Abstract
Open reading frame 45 (ORF45) of Kaposi's sarcoma-associated herpesvirus 8 (KSHV) is an immediate-early phosphorylated tegument protein and has been shown to play important roles at both early and late stages of viral infection. Homologues of ORF45 exist only in gammaherpesviruses, and their homology is limited. These homologues differ in their protein lengths and subcellular localizations. We and others have reported that KSHV ORF45 is localized predominantly in the cytoplasm, whereas its homologue in murine herpesvirus 68 is localized exclusively in the nucleus. We observed that ORF45s of rhesus rhadinovirus and herpesvirus saimiri are found exclusively in the nucleus. As a first step toward understanding the mechanism underlying the distinct intracellular distribution of KSHV ORF45, we identified the signals that control its subcellular localization. We found that KSHV ORF45 accumulated rapidly in the nucleus in the presence of leptomycin B, an inhibitor of CRM1 (exportin 1)-dependent nuclear export, suggesting that it could shuttle between the nucleus and cytoplasm. Mutational analysis revealed that KSHV ORF45 contains a CRM1-dependent, leucine-rich-like nuclear export signal and an adjacent nuclear localization signal. Replacement of the key residues with alanines in these motifs of ORF45 disrupts its shuttling between the cytoplasm and nucleus. The resulting ORF45 mutants have restricted subcellular localizations, being found exclusively either in the cytoplasm or in the nucleus. Recombinant viruses were reconstituted by introduction of these mutations into KSHV bacterial artificial chromosome BAC36. The resultant viruses have distinct phenotypes. A mutant virus in which ORF45 is restricted to the cytoplasm behaves as an ORF45-null mutant and produces 5- to 10-fold fewer progeny viruses than the wild type. In contrast, mutants in which the ORF45 protein is mostly restricted to the nucleus produce numbers of progeny viruses similar to those produced by the wild type. These data suggest that the subcellular localization signals of ORF45 have important functional roles in KSHV lytic replication.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a DNA tumor virus and the causative agent of several human cancers, including Kaposi's sarcoma (KS), primary effusion lymphoma, and multicentric Castleman's disease (3, 6). Like all herpesviruses, KSHV has two alternative life cycles, a latent and a lytic cycle. During latency, only a few viral genes are expressed, and no progeny viruses are produced. Under appropriate conditions, latent viral genomes are activated, initiate lytic replication, and express a full panel of viral genes, in a process that leads to viral assembly, release of progeny virus particles, and de novo infection of naïve cells (3, 6). KSHV establishes latent infection in the majority of infected cells in cases of KS, primary effusion lymphoma, and multicentric Castleman's disease, but lytic replications occur in a small fraction. The recurrent and periodic lytic cycles of KSHV are believed to play critical roles in viral pathogenesis (6, 7).
Open reading frame 45 (ORF45) is a KSHV-encoded gene product that plays a critical role in the viral lytic cycle. It is an immediate-early protein and is also present in viral particles as tegument protein (26, 27, 30). Disruption of ORF45 has no significant effect on overall viral lytic gene expression or DNA replication in BAC36-reconstituted 293T cells induced with both tetradecanoyl phorbol acetate (TPA) and sodium butyrate together, but the ORF45-null mutant produces 5- to 10-fold fewer progeny viruses than the wild type and the mutant virus has dramatically reduced infectivity, suggesting that ORF45 plays important roles at both early and late stages of viral infection (29). In addition to its roles as a tegument component, which are possibly involved in viral ingress and egress processes, KSHV ORF45 interacts with cellular proteins and modulates the cellular environment. At least two such functions have been described. First, KSHV ORF45 inhibits activation of interferon regulatory factor 7 (IRF-7) and therefore antagonizes the host innate antiviral response (28). Second, KSHV ORF45 interacts with p90 ribosomal kinase 1 and 2 (RSK1/RSK2) and modulates the extracellular signal-regulated kinase/RSK signaling pathway, which is known to play essential roles in KSHV reactivation and lytic replication (12). All of these data suggest that KSHV ORF45 is a multifunctional protein.
ORF45 is unique to the gammaherpesviruses; it has no homologue in the alpha- or betaherpesviruses. ORF45 homologues have been identified as virion protein components in other gammaherpesviruses, such as Epstein-Barr virus (EBV), rhesus rhadinovirus (RRV), and murine herpesvirus 68 (MHV-68), suggesting that certain tegument functions of ORF45 are conserved (2, 11, 18). ORF45 homologues differ in protein length. KSHV ORF45 is the longest, at 407 amino acids (aa); RRV, EBV, MHV-68, and herpesvirus saimiri (HVS) have proteins of 353, 217, 206, and 257 aa, respectively. The limited homologies lie mostly at the amino- and carboxyl-terminal ends. The middle portion of KSHV ORF45 diverges from those of its homologues. The homologues differ in subcellular localization. We and others have reported previously that KSHV ORF45 is found predominantly in the cytoplasm (1, 21, 28, 30), whereas ORF45 of MHV-68 is found exclusively in the nucleus (9). Recently, we found KSHV ORF45 also present in the nuclei of BCBL-1 cells in what resembled viral replication compartments, suggesting that ORF45 could shuttle into the nucleus (12).
Nucleocytoplasmic trafficking of proteins across the nuclear membrane occurs through nuclear pore complexes. Small molecules of up to approximately 9 nm in diameter, corresponding to a globular protein of approximately 40 to 60 kDa, can in principle enter or leave the nucleus by diffusion through nuclear pores (15, 17, 24). Large molecules are transported with the aid of a related family of transport factors, importins and exportins, which recognize nuclear localization sequence (NLS)-containing or nuclear export sequence (NES)-containing proteins (15, 17, 23). CRM1 (exportin 1) has been identified as a common export receptor that recognizes human immunodeficiency virus Rev-like leucine-rich NES sequences and is responsible for the export of such NES-containing proteins (4, 5, 19, 22). CRM1-dependent nuclear export is specifically inhibited by a pharmacological compound, leptomycin B (LMB), that interacts with CRM1 and thus blocks such NES-mediated protein export (4).
To understand the mechanism underlying the distinct intracellular distribution of KSHV ORF45, we attempted to locate the signals that control its subcellular localization. In the research reported here, we identified a leucine-rich NES and an adjacent basic NLS in KSHV ORF45. We demonstrated that the regulated intracellular trafficking of ORF45, especially its translocation into the nucleus, is important for KSHV lytic replication.
MATERIALS AND METHODS
Plasmids.
We generated various enhanced green fluorescent protein (EGFP)-ORF45 fusion expression vectors by cloning PCR-amplified fragments in frame into the pEGFP-C vector (Clontech). In each construct, the coding sequence for each of the ORF45s from different gammaherpesviruses was fused to the coding sequence for the C-terminal end of GFP. The template for amplifying KSHV ORF45 is pCR3.1-ORF45, which has been described previously (28). EBV ORF45 was amplified from cDNA of butyrate-induced BC-1 cells (27). RRV ORF45 was amplified from a cosmid provided by Scott Wong, and HVS ORF45 was amplified from HVS viral DNA provided by Jae Jung. Similarly, the PCR product of each ORF45 was also cloned into pKH3, a mammalian expression vector kindly provided by Ian Macara, to generate three-hemagglutinin (3×HA)-tagged ORF45 expression plasmids. Various KSHV ORF45 truncation mutants were also created by PCR amplification and cloning into pEGFP-C. Diagrams of these constructs are shown in Fig. 2A. The p2×EGFP-C2 vector with two tandem EGFP sequences was constructed by insertion of a Klenow-blunted Ncol/BglII EGFP fragment into the SmaI site of pEGFP-C2 and then removal of the extra A (adenosine) upstream of the SalI site by QuikChange mutagenesis (Stratagene, La Jolla, CA). Annealed oligonucleotides encoding the NES (NES-BglII, 5′-GATCGAAGTATTGAGTCAGAGAATCGGGCTCATGGACG-3′; and NES-SalI, 5′-TCGACGTCCATGAGCCCGATTCTCTGACTCAATACTTC-3′), NES/NLS (NES/NLS-BglII, 5′-GATCGAAGTATTG AGTCAGAGAATCGGGCTCATGGACGTGGGCCAGAAGCGCAAAAGG G-3′; and NES/NLS-SalI, 5′-TCGACCCTTTTGCGCTTCTGGCCCACGTCCATGAGCCCGATTCTCTGACTCAATACTTC-3′), or NLS (NLS-BglII, 5′-GATCCAGAAGCGCAAAAGGCAGG-3′; and NLS-SalI, 5′-TCGACCTGCCTTTTGCGCTTCTG-3′) were inserted into p2×EGFP-C2 at the BglII and SalI sites to generate plasmids p2×EGFP-NES, p2×EGFP-NLS/NES, and p2×EGFP-NLS, respectively. Plasmids p2×EGFP-284-383 and p2×EGFP-238-283 were generated by cloning the corresponding PCR fragments into p2×EGFP-C2. Site-directed mutagenesis of ORF45 was carried out with a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Primer sequences used for cloning and mutagenesis are available upon request. All clones were verified by DNA sequencing.
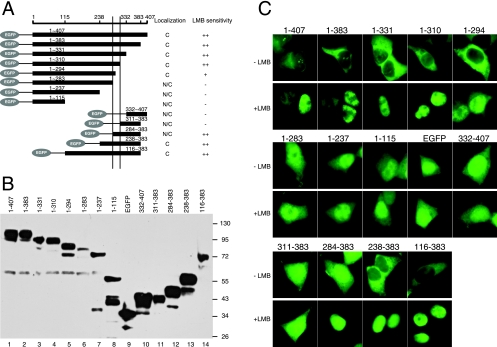
FIG. 2.
Mapping of subcellular localization signals of KSHV ORF45. (A) Schematic representation of KSHV ORF45 full-length protein and truncation mutants. HEK293T cells were transfected with EGFP-ORF45 fusion constructs and treated with LMB for 30 min 24 h after transfection. GFP was then visualized with fluorescence microscopy. The subcellular localizations and LMB sensitivities of EGFP-ORF45 mutants are summarized on the right. C, predominantly cytoplasmic; N, predominantly nuclear; ++, translocates into the nucleus within 30 min of LMB treatment; +, remains evenly distributed in both the cytoplasm and nucleus after 120 min of LMB treatment. (B) Western blot of the full-length and truncated EGFP-ORF45 fusion proteins. Cell lysates of HEK293T cells transfected with EGFP-ORF45 fusion constructs were analyzed by Western blotting with anti-GFP antibody. The sizes of molecular weight standards are shown on the right. (C) Representative images of full-length and truncated EGFP-ORF45 fusion proteins expressed in 293T cells in the presence (+LMB) and absence (−LMB) of LMB treatment.
Cell culture, transfection, and live fluorescence microscopy.
293T cells were cultured under 5% CO2 at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. Transient transfection of EGFP fusion plasmids was performed in 24-well plates with Effectene transfection reagent (Qiagen, Valencia, CA). Twenty-four hours after transfection, cells expressing EGFP-fused proteins were observed under a fluorescence microscope. In some cases, 10 ng/ml LMB (Sigma, St. Louis, MO) was added to the culture medium, and images were taken at the times indicated in Fig. 1, 2, and 3.
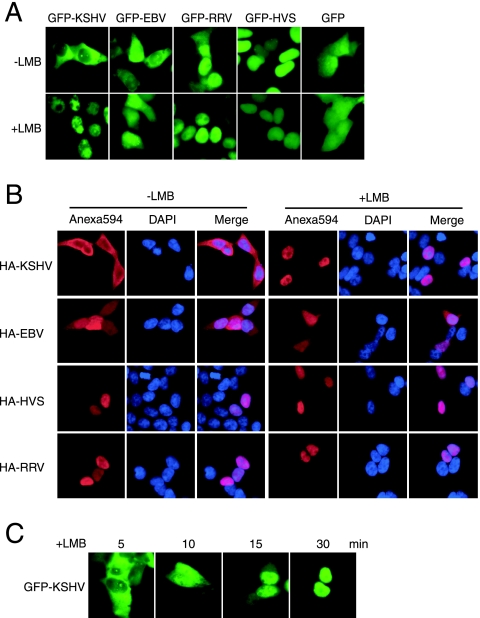
FIG. 1.
Different subcellular localization of KSHV ORF45 from that of its homologues in other gammaherpesviruses. (A) Subcellular localizations of KSHV ORF45 and its homologues. EGFP-fused ORF45 expression vectors were transfected into 293T cells. Green fluorescence was visualized 24 h after transfection. As indicated, some transfected cells were treated with 10 ng/ml LMB, which specifically inhibits CRM1-dependent nuclear export. (B) HA-tagged ORF45 expression vectors were transfected into 293T cells. The transfected cells were fixed, permeabilized, and immunofluorescently stained with anti-HA antibodies (red). The cells were also counterstained with DAPI to show the nucleus (blue). (C) KSHV ORF45 rapidly accumulates in the nucleus after LMB treatment. EGFP-fused KSHV ORF45 was visualized at different times, as indicated, after LMB treatment.
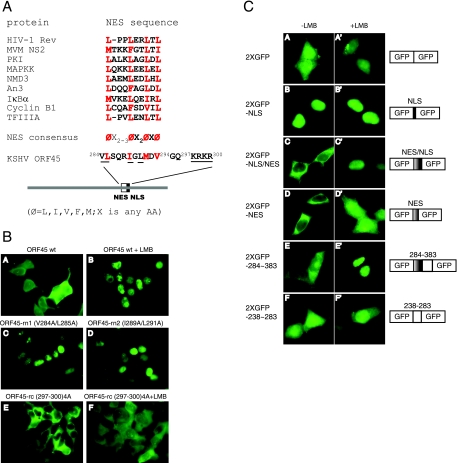
FIG. 3.
Identification of functional NES and adjacent NLS of KSHV ORF45. (A) Comparison of the putative ORF45 NES with other leucine-rich HIV-1 Rev-like NESs (13). Conserved hydrophobic residues (primarily leucines) are shown in red. In the NES consensus sequence, X is any amino acid and Φ is any hydrophobic residue, as indicated. A schematic representation of the ORF45 bipartite NES/NLS motif is shown at the bottom, with key amino acids highlighted. (B) Subcellular localization of EGFP-ORF45 wild type and mutants with altered NES or NLS sequences. The responses to LMB are also shown, as indicated for ORF45 wt and for ORF45-rc, whose NLS has been mutated. (C) Subcellular localization of 2×EGFP construct fused with KSHV ORF45 NLS and/or NES. The KSHV-ORF45 NLS and NES sequences were inserted in frame between two EGFPs. The constructs were transfected into HEK293T cells. Twenty-four hours after transfection, the cells were treated with LMB for 30 min. GFP was then visualized with fluorescence microscopy. Representative images and schematic illustration of these constructs are shown.
Indirect immunofluorescence staining.
Cells cultured on coverslips in 24-well plates were transfected with 3×HA-tagged ORF45 expression vectors. The transfected cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, permeabilized with 0.5% Triton X-100 for 5 to 10 min, blocked with 2% bovine serum albumin in PBS for 30 min, and then incubated with primary antibody (1 μg/ml) for 1 h. After three washes with PBS with 0.1% Triton X-100, the cells were incubated with Alexa 594-labeled secondary antibodies (Invitrogen, Carlsbad, CA) for 1 h. The cells were also counterstained with DAPI (4′,6-diamidino-2-phenylindole; Sigma, St. Louis, MO) to detect nuclei. The cells were then mounted in antifade agent (Invitrogen, Carlsbad, CA) and visualized with a fluorescence microscope.
Genetic manipulation of BAC36 with recombineering technology.
The strategy for introducing point mutations into BAC36 by homogeneous recombination has been described previously (29). The Escherichia coli strain EL350, carrying BAC-del45, in which the ORF45 coding sequence has been replaced with a Kan/SacB double selection cassette, was used as starting material. The EL350 strain contains a defective λ prophage that harbors the recombination genes exo, beta, and gam under tight control of a temperature-sensitive cl857 repressor. Recombination functions can be supplied transiently by shifting the culture to 42°C for 15 min (14). To generate a mutant with a desired mutation in the ORF45 coding sequence, we replaced the Kan/SacB cassette of BAC-del45 with the ORF45 coding sequence by homologous recombination. Two primers, the 78-bp primer ORF45wt5′ (5′-TTTC CGCCCCTAGCGGTCAACCCCGTACAAGGCCATGGCGATGTTTGT GAGGACCTCGTCTAGCACACACGATGAAGA-3′) and the 76-bp primer ORF45wt3′II (5′-GCATGAGACTTGACACCTATAATGGTCTGTATTGACACCATTCTTTTATTTATCAGTCCAGCCACGGCCAGTTATA-3′), were used to amplify the ORF45 coding region, with plasmids carrying the desired mutations as templates. PCR was carried out at 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min for 30 cycles, using Expand high-fidelity Taq polymerase (Roche, Indianapolis, IN). The PCR product was digested with DpnI to remove the plasmid template. The digested product was gel purified and electroporated into BAC36-del45-containing EL350 cells that had been induced at 42°C for 15 min. The parameters for electroporation were set at 1.75 kV, 276 Ω, and 50 μF in a 1-mm cuvette (BTX). After electroporation, all transformants were spread equally on 10 150-mm LB plates containing 12.5 μg/ml of chloramphenicol and 7% sucrose. The plates were incubated at 32°C overnight. Presumably as a result of repetitive sequences in the viral genome, many of the Kans sucrose-resistant colonies grew, but the vast majority of them had unexpected recombinations. To identify the clones with desired recombinations, we performed in situ colony hybridization to screen all colonies (about 50,000 or more), using the wild-type ORF45 coding sequence as a probe. We usually obtained fewer than 10 colonies with positive hybridization signals. The positive clones were expanded, and the bacterial artificial chromosome (BAC) DNAs were extracted and analyzed by PCR, restriction enzyme digestion, Southern blotting, and sequencing analyses. The BAC DNAs with the proper recombinations were prepared from overnight cultures with a Large-Construct kit (Qiagen, Valencia, CA).
Reconstitution of recombinant KSHVs.
Briefly, 293T cells seeded in a 60-mm dish were transfected with 2 μg of BAC DNAs, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Two days after transfection, cells were subcultured into a T150 flask with fresh medium containing 200 μg/ml hygromycin. After 8 days of selection, hygromycin-resistant colonies were trypsinized, pooled, and subcultured, with 1:3 dilution, every 3 days. To induce viral lytic replication, we seeded 3 × 107 BAC-containing 293T cells into a T150 flask, and 2 days later, we replaced the medium with fresh medium containing 20 ng/ml TPA and 0.3 mM butyrate.
Virus stock preparation and infection.
Usually, six or more T150 flasks of cells were induced for 4 to 5 days, and viruses were concentrated from the supernatant. The induced medium was collected and centrifuged to remove cell debris. The cleared supernatant was filtered through a 0.45-μm filter, and virions were pelleted at 100,000 × g for 1 h on a 25% sucrose cushion with a Beckman SW28 rotor. The virus pellets were dissolved in 1% of the original volume of PBS or Dulbecco's modified Eagle's medium and stored at −80°C. The viral genome copy number of stock virus was then quantified by real-time quantitative PCR. Infection was carried out as previously described (29). Briefly, 293T cells plated in 24-well plates were incubated with concentrated virus plus Polybrene (4 μg/ml) and spun at 800 × g for 1 h at room temperature. The plates were then incubated at 37°C for another 2 h, and the inocula were then removed and replaced with fresh medium with 5% FBS. The next day, the medium was replaced with fresh medium containing 1% FBS. The plates were examined with an inverted fluorescence microscope 48 h after infection for cells expressing GFP.
Real-time quantitative PCR analysis of virion DNA.
After TPA/butyrate induction, medium from induced BAC-293T cells was collected, centrifuged, and passed through a 0.45-μm filter to clear cell debris. Treatment of 200 μl of the cleared supernatant with 10 units of Turbo DNase (Ambion, Austin, TX) at 37°C for 1 h degraded extravirion DNAs. The reaction was stopped by the addition of EDTA followed by heat inactivation at 70°C. Twenty microliters of proteinase K solution and 200 μl of buffer AL from a DNeasy kit (Qiagen, Valencia, CA) were then added. The mixture was kept at 70°C for 15 min and then extracted with phenol-chloroform. The DNA was ethanol precipitated with glycogen as a carrier, and the DNA pellet was dissolved in 40 μl of Tris-EDTA buffer. Two microliters of such DNA was used in a real-time quantitative PCR with a LightCycler FastStart DNA Master Plus SYBR green kit as we described previously (29). Viral DNA copy numbers were calculated with external standards of known concentrations of BAC36 DNA.
Western blotting.
Cells were washed with PBS and lysed with whole-cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate [Na3VO4], 40 mM β-glycerophosphate, 30 mM sodium fluoride, 10% glycerol, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml of aprotinin, 5 μg/ml of leupeptin, 5 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride) to produce whole-cell lysates for most Western blot analyses. For the Western blot experiments described for Fig. 4C, the cytoplasmic and nuclear extracts were used instead. Briefly, HEK293T cells reconstituted with BAC36 or mutant BACs were induced with TPA and sodium butyrate together for 3 days. Cells were washed with cold PBS, resuspended in hypotonic lysis buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, protease inhibitor cocktail), and incubated on ice with gentle mixing from time to time for 15 min. The resulting suspensions were centrifuged at 1,500 × g for 15 min, and the supernatants were collected as cytoplasmic extracts. The pellets were washed once with hypotonic lysis buffer, resuspended in whole-cell lysis buffer, and cleared by high-speed centrifugation to produce nuclear extracts. The protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce Biotechnology, Rockford, IL). About 50 μg of cell extract of each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were blocked in 5% dried milk in PBS plus 0.2% Tween 20 and then incubated with diluted primary antibodies for 2 h at room temperature or 4°C overnight. Anti-rabbit or anti-mouse immunoglobulin G antibodies conjugated to horseradish peroxidase (Pierce Biotechnology, Rockford, IL) were used as the secondary antibodies. A Supersignal chemiluminescence system (Pierce Biotechnology, Rockford, IL) was used for detection of antibody-antigen complexes. Mouse antibodies against KSHV ORF45, K8, and ORF21 have been described before (26). Mouse monoclonal anti-RTA and anti-ORF65 were gifts from K. Ueda and S.-J. Gao, respectively. Rat anti-LANA antibody was purchased from Advanced Biotechnologies, Inc. (Columbia, MD). Mouse monoclonal anti-EGFP was purchased from Clontech Inc. (Mountain View, CA). Anti-HA and anti-β-actin were purchased from Sigma (St. Louis, MO).
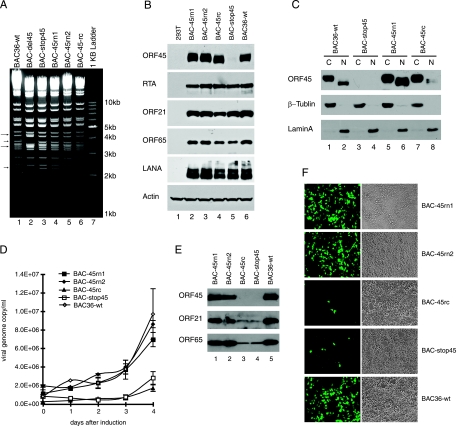
FIG. 4.
Role of ORF45 NES and NLS in KSHV replication. (A) Recombinant KSHV BACs with a mutated NES or NLS sequence in ORF45. The wild-type BAC36 and mutant KSHV BAC DNAs were constructed and prepared as described in Materials and Methods. The BAC DNAs were digested with KpnI, resolved in a 0.8% agarose gel, and stained with ethidium bromide. The arrows on the left mark the differences between NES/NLS mutants and their parental BAC-del45 construct, in which the ORF45 coding sequence is replaced with the double selection marker Kan/SacB. The sizes of molecular markers are shown on the right. (B) Expression of viral proteins. Recombinant viruses were reconstituted in 293T cells, and lytic replication was induced by TPA and sodium butyrate. Cell lysates were analyzed by Western blotting with specific antibodies against KSHV viral proteins. The blot was also probed with anti-β-actin as a loading control. (C) Subcellular localization of ORF45 in virus-infected cells. HEK293T cells reconstituted with wild-type BAC36 or mutant BACs were induced with TPA and sodium butyrate for 3 days. The nuclear and cytoplasmic extracts were prepared and analyzed by Western blotting to detect ORF45. The blot was also probed to detect lamin A and β-tubulin as nuclear and cytoplasmic fraction markers, respectively. (D) Growth curves for recombinant KSHV BAC viruses. Supernatants of induced medium were collected at different time points as indicated. Viral genome copies in the medium were determined by real-time PCR and were plotted against the time since induction. (E) Western blot analysis of virion lysates. The virions secreted into the medium 4 days after induction were collected, purified, and analyzed by Western blotting for virion proteins ORF45, ORF21 (tegument), and ORF65 (capsid). (F) Infection of 293T cells by reconstituted recombinant viruses. Concentrated extracellular viruses were used to infect 293T cells. The infected cells expressed GFP and appeared green when examined by fluorescence microscopy.
RESULTS
Difference in subcellular localization of KSHV ORF45 from its homologues in other gammaherpesviruses.
We and others have observed that the KSHV ORF45 protein is localized primarily in the cytoplasm (1, 21, 28, 30) (Fig. 1A), whereas its homologue MHV-68 ORF45 is seen mostly in the nucleus (9). To examine the subcellular localization of ORF45s encoded by other gammaherpesviruses, we tagged each ORF45 with EGFP and examined the fusion protein in living transfected 293T cells by fluorescence microscopy. Both RRV and HVS ORF45s were seen exclusively in the nucleus, whereas the EBV ORF45 homologue BKRF4 was present in both the cytoplasm and nucleus (Fig. 1A). Fusion of EGFP to either the N or C terminus of KSHV ORF45 resulted in the same subcellular localization of the native protein, as revealed by immunofluorescence assay with specific anti-KSHV ORF45 antibodies (28, 30). Because antibodies to other ORF45s were not available, each ORF45 from different gammaherpesviruses was tagged with HA and its subcellular localization was examined by immunofluorescence assay with anti-HA antibody. In all cases, HA-tagged and EGFP-tagged proteins showed identical localizations, suggesting that the distinct subcellular localization is an intrinsic characteristic of the ORF45 protein itself and is not affected by the EGFP or HA tag (Fig. 1B).
Recently, we observed that KSHV ORF45 in induced BCBL-1 cells was localized predominantly in the cytoplasm but that a portion accumulated in the nucleus (12). We therefore speculated that KSHV ORF45 could shuttle between the nucleus and cytoplasm. To determine whether it can, we treated EGFP-ORF45-transfected cells with LMB, which specifically inhibits CRM1-dependent nuclear export. We found that KSHV ORF45 accumulated rapidly in the nucleus after LMB treatment (Fig. 1C). This result indicates that KSHV ORF45 can shuttle between the cytoplasm and nucleus and possibly contains a CRM1-dependent NES.
Control of subcellular localization by signals located between aa 284 and 310 of KSHV ORF45.
To identify the sequence elements that control KSHV ORF45 subcellular localization, we constructed a series of EGFP-fused ORF45 truncation mutants (Fig. 2A). Western blot analyses revealed that all constructs were expressed and were largely in line with their expected sizes (Fig. 2B). Progressive truncations up to aa 310 from the carboxyl terminus had no effect on the cellular localization of EGFP-ORF45 fusion proteins. All of these EGFP fusion mutants, including those with aa 1 to 383, 1 to 331, and 1 to 310, were predominantly cytoplasmic in untreated cells. After LMB treatment, these truncated EGFP-ORF45 fusion mutants accumulated in the nucleus (Fig. 2A and C), indicating that the putative NES element was localized to the N-terminal 310 aa. Truncation from aa 310 to 294 did not affect the cytoplasmic localization, suggesting that the NES remained intact, but the nuclear accumulation of the remaining residues, i.e., aa 1 to 294, became less sensitive to LMB treatment (Fig. 2A and C). Further truncation from aa 294 to 284 drastically changed the subcellular localization, and the resultant mutant with aa 1 to 283 was not restricted to the cytoplasm but evenly distributed in both the cytoplasm and nucleus, like EGFP itself. Smaller truncation mutants with aa 1 to 237 and 1 to 115 displayed a similar pattern to that for aa 1 to 283. These results suggest that the region between aa 284 and 294 confers ORF45's cytoplasmic localization and possibly contains an NES. The region between aa 294 and 310 seems to increase nuclear localization, and deletion of this region results in less efficient nuclear accumulation. These results could be explained by the existence of an NLS in this region. Results from N-terminal progressive truncations support this interpretation. Deletion of up to 238 aa did not affect ORF45's predominant cytoplasmic localization. EGFP-ORF45 fusion mutants containing aa 116 to 383 and 238 to 383 were both found mostly in the cytoplasm and were highly sensitive to LMB treatment, so they may both contain a functional NES and NLS. The mutant with aa 284 to 383 was seen in both the cytoplasm and the nucleus even though it contains a putative NES, presumably because its small size enables it to pass through nuclear pores. Indeed, when it was fused with two tandem EGFP molecules, the fusion protein became localized predominantly in the cytoplasm (Fig. 3C). Although other possibilities might exist, for example, the neighboring sequence could regulate the NES activity, its high sensitivity to LMB suggests that the region of aa 284 to 383 preserves a functional NES. Other mutants that do not have the region between aa 284 and 310, including mutants with aa 311 to 383 and 332 to 407, show diffusive patterns like that of EGFP and are not sensitive to LMB treatment. In conclusion, the region of aa 284 to 310 of KSHV ORF45 contains important signals that control its subcellular localization.
Identification of a leucine-rich NES and an adjacent NLS in KSHV ORF45.
Inspection of the sequence in the region revealed that residues 284VLSQRIGLMDV294 contain a stretch of hydrophobic amino acids resembling an NES consensus (ΦX2-3ΦX2ΦXΦ [Φ represents any hydrophobic amino acid, such as leucine, isoleucine, valine, tryptophan, or methionine, whereas X represents any amino acid]) (Fig. 3A) (13, 23), and the region of aa 297 to 300 contains a typical basic-residue-rich NLS signal, 297KRKR300 (16). To determine whether residues aa 284 to 294 are a functional NES, we introduced point mutations into this region to replace the conserved hydrophobic residues with alanines. Replacement of valine 284 and leucine 285 with alanines within the context of full-length ORF45 led to nuclear accumulation of the mutated EGFP-ORF45 (V284A/L285A) fusion protein (Fig. 3B, panel C). Similarly, changing isoleucine 289 and leucine 291 to alanines had a similar result, and the resultant EGFP-ORF45 (I289A/L291A) protein was seen exclusively in the nucleus (Fig. 3B, panel D). These results indicate that these hydrophobic residues are important for nuclear export of ORF45. These two mutants were designated ORF45-rn1 (restricted to the nucleus) and ORF45-rn2. To determine whether the adjacent basic residues (297KRKR300) are a functional NLS, we replaced the basic residues with alanines. We found that the resulting fusion protein was restricted to the cytoplasm and diffusely distributed in both the cytoplasm and nucleus even after LMB treatment (Fig. 3B, bottom panels). This mutant was designated ORF45-rc (restricted to the cytoplasm).
To determine whether 297KRKR300 and 284VLSQRIGLMDV294 function as an authentic NLS and NES, respectively, we fused corresponding coding sequences in frame between two EGFPs and examined the subcellular localization of the fusion protein. The 2×EGFP construct alone was present in both the nucleus and cytoplasm (Fig. 3C, panel A). The addition of the NLS 297KRKR300 to the 2×EGFP construct resulted in a 2×EGFP-NLS fusion protein that was exclusively nuclear (Fig. 3C, panel B), suggesting that 297KRKR300 is a functional NLS. Fusion of the NES to the 2×EGFP construct resulted in a predominantly cytoplasmic 2×EGFP-NES fusion protein, suggesting that 284VLSQRIGLMDV294 is a functional NES (Fig. 3C, panel D). Inclusion of both the NES and NLS resulted in a mostly cytoplasmic 2×EGFP-NES/NLS fusion protein that rapidly accumulated in nuclei after LMB treatment, suggesting that 284VLSQRIGLMDV294 has authentic NES activity that counteracts the NLS activity (Fig. 3C, panel C). Fusion of KSHV ORF45 aa 284 to 383, which contain the NES and NLS signals, to the 2×EGFP construct gave similar results (Fig. 3C, compare panels E and C). However, insertion of aa 238 to 283, which lack the NES or NLS signal, into the 2×EGFP construct resulted in a pancellular fusion protein that was not responsive to LMB treatment (Fig. 3C, panel F).
Functional roles of ORF45 NES and NLS in the KSHV lytic life cycle.
Next, we examined the effect of alteration of ORF45 subcellular localization on KSHV lytic replication. Mutations were introduced into the KSHV viral genome BAC36 by recombineering as previously described (25, 29). The clones with expected recombination were selected on a kanamycin- and sucrose-containing plate and then subjected to in situ hybridization and further analyzed by restriction enzyme analysis (Fig. 4A). All of these mutants showed KpnI digestion patterns similar to that of wild-type BAC36, which generated two fragments, of 3.7 kb and 2.2 kb, but distinct from that of parental BAC-del45, which yielded two upshifted bands, of 4.3 kb and 3.4 kb (Fig. 4A), so proper recombination was present at the expected locus. Direct sequencing of each clone also confirmed the expected mutations (data not shown). These BAC DNAs were transfected into 293T cells, which were then subjected to hygromycin selection. The pooled hygromycin-resistant and GFP-positive cells were induced with TPA/butyrate, which initiated lytic viral replication. Whole-cell lysates were prepared and analyzed for expression of several viral proteins. Except for ORF45-null BAC-stop45 cells, which did not express ORF45 as expected, other ORF45 NES or NLS mutants expressed comparable levels of viral proteins, including ORF45, RTA (replication and transcription activator), ORF21, ORF65, and LANA (latency-associated nuclear antigen), to those in BAC36-wt cells (Fig. 4B). We next examined whether the mutant viruses had the expected ORF45 subcellular localizations. Because the KSHV BAC-reconstituted 293T cells adhered very poorly to the culture surface, we had difficulty in performing immunofluorescence staining to visualize the subcellular localization of ORF45. Instead, we isolated cytoplasmic and nuclear extracts of the induced cells and analyzed them by Western blotting. As shown in Fig. 4C, a stronger signal was detected in the cytoplasmic fraction of BAC36-wt 293T cells than in the nuclear one, confirming the mainly cytoplasmic localization of KSHV ORF45 (Fig. 4C, compare lanes 2 and 1). Also, as expected, ORF45 was not expressed in the BAC-stop45 293T cells (Fig. 4C, lanes 3 and 4). ORF45 was equally detected in both cytoplasmic and nuclear fractions of cells reconstituted with BAC-45rn1 (Fig. 4C, compare lanes 5 and 6) or BAC-45rn2 (data not shown), suggesting that the BAC-45rn mutants became more nuclear than the wild type because of the loss of a functional NES. In contrast, the signal of ORF45 was much stronger in the cytoplasmic extract than in the nuclear one for cells reconstituted with BAC-45rc, in which the NLS was destroyed, confirming its more cytoplasmic phenotype. These results largely confirmed the expected subcellular localizations of wild-type ORF45 and its mutants. Interestingly, the nuclear and cytoplasmic ORF45 proteins apparently differ in mobility in sodium dodecyl sulfate-polyacrylamide gels, suggesting that they are differently posttranslationally modified. The difference in electrophoretic mobility was caused at least partially by phosphorylation (unpublished data). Having confirmed the expression and subcellular localization of each ORF45 mutant in the reconstituted cells, we next examined the growth curve of each virus. Extracellular viral particles were collected from induced cells, and the viral genome copies were determined by real-time PCR. Single-step growth curve experiments showed that the BAC-45rn1 and BAC-45rn2 mutants, in which the NES was nonfunctional and the ORF45 protein was therefore restricted to the nucleus, had growth curves similar to that of BAC36-wt. In contrast, the BAC-45rc mutant, in which the NLS was destroyed, appeared to have a growth defect like that of ORF45-null BAC-stop45 (Fig. 4D). Viruses collected 4 days after induction were analyzed by Western blotting for virion proteins ORF45 (tegument), ORF21 (tegument), and ORF65 (capsid), and significantly less virion protein was found in the BAC-stop45 and BAC-45rc preparations (Fig. 4E), confirming the lower yields of these two mutants. The viruses collected were also used to infect 293T cells, and infectivity was judged by the appearance of green fluorescent cells upon inoculation of fresh 293T cells. In agreement with the titration by real-time PCR and Western blotting of virion proteins, we found that BAC-45rn1 and BAC-45rn2 produced numbers of infectious viruses similar to that for wild-type BAC36. However, we found that BAC-45rc as well as ORF45-null BAC-stop45 produced significantly fewer infectious viruses (Fig. 4F). These results suggested that the NES and NLS have functional roles in KSHV lytic replication.
DISCUSSION
The difference between the subcellular localizations of KSHV ORF45 (mainly cytoplasmic) and MHV-68 ORF45 (mainly nuclear) prompted us to examine ORF45s of other gammaherpesviruses. We tagged several ORF45 homologues with EGFP or HA and determined the subcellular localizations of the EGFP fusion proteins in 293T cells. We found only KSHV ORF45 to be exclusively cytoplasmic; its homologues from other gammaherpesviruses were either exclusively nuclear (RRV and HVS) or evenly distributed in both the cytoplasm and nucleus (EBV). We recently observed a portion of KSHV ORF45 in the nucleus in BCBL-1 cells (12). Presence in the nucleus therefore seems to be common for ORF45s of gammaherpesviruses. In order to understand the mechanism of the distinct subcellular localization of KSHV ORF45, we performed detailed mutational analysis to identify the signals that control its subcellular localization. Analysis of the progressive truncation mutants revealed that the region between aa 284 and 310 is critical for KSHV ORF45 subcellular localization. Examination of the sequence revealed a leucine-rich-like NES (284VLSQRIGLMDV294) and an adjacent NLS (297KRKR300) in this region. The exclusively cytoplasmic localization of KSHV ORF45 is mediated mainly by this CRM1-dependent NES. Treatment with the CRM1-specific inhibitor LMB or replacement of the NES consensus hydrophobic amino acids with alanines disrupted KSHV ORF45 nuclear export and resulted in its rapid nuclear accumulation. This NES is transferable because it can direct nuclear export of 2×GFP (Fig. 3C, compare panels D and A). The adjacent sequence (297KRKR300) seems to be an authentic NLS signal. It is also transferable and is sufficient to convert 2×GFPs from pancellular to completely nuclear localization (Fig. 3C, compare panels B and A). This NLS is important for nucleus-to-cytoplasm shuttling of ORF45 because replacement of the basic residues with alanines resulted in drastically reduced efficiency of nuclear accumulation in the presence of LMB (Fig. 3B, panels E and F).
The common nuclear presence of KSHV ORF45 and its homologues suggests a conserved function in the nucleus. This nuclear function seems to be important for viral replication, possibly at the late stage, because both BAC-45rn1 and BAC-45rn2 had growth curves similar to that of BAC36-wt, but BAC-45rc grew similarly to the ORF45-null BAC-stop45. Previous studies have shown that KSHV ORF45 has important functions at both early and late stages of viral replication, but its exact roles remained to be elucidated (29). Among all other gammaherpesvirus ORF45s, only MHV-68 ORF45 has been characterized (9, 10). By small interfering RNA-based ablation of gene expression or BAC-based mutagenesis, Jia et al. (9, 10) demonstrated that MHV-68 ORF45 is essential for MHV-68 replication, but again the exact function was not demonstrated. Interestingly, they showed that ectopic expression of an NLS deletion mutant of ORF45 could partially rescue ORF45-null MHV-68, suggesting that the NLS is not absolutely essential for MHV-68 ORF45. This result seems to contradict our observation that a functional KSHV ORF45 NLS is important for viral replication. Because the MHV-68 ORF45 protein is so small (206 aa), it could possibly cross the nuclear pore complexes by diffusion even in the absence of a functional NLS. Importantly, Jia et al. (9) also demonstrated that KSHV ORF45 could partially rescue an MHV-68 ORF45-deficient mutant, suggesting that a certain nuclear function is conserved among gammaherpesviruses.
What is the possible function of KSHV ORF45 in the nucleus? It is a relatively abundant tegument protein located between the capsid and virion envelope in the virion particle. Early evidence suggests that KSHV ORF45 is located in the inner layer of the tegument and is tightly associated with the capsid (26, 30). A recent virion-wide interaction study determined that ORF45 interacts with capsid protein ORF62, a triplex component (20). Genetic analysis revealed that ORF45 could have a role in the late stage of virion maturation that takes place after viral DNA replication (29). In BCBL-1 cells, a portion of ORF45 was found to be colocalized with the viral replication compartment where viral DNA replication and initial capsid assembly occur (12). These observations suggest that ORF45 could be one of the tegument proteins that are acquired in the nucleus soon after capsid assembly. The association of ORF45 with the capsid is probably important to subsequent virus maturation processes. Further studies are needed to clarify the role of ORF45 in this late stage of viral replication.
In addition to its viral functions as a tegument component, KSHV ORF45 interacts with a number of cellular proteins and has important roles in modulation of the host cellular environment. Two cellular functions have been reported for KSHV ORF45, including (i) inhibition of IRF-7 activation and evasion of the host antiviral response (28) and (ii) activation of RSK1/RSK2 and modulation of the extracellular signal-regulated kinase mitogen-activated protein kinase signaling pathway (12). Our preliminary data suggested that alternation of the subcellular localization of KSHV ORF45 has little effect on its binding to IRF-7 or to RSK1/RSK2. All NES and NLS mutants were still able to inhibit IRF-7 activation in a luciferase reporter assay (data not shown). These mutants retained the ability to activate RSK1 and RSK2 when they were overexpressed in cells (data not shown), but a change of subcellular localization may result in a difference in the local concentration of ORF45 that could have a dramatic functional consequence and may not be revealed by gross activity assays.
KSHV ORF45 is the only one of the gammaherpesvirus ORF45 proteins that appears exclusively in the cytoplasm, suggesting that a relatively strong NES exists in the KSHV ORF45 sequence. Mutation of the key hydrophobic residues in the KSHV ORF45 NES drastically changes its predominantly cytoplasmic localization to a nuclear localization. Fusion of KSHV ORF45 NES/NLS or NES to 2×GFP resulted in an exclusively cytoplasmic localization (Fig. 3C), further confirming that the 284VLSQRIGLMDV294 sequence possesses authentic and transferable NES activity. The leucine-rich NES is known to be subject to various regulations (13). The mechanism by which subcellular localization is regulated and whether it is regulated over the course of KSHV infection will be interesting to determine. Upon KSHV infection of human fibroblast cells followed by RTA-adenovirus infection, ORF45 was seen throughout the cell at 8 h postinfection and then was predominantly nuclear at 24 h, suggesting that ORF45 relocates over the course of infection (Britt Glaunsinger, personal communication). Phosphorylation of residues in the vicinity of the NES and NLS may regulate the intracellular distribution of a protein (8, 13). ORF45 is rich in serine and threonine residues and is heavily phosphorylated in cells (30). The slower electrophoretic mobility of the cytoplasmic fraction suggests that ORF45 is posttranslationally modified (including possible phosphorylation) more extensively in the cytoplasm than in the nucleus. Whether the subcellular localization of ORF45 is regulated by phosphorylation or other modifications remains to be determined. The only known kinases that phosphorylate ORF45 are RSK1 and RSK2, but the major phosphorylation sites of RSKs lie in the region of aa 1 to 115, which is dispensable for the cytoplasmic-dominant localization (12), so the RSKs are unlikely to be involved directly in the regulation of ORF45 subcellular localization.
KSHV ORF45 is larger in size than its homologues. The middle portion of the KSHV ORF45 amino acid sequence, including the NES/NLS and the possible regulatory region, seems to be “extra” and has no homology to other proteins of gammaherpesviruses. The putative basic-residue-rich NLS can be found at different positions in ORF45s of all gammaherpesviruses, suggesting that a common nuclear function is conserved and possibly required for efficient viral lytic replication and progeny virus production. Although the NES seems dispensable for KSHV lytic replication, it might be important for certain ancillary functions gained by KSHV during evolution that could facilitate its interaction with the host. The different modifications of nuclear and cytoplasmic fractions further support the hypothesis that ORF45 has distinct functions in the nucleus and the cytoplasm. Nevertheless, identification of the subcellular localization signals of ORF45 opens an avenue to further characterize the functions of this protein in the KSHV life cycle.
Acknowledgments
We thank Shou-Jiang Gao, Jae Jung, Ian Macara, Robert Ricciardi, Keiji Ueda, Scott Wong, and Yan Yuan for kindly providing reagents. We thank Joseph Gillen and other members of the Zhu laboratory for critical readings of the manuscript and for helpful discussions. We also thank Anne B. Thistle at the Florida State University for excellent editorial assistance.
This work was supported by National Institutes of Health grant R01DE016680, a Bankhead-Coley bridge grant, an FSU setup fund, and a planning grant to F.Z.
Footnotes
Published ahead of print on 30 December 2008.
REFERENCES
- 1.Abada, R., T. Dreyfuss-Grossman, Y. Herman-Bachinsky, H. Geva, S. R. Masa, and R. Sarid. 2008. SIAH-1 interacts with the Kaposi's sarcoma-associated herpesvirus-encoded ORF45 protein and promotes its ubiquitylation and proteasomal degradation. J. Virol. 822230-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bortz, E., J. P. Whitelegge, Q. Jia, Z. H. Zhou, J. P. Stewart, T. T. Wu, and R. Sun. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 7713425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390308-311. [DOI] [PubMed] [Google Scholar]
- 6.Ganem, D. 2007. Kaposi's sarcoma-associated herpesvirus, p. 2847-2888. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1273-296. [DOI] [PubMed] [Google Scholar]
- 8.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76651-685. [DOI] [PubMed] [Google Scholar]
- 9.Jia, Q., V. Chernishof, E. Bortz, I. McHardy, T. T. Wu, H. I. Liao, and R. Sun. 2005. Murine gammaherpesvirus 68 open reading frame 45 plays an essential role during the immediate-early phase of viral replication. J. Virol. 795129-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia, Q., T. T. Wu, H. I. Liao, V. Chernishof, and R. Sun. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J. Virol. 786610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuang, E., Q. Tang, G. G. Maul, and F. Zhu. 2008. Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi's sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 821838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutay, U., and S. Guttinger. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15121-124. [DOI] [PubMed] [Google Scholar]
- 14.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 15.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67265-306. [DOI] [PubMed] [Google Scholar]
- 16.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 2434-36. [DOI] [PubMed] [Google Scholar]
- 17.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386779-787. [DOI] [PubMed] [Google Scholar]
- 18.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 801574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278141-144. [DOI] [PubMed] [Google Scholar]
- 20.Rozen, R., N. Sathish, Y. Li, and Y. Yuan. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 824742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. Schulz, F. Neipel, and M. Sturzl. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 821908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 901041-1050. [DOI] [PubMed] [Google Scholar]
- 23.Tran, E. J., T. A. Bolger, and S. R. Wente. 2007. SnapShot: nuclear transport. Cell 131420. [DOI] [PubMed] [Google Scholar]
- 24.Wang, R., and M. G. Brattain. 2007. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 5813164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 766185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 735556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 995573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu, F. X., X. Li, F. Zhou, S. J. Gao, and Y. Yuan. 2006. Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J. Virol. 8012187-12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu, F. X., and Y. Yuan. 2003. The ORF45 protein of Kaposi's sarcoma-associated herpesvirus is associated with purified virions. J. Virol. 774221-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]