infected animals on days 3, 6, and 9 postchallenge. (b) Mice were treated with 10 mg/kg, 5 mg/kg, or 0 mg/kg (PBS) of MAb 1C9 or 1B12 1 day after challenge with mouse-adapted Indonesian HPAI H5N1 (A/TLL013/06 [clade 2.1]) virus. The viral loads were measured in the lungs of the infected animals on days 3, 6, and 9 postchallenge. The results are expressed in terms of mean log number of copies/500 ng of RNA ± standard deviation. #, no survival of any animals in the group; &, undetectable viral numbers. (c) Measurement of viral infectivity titers in the lungs of infected animals challenged with mouse-adapted Indonesian HPAI H5N1 (A/TLL013/06 [clade 2.1]) virus and treated with 10 mg/kg, 5 mg/kg, or 0 mg/kg (PBS) of MAb 1C9 or 1B12 1 day after challenge. The viral loads were measured in the lungs of the infected animals on days 3, 6, and 9 postchallenge. The results are expressed in terms of mean log TCID50/ml ± standard deviation. #, no survival of any animals in the group; &, undetectable viral titers. The lower limit of detection was 1.5 log10 TCID50/ml.
Humoral immune response to influenza virus infection.
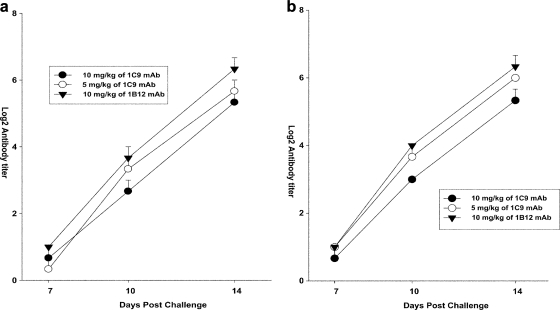
On days 7, 10, and 14 postchallenge, the HI titers of the serum samples were measured for the treated mice who survived the infection. The HI titers for both the prophylactic group (Fig. 7a) and the therapeutic group (Fig. 7b) were measured. The antibody response in the mice started on day 10 and reached a titer of >25 log irrespective of the dose of MAb treatment. The mice who survived the infection mounted an effective immune response by day 14.
FIG. 7.
Determination of humoral immune response in treated mice. (a) Mice were pretreated (24 h before challenge) with 5 mg/kg and 10 mg/kg of MAb 1C9 and 10 mg/kg of MAb 1B12 and challenged with the clade 2.1 virus. (b) Mice were challenged with the clade 2.1 virus and treated (24 h after challenge) with 5 mg/kg and 10 mg/kg of MAb 1C9 and 10 mg/kg of MAb 1B12. The HI titers of the serum samples (n = 3) were measured on days 7, 10, and 14 after viral challenge in mice surviving the viral infection. The results are expressed in terms of mean log2 HI titer ± standard error.
DISCUSSION
The HA2 N-terminal fusion peptide is the most highly conserved region in HA among all influenza A virus subtypes (7). Part of this region is exposed as a surface loop in the precursor molecule (4, 34). Since most HA subtypes are cleaved by extracellular enzymes, this surface loop is accessible to antibody on HA0 expressed in the plasma membranes of infected host cells (1). Previous studies found that mice vaccinated with a peptide spanning the HA1-HA2 joining region exhibited less illness and fewer deaths upon virus challenge (13). Hence, the HA1-HA2 joining region seems to be a promising candidate for a universal vaccine and therapeutic agent against influenza A viruses. MAbs against the HA2 fusion peptide have attracted some interest in the recent past, as they are known to neutralize the infectivity of the influenza virus by interfering with the low-pH-induced conformational change in the HA molecule, resulting in the inhibition of fusion during viral replication (28, 29). These MAbs can also block fusion at the cell membrane at the postbinding/prefusion stage (19). It has been known that anti-HA2 MAbs lack HI activity. However, a lack of in vitro HI activity does not rule out protective activity in vivo, because the binding of antibody to native HA expressed on infected cells and infectious virus could mediate protective activity by targeting Fc receptor-expressing cells or complement deposition to these structures (7).
We describe here the cross-clade protection of mice against lethal infection, mediated by a MAb against the fusion peptide of the HA2 glycoprotein. The epitope of MAb 1C9 was against the more significant HA1-HA2 joining region, specifically against a part of the highly conserved fusion peptide. MAb 1C9 was analyzed for its fusion inhibition activity by an in vitro cell-cell fusion assay. It was observed that MAb 1C9 could completely inhibit fusion at a concentration of 100 μg/ml, and even at 50 μg/ml, moderate fusion inhibition activity was observed. Although fusion inhibition characteristics are not synonymous with virus neutralization, the fact that the MAb inhibits fusion indicates that it can prevent replication of the virus. Previous studies reported inhibition of fusion activity of influenza virus HA by 100 μg/ml HA2-specific MAbs recognizing amino acids 1 to 35 of HA2 (30). These MAbs were found to contribute to the protection of infected mice and to mediate more effective recovery from influenza virus H3N2 infection (9).
In our studies, we observed that the anti-fusion peptide MAb 1C9 provided 100% protection against two different clades, clades 1.0 and 2.1, at 10 mg/kg body weight when it was administered as a prophylactic as well as a therapeutic agent at 1 day pre- or postchallenge. Delayed treatment, at 3 days postchallenge, with 10 mg of MAb 1C9 afforded partial protection (75%) against 5 MLD50 of H5N1 virus infection. This lesser protection was due to the advanced stage of the viral infection at 3 days postchallenge.
MAb 1C9 conferred more protection than MAb 1B12. This may be attributable to the different epitopes recognized by the antibodies. MAb 1C9 recognizes the epitope GLFGAIAGF and hence targets and binds to the fusion peptide of HA. Due to this, it could block the pH-dependent conformational change and/or the requisite interactions between the viral and endosomal membrane proteins, which would delay or prevent the penetration of the viral core into the target cell cytoplasm (14, 21). As a result, it could prevent further replication of the virus and hence mediate more protection against influenza A virus infection. On the other hand, 1B12 was found to be against the epitope WYGYHHSN, which is also at the N terminus of HA2, but away from the region of the fusion peptide. Hence, it may bind to the respective epitope and block the necessary interactions between the viral and endosomal membranes. Also, mere binding of antibody to native HA expressed on infected cells and infectious virus could mediate protective activity by targeting Fc receptor-expressing cells or complement deposition to these structures (7, 25). Therefore, even MAb 1B12 could mediate at least some protection at higher concentrations.
In our study, therapy with MAb 1C9 probably helped to control the initial course of infection, thus allowing the animal to mount an effective immune response. It is possible that passive antibody could act at the level of the infected cell, as well as with free virus particles, to help control infection-related tissue damage (22). Development of peak virus titers in the lung within 4 days of infection probably restricts the time frame for successful therapy. We observed that some mice succumbed to the infection even before the commencement of treatment 3 days after viral challenge. However, we were able to demonstrate that passive antibody therapy with MAb 1C9 protected mice from lethal viral infection even at 5 mg/kg. A single dose of the MAb was sufficient to protect the mice for a few days at the onset of the viral infection, giving the animal enough time to mount an effective immune response to the infection. This was evident by the HI titers in sera 14 days after viral challenge.
MAb 1C9 in a single dose offered good protection against infection, but the animals showed reductions in body weight, and some symptoms of the disease were seen in the early stages. Multiple doses of MAb 1C9 could be more effective. Inclusion of another neutralizing antibody in combination with this antibody could prove to be efficient treatment, as the neutralizing antibody would effect an even earlier clearance of the virus and hence a lesser reduction in body weight and the animal would have a chance at a much earlier recovery from the infection.
Currently available MAbs for therapy against influenza include the neutralizing anti-HA1 MAbs and MAbs against the conserved ectodomain of the matrix protein (M2e). Neutralizing MAbs against HA1 confer effective protection against influenza virus infections, but the risk of escape mutants arising from this approach is currently an issue (15). Furthermore, the fact that HA1 is not conserved indicates that any MAb against HA1 will not be effective against all H5N1 strains, let alone all subtypes. Although M2e is a more conserved protein than HA1, recent studies have shown the formation of escape mutants against the anti-M2e MAbs (42). This can be a significant drawback in the long run. Though the HA2 gp is relatively conserved, the fusion peptide of HA2 is comparatively more conserved than any other antigen of influenza A virus. We describe the protective efficacy of MAbs against the highly conserved fusion peptide of HA. We believe that since the fusion peptide is almost invariant in all strains of influenza A viruses, these MAbs could confer protection against all influenza virus infections, irrespective of the subtype. The inclusion of these MAbs against the conserved fusion peptide, in combination with virus-neutralizing MAbs, could be an effective approach to block the further replication of any escape mutants formed from anti-HA1 MAbs.
In conclusion, our results suggest a prospective universal passive immunotherapy for cross-protection against H5N1 human virus infection. The clinical application of this approach can be ascertained by humanization of this antibody and its evaluation as a therapeutic agent in nonhuman primates challenged with influenza A viruses. We hope that the inclusion of this MAb along with other neutralizing MAbs in a cocktail could be employed as an effective treatment of H5N1 influenza in the future.
Acknowledgments
We are grateful for financial support received from Temasek Life Science Laboratory, Singapore.
We thank the Ministry of Health, Indonesia, for technical support and collaboration. We also thank Ruben Donis, Influenza Division, Centers for Disease Control and Prevention, Atlanta, GA, for providing the plasmids for reverse genetics.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Atassi, M. Z., and R. G. Webster. 1983. Location, synthesis and activity of an antigenic site on influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 180840-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beigel, J. H., J. Farrar, A. M. Han, F. G. Hayden, R. Hyer, M. D. de Jong, S. Lochindarat, T. K. Nguyen, T. H. Nguyen, T. H. Tran, A. Nicoll, S. Touch, K. Y. Yuen, and Writing Committee of the W. H. O. Consultation on Human Influenza A/H5. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 3531374-1385. [DOI] [PubMed] [Google Scholar]
- 3.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295891-894. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95409-417. [DOI] [PubMed] [Google Scholar]
- 5.de Jong, M. D., and T. T. Hien. 2006. Avian influenza A (H5N1). J. Clin. Virol. 352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher, S., S. E. Winston, S. A. Fuller, and J. G. Hurrell. 2004. Immunoblotting and immunodetection, p. 10.8.1-10.8.24. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, et al. (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Newcastle, United Kingdom.
- 7.Gerhard, W., K. Mozdzanowska, and D. Zharikova. 2006. Prospects for universal influenza virus vaccine. Emerg. Infect. Dis. 12569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gocnik, M., T. Fislova, T. Sladkova, V. Mucha, F. Kostolansky, and E. Vareckova. 2007. Antibodies specific to the HA2 glycopolypeptide of influenza A virus with fusion-inhibition activity contribute to the protection of mice against lethal infection. J. Gen. Virol. 88951-955. [DOI] [PubMed] [Google Scholar]
- 9.Gocnik, M., T. Fislova, V. Mucha, T. Sladkova, G. Russ, F. Kostolansky, and E. Vareckova. 2008. Antibodies induced by the HA2 glycopolypeptide of influenza virus haemagglutinin improve recovery from influenza A virus infection. J. Gen. Virol. 89958-967. [DOI] [PubMed] [Google Scholar]
- 10.Godley, L., J. Pfeifer, D. Steinhauer, B. Ely, G. Shaw, R. Kaufmann, E. Suchanek, C. Pabo, J. J. Skehel, D. C. Wiley, and S. Wharton. 1992. Introduction of intersubunit disulphide bonds in the membrane distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68635-645. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein, M. A., and N. M. Tauraso. 1970. Effect of formalin, beta-propiolactone, merthiolate, and ultraviolet light upon influenza virus infectivity chicken cell agglutination, hemagglutination, and antigenicity. Appl. Microbiol. 19290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson, J. H., C. M. Boon, P. C. Lim, A. Webb, E. E. Ooi, and R. J. Webby. 2006. Passive immunoprophylaxis and therapy with humanized monoclonal antibody specific for influenza A H5 hemagglutinin in mice. Respir. Res. 7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath, A., G. K. Toth, P. Gogolak, Z. Nagy, I. Kurucz, I. Pecht, and E. Rajnavolgyi. 1998. A hemagglutinin-based multipeptide construct elicits enhanced protective immune response in mice against influenza A virus infection. Immunol. Lett. 60127-136. [DOI] [PubMed] [Google Scholar]
- 14.Imai, M., K. Sugimoto, K. Okazaki, and H. Kida. 1998. Fusion of influenza virus with the endosomal membrane is inhibited by monoclonal antibodies to defined epitopes on the hemagglutinin. Virus Res. 53129-139. [DOI] [PubMed] [Google Scholar]
- 15.Kaverin, N. V., I. A. Rudneva, E. A. Govorkova, T. A. Timofeeva, A. A. Shilov, K. S. Kochergin-Nikitsky, P. S. Krylov, and R. G. Webster. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J. Virol. 8112911-12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakadamyali, M., M. J. Rust, and X. Zhuang. 2004. Endocytosis of influenza viruses. Microbes Infect. 6929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 4371108. [DOI] [PubMed] [Google Scholar]
- 18.Liu, W., P. Zou, and Y. H. Chen. 2004. Monoclonal antibodies recognizing EVETPIRN epitope of influenza A virus M2 protein could protect mice from lethal influenza A virus challenge. Immunol. Lett. 93131-136. [DOI] [PubMed] [Google Scholar]
- 19.Marasco, W. A., and J. Sui. 2007. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 251421-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Institutes of Health and Centers for Disease Control and Prevention. 1999. Biosafety in microbiological and biomedical laboratories, 4th ed. U.S. Government Printing Office, Washington, D.C.
- 21.Outlaw, M. C., and N. J. Dimmock. 1993. IgG neutralization of type A influenza viruses and the inhibition of the endosomal fusion stage of the infectious pathway in BHK cells. Virology 195413-421. [DOI] [PubMed] [Google Scholar]
- 22.Parren, P. W. H. I., T. W. Gieisbert, T. Maruyama, P. B. Jahrling, and D. R. Burton. 2002. Pre- and post-exposure prophylaxis for Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J. Virol. 766408-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabakaran, M., S. Velumani, F. He, A. K. Karuppannan, G. Y. Geng, L. K. Yin, and J. Kwang. 2008. Protective immunity against influenza H5N1 virus challenge in mice by intranasal co-administration of baculovirus surface-displayed HA and recombinant CTB as an adjuvant. Virology 380412-420. [DOI] [PubMed] [Google Scholar]
- 24.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 25.Saikh, K. U., J. D. Martin, A. H. Nishikawa, and S. B. Dillon. 1995. Influenza A virus-specific H2d restricted cross-reactive cytotoxic T lymphocyte epitope(s) detected in the hemagglutinin HA2 subunit of A/Udorn/72. Virology 214445-452. [DOI] [PubMed] [Google Scholar]
- 26.Sawyer, L. A. 2000. Antibodies for the prevention and treatment of viral diseases. Antivir. Res. 4757-77. [DOI] [PubMed] [Google Scholar]
- 27.Simmons, C. P., N. L. Bernasconi, A. L. Suguitan, Jr., K. Mills, J. M. Ward, N. V. Chau, T. T. Hien, F. Sallusto, Q. Ha Do, J. Farrar, M. D. de Jong, A. Lanzavecchia, and K. Subbarao. 2007. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 4928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Styk, B., G. Russ, and K. Polakova. 1979. Antigenic glycopeptides HA1 and HA2 of influenza hemagglutinin. IV. Immunogenic properties of separated hemagglutinin glycopolypeptides. Acta Virol. 239-20. [PubMed] [Google Scholar]
- 29.Vanlandschoot, P., E. R. Beirnaert, B. Barrere, L. Calder, B. Millar, S. Wharton, W. M. Jou, and W. Fiers. 1998. An antibody which binds to the membrane-proximal end of influenza virus hemagglutinin (H3 subtype) inhibits the low-pH-induced conformational change and cell-cell fusion but does not neutralize virus. J. Gen. Virol. 791781-1791. [DOI] [PubMed] [Google Scholar]
- 30.Vareckova, E., S. A. Wharton, V. Mucha, M. Gocnik, and F. Kostolansky. 2003. A monoclonal antibody specific to the HA2 glycoprotein of influenza A virus hemagglutinin that inhibits its fusion activity reduces replication of the virus. Acta Virol. 47229-236. [PubMed] [Google Scholar]
- 31.Veits, J., A. Romer-Oberdorfer, D. Helferich, M. Durban, et al. 2008. Protective efficacy of several vaccines against highly pathogenic H5N1 avian influenza virus under experimental conditions. Vaccine 261688-1696. [DOI] [PubMed] [Google Scholar]
- 32.Velumani, S., Q. Du, B. J. Fenner, M. Prabakaran, L. C. Wee, L. Y. Nuo, and J. Kwang. 2008. Development of an antigen-capture ELISA for detection of H7 subtype avian influenza from experimentally infected chickens. J. Virol. Methods 147219-225. [DOI] [PubMed] [Google Scholar]
- 33.Wang, R., A. Song, J. Levin, D. Dennis, N. J. Zhang, H. Yoshida, L. Koriazova, L. Msadura, L. Shapiro, A. Matsumoto, H. Yoshida, T. Mikayama, R. T. Kubo, S. Sarawar, H. Cheroutre, and S. Kato. 2008. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antivir. Res. 80168-177. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289366-373. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2004. Laboratory biosafety manual, 3rd ed. World Health Organization, Geneva, Switzerland.
- 36.World Health Organization. 2005. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 111515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. 10 September 2007, posting date. Cumulative number of confirmed human cases of avian influenza A/ (H5N1). World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_01_03/en/index.html. Accessed 3 January 2008.
- 38.World Health Organization. 1995. Laboratory biosafety manual. Ann. Ist Super Sanita 311-121. [PubMed] [Google Scholar]
- 39.World Health Organization. 2006. Epidemiology of W. H. O. confirmed human cases of avian influenza A (H5N1) infection. Wkly. Epidemiol. Rec. 81249-257. [PubMed] [Google Scholar]
- 40.Yen, H. L., A. S. Monto, R. G. Webster, and E. A. Govorkova. 2005. Virulence may determine the necessary duration and dosage of oseltamivir treatment for highly pathogenic A/Vietnam/1203/04 influenza virus in mice. J. Infect. Dis. 192665-672. [DOI] [PubMed] [Google Scholar]
- 41.Yokoyama, W. M. 2001. Production of monoclonal antibody, p. 2.5.1-2.5.17. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., Newcastle, United Kingdom.
- 42.Zharikova, D., K. Mozdzanowska, J. Feng, M. Zhang, and W. Gerhard. 2007. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J. Virol. 796644-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]