Abstract
Although there is increasing evidence that individuals already infected with human immunodeficiency virus type 1 (HIV-1) can be infected with a heterologous strain of the virus, the extent of protection against superinfection conferred by the first infection and the biologic consequences of superinfection are not well understood. We explored these questions in the simian immunodeficiency virus (SIV)/rhesus monkey model of HIV-1/AIDS. We infected cohorts of rhesus monkeys with either SIVmac251 or SIVsmE660 and then exposed animals to the reciprocal virus through intrarectal inoculations. Employing a quantitative real-time PCR assay, we determined the replication kinetics of the two strains of virus for 20 weeks. We found that primary infection with a replication-competent virus did not protect against acquisition of infection by a heterologous virus but did confer relative control of the superinfecting virus. In animals that became superinfected, there was a reduction in peak replication and rapid control of the second virus. The relative susceptibility to superinfection was not correlated with CD4+ T-cell count, CD4+ memory T-cell subsets, cytokine production by virus-specific CD8+ or CD4+ cells, or neutralizing antibodies at the time of exposure to the second virus. Although there were transient increases in viral loads of the primary virus and a modest decline in CD4+ T-cell counts after superinfection, there was no evidence of disease acceleration. These findings indicate that an immunodeficiency virus infection confers partial protection against a second immunodeficiency virus infection, but this protection may be mediated by mechanisms other than classical adaptive immune responses.
Superinfection with human immunodeficiency virus type 1 (HIV-1) is the infection of an HIV-seropositive individual with a second heterologous strain of the virus after infection with the first infecting strain is established. There is accruing evidence for HIV-1 intra- and intersubtype superinfection in settings of intravenous drug use, structured treatment interruptions, and with strains that are resistant to antiretroviral drugs (2, 4, 6, 22, 26, 28, 32, 39, 42, 43, 52, 60, 66). Epidemiologic studies have suggested that the frequency of superinfection ranges from rare to as high as 5% per year in high-risk populations (9, 10, 15, 20, 24, 27, 31, 40, 41, 51, 59, 65, 67). However, it remains unclear how readily superinfections occur after exposure of an infected individual to a heterologous strain of virus. Furthermore, the variables that may contribute to susceptibility or resistance to superinfection, such as the timing of exposure to a second virus or the immunologic status of the exposed individual, have not been well defined. It is also uncertain whether superinfection is invariably associated with the loss of HIV containment and clinical deterioration (8, 17, 21, 23, 26, 27, 30, 60). Understanding the risks for and the biological consequences of HIV superinfection will not only clarify an important clinical problem, it may also provide important insights into the nature of the immune responses that may confer protection against the initial acquisition of HIV.
The nonhuman primate model provides an ideal means of studying the pathogenesis of HIV-1 superinfection. This system allows for control of many important variables, including the dose, strain, route, and timing of infection. However, there have only been a few animal studies that have attempted to explore the biology of superinfection. The implications of these studies are uncertain because they have been done in models in which infected monkeys do not develop AIDS and the viruses used are either replication incompetent or replicate at low levels (11-13, 18, 36-38, 46-48, 53, 56-58, 61-64). Therefore, it is unclear whether we can extrapolate from these studies the frequency HIV-1 superinfection, the implications of superinfection on HIV pathogenesis, and the feasibility of inducing broadly cross-protective immune responses.
In the present study, we have developed a rhesus monkey model of mucosal superinfection to examine whether infection with replication-competent simian immunodeficiency virus (SIV) confers a relative resistance to superinfection and elucidate the factors that influence the clinical course of infection with a second virus. We show that although prior infection with SIV does not protect against subsequent mucosal challenge with a heterologous SIV isolate, the primary infection does attenuate the replication capacity of the second virus.
MATERIALS AND METHODS
Animals.
Fourteen adult rhesus monkeys (Macaca mulatta) were used in this study. All animals were housed at Bioqual (Rockville, MD) and maintained in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care guidelines at the National Institutes of Health.
SIV challenge stocks.
The viruses used in this study included cell-free uncloned pathogenic SIVmac251 and pathogenic SIVsmE660 (kindly provided by Vanessa Hirsch, NIAID/NIH). The stock of SIVmac251 was expanded on human peripheral blood mononuclear cells (PBMCs), and the stock of SIVsmE660 was expanded on rhesus monkey PBMCs. To initiate intravenous infections, 2.1 × 105 RNA copies of SIVmac251 and SIVsmE660 were used. Doses of 6.3 × 107 RNA copies of SIVmac251 and 4.3 × 108 copies of SIVsmE660 were used for the intrarectal exposures. These were doses that were previously shown to reproducibly initiate mucosal infections in rhesus monkeys (29).
qRT-PCR.
Plasma SIVmac251 and SIVsmE660 RNA levels were determined using a two-step quantitative real-time reverse transcription-PCR (qRT-PCR) assay. Four sets of strain-specific probes and primers for gag and env were used to distinguish and quantify SIVmac251 and SIVsmE660. Viral RNA was extracted and purified from plasma using the QIAmp viral RNA minikit (Qiagen, Valencia, CA). RNAs were subjected to RT with MultiScribe reverse transcriptase (Applied Biosystems, Foster City, CA) to generate cDNA products for quantitative PCR using the env RT primer, 5′-GAACCCTAGCACAAAGACCCC-3′, and the gag RT primer, 5′-GGTGCAGCAAATCCTCT-3′. These primers were designed to anneal to conserved regions of gag and env that are shared by the two viral strains.
The subsequent qRT-PCRs were set up using TaqmanGold Mastermix (Applied Biosystems, Foster City, CA). cDNAs were amplified with SIVsmE660 TaqMan env and gag probes that were labeled with 6-carboxyfluorescein (FAM) and quencher dye BHQTM1, while the SIVmac251 env and gag TaqMan probes were labeled with Quasar 670 and quencher dye BHQTM2 (Biosearch Technologies, Novato, CA). For each sample, analyses for SIVmac251 and SIVsmE660 were conducted separately for both env and gag. The sequences and annealing temperatures for primers and probes are outlined in Table 1.
TABLE 1.
Primers and probes for quantitation of viral load by PCR
| Strain and gene | Primer or probe | Sequence | Temp (°C) |
|---|---|---|---|
| SIVmac251 | |||
| gag | Forward primer | 5′-TTCGGTCTTAGCTCCATTAGTG-3′ | 62 |
| Reverse primer | 5′-AGTTACCACCTATTTGTTGTACTG-3′ | ||
| Probe | 5′-(Quasar)CTCCTCTGCCGCTAGATGGTGCTG(BHQTM2)-3′ | ||
| env | Forward primer | 5′-CCAAGAGAGGGAGACCTCA-3′ | 56 |
| Reverse primer | 5′-CCAAGCCAATCGGAGTGAT-3′ | ||
| Probe | 5′-(Quasar)ACTCCACAGTGACCAGTCTCATAGCA(BHQTM2)-3′ | ||
| SIVsmE660 | |||
| gag | Forward primer | 5′-CAAGGGTCTGGGTATGAATCC-3′ | 62 |
| Reverse primer | 5′-TCAATGCTTCTGCCATTAATCTAG-3′ | ||
| Probe | 5′-(FAM)TCCTGGCCCTCCTATTCCCTGACA(BHQTM1)-3′ | ||
| env | Forward primer | 5′-AAACTGAGACAGATAGGTGGG-3′ | 58 |
| Reverse primer | 5′-CCTGTTCCAAGCCTGCAC-3′ | ||
| Probe | 5′-(FAM)ACAAGGAACGCAGGGACAACAACA(BHQTM1)-3′ |
The assembled reactions were run on a Stratagene Mx4000 multiplex quantitative PCR system (Stratagene, La Jolla, CA). Thermal cycling conditions consisted of 10 min at 95°C for AmpliTaq activation, followed by 45 cycles of 30 s at 95°C, 35 s at gene- and strain-specific annealing temperatures as described above, and 30 s at 70°C. Triplicate test reactions were performed for each sample. The nominal copy numbers for test samples were determined by interpolation onto standard curves of RNA standards (duplicate reactions for log10 dilutions of 101 to 106 copies eq/ml). All data analysis was performed with the Mx4000 v3.00 software (Stratagene, La Jolla, CA). The threshold sensitivity of this assay is 100 copies eq/ml of plasma. Because a low level of cross-reactivity of probes between the two strains for SIV could not be eliminated, the baseline signal for the heterologous strain was subtracted for all tested samples.
Infection.
For intrarectal exposure to SIV, animals were placed in a sternal position after anesthesia (10 mg/kg intramuscular [i.m.] Ketamine and 0.5 mg/kg i.m. Xylazine) with the pelvis propped up at approximately a 45° angle. A lubricated infant feeding catheter was inserted gently into the rectum of the animal approximately 4 to 6 in. without causing any injury. First, 5 ml of diluent (phosphate-buffered saline [PBS], 0.5% human serum albumin) was gently flushed through the catheter and then 1 ml of the virus was injected through the catheter, followed by a 5-ml flush with diluent. The animal was returned to its cage and kept tilted at a 45° angle until it fully recovered from anesthesia. Six weekly, intrarectal challenges were carried out with the heterologous virus.
Antibodies.
The antibodies used for surface staining of memory-associated molecules and in the intracellular cytokine staining were purchased from BD Biosciences (BD) and Beckman Coulter (BC). All reagents were validated and titers determined using rhesus monkey PBMCs. The antibodies and conjugates used in memory staining were anti-CD3-peridinin chlorophyll protein (PerCP)-Cy5.5 (SP34.2 from BD), anti-CD4-fluorescein isothiocyanate (19Thy5D7 from BC), anti-CD95-allophycocyanin (APC) (DX2 from BD), and anti-CD28-phycoerythrin (PE) (CD28.2 from BC). For intracellular cytokine staining, the antibodies and conjugates used were anti-tumor necrosis factor alpha (TNF-α)-fluorescein isothiocyanate (FITC) (Mab11 from BD), anti-CD95-PE (DX2 from BD), anti-gamma interferon (IFN-γ)-PE-Cy7 (B27 from BD), anti-CD28-PerCP-Cy5.5 (L293 from BD), anti-interleukin-2 (IL-2)-APC (MQ1-17H12 from BD), anti-CD4-AmCyan (L200 from BD), anti-CD3-Alexa Fluor 700 (SP34.2 from BD), and anti-CD8α-APC-cy7 (SK1 from BD).
CD4+ T-lymphocyte counts and CD4+ memory subsets.
Whole blood collected in EDTA was surface stained with anti-CD3-PerCP-Cy5.5, anti-CD4-FITC, anti-CD95-APC, and anti-CD28-PE. Peripheral blood CD4+ T-lymphocyte counts were calculated by multiplying the percentage of CD3+ CD4+ T lymphocytes by the total lymphocyte counts. The percentages of central, naïve, and effector memory cells were calculated by multiplying the percentages of CD28+ CD95+, CD28+ CD95−, and CD28− CD95+ T lymphocytes by the total lymphocyte counts.
IFN-γ ELISPOT assays.
Multiscreen 96-well plates were coated overnight with 100 μl per well of 5 μg/ml anti-human IFN-γ antibody (B27; BD Pharmingen) in endotoxin-free Dulbecco's PBS (D-PBS). The plates were then washed three times with D-PBS containing 0.25% Tween 20, blocked for 2 h with D-PBS containing 5% fetal bovine serum to remove the Tween 20, and incubated with peptide pools and 2 × 105 PBMCs in triplicate in 100-μl reaction mixture volumes. The peptide pool used in this study spanning the SIVmac239 Gag protein was comprised of 15-amino-acid peptides overlapping by 11 amino acids. Each peptide in a pool was present at a 1-μg/ml concentration. Following an 18-h incubation at 37°C, the plates were washed nine times with D-PBS containing 0.25% Tween 20 and once with distilled water. The plates were then incubated with 2 μg/ml biotinylated rabbit anti-human IFN-γ (Biosource) for 2 h at room temperature, washed six times with Coulter wash (Beckman Coulter), and incubated for 2.5 h with a 1:500 dilution of streptavidin-alkaline phosphatase (Southern Biotechnology). After five washes with Coulter wash and one with D-PBS, the plates were developed with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) chromogen (Pierce). The process was stopped by washing with tap water, and the plates were air dried and read with an enzyme-linked immunospot (ELISPOT) reader (Hitech Instruments) using Image-Pro Plus image-processing software (version 4.1) (Media Cybernetics, Des Moines, IA).
PBMC stimulation and intracellular cytokine staining.
Purified PBMCs were isolated from EDTA-anticoagulated blood and incubated at 37°C in a 5% CO2 environment for 6 h in the presence of RPMI 1640-10% fetal calf serum alone (unstimulated), a pool of 15-mer Gag peptides (5 μg/ml [each peptide]), or staphylococcal enterotoxin B (5 μg/ml; Sigma-Aldrich) as a positive control. All cultures contained monensin (GolgiStop; BD Biosciences) as well as 1 μg/ml of anti-CD49d (BD Biosciences). The cultured cells were stained with monoclonal antibodies specific for cell surface molecules (CD3, CD4, CD8, CD28, and CD95) and with an amine dye (Invitrogen) to discriminate live from dead cells. After being fixed with Cytofix/Cytoperm solution (BD Biosciences), cells were permeabilized and stained with antibodies specific for IFN-γ, TNF-α, and IL-2. Labeled cells were fixed in 1.5% formaldehyde-PBS. Samples were collected on an LSR II instrument (BD Biosceiences) and analyzed using FlowJo software (Tree Star). Approximately 200,000 to 1,000,000 events were collected per sample. The background level of cytokine staining varied within different samples and different cytokine patterns but was typically <0.01% of the level of the CD4+ T cells (median, 0%) and <0.05% of the level of the CD8+ T cells (median, 0.01%). All data are reported after background correction. The only samples considered positive were those in which the percentage of cytokine-staining cells was at least twice that of the background.
Virus neutralization assay.
Plasma samples were collected from all 14 infected animals immediately prior to intrarectal exposure to the second virus. Neutralizing antibodies were measured in a luciferase reporter gene assay that utilized either TZM-bl or 5.25.EGFP.Luc.M7 (M7-Luc) cells as described previously (33). The 50% inhibitory dose (ID50) was defined as the plasma dilution that resulted in a 50% reduction in relative luminescence units (RLU) compared to virus control wells after subtraction of background RLU. Assay stocks of uncloned SIVsmE660 were generated in CEMx174 cells. Assay stocks of the Env-pseudotyped virus, SIVmac251/CS.41, were generated by cotransfection of a SIVmac251CS Env plasmid and an Env-deficient HIV backbone plasmid (pSG3ΔEnv) in 293T cells. Both viral stocks were made cell free by filtration through 0.45-μm pores and stored at −70°C until use.
Statistical analyses.
Statistical analyses and graphical presentations were computed with GraphPad Prism, using nonparametric Wilcoxon rank sum tests and Mann-Whitney U test. P values of <0.05 were considered significant.
RESULTS
SIVmac251 and SIVsmE660 differ by typical intraclade HIV-1 distance.
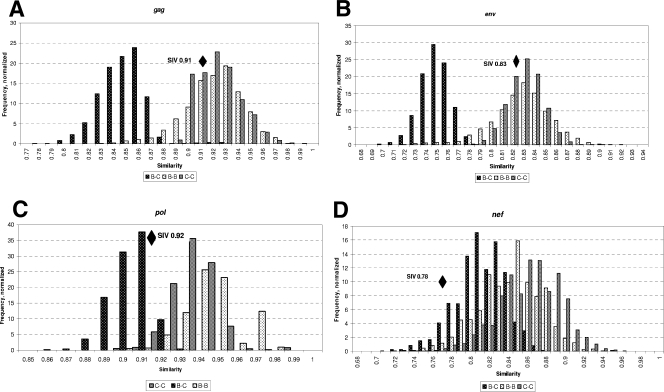
To evaluate the genetic relatedness of two isolates of SIV that are frequently used in nonhuman primate studies, we compared the genetic distance between SIVmac251 and SIVsmE660 to intraclade and interclade HIV-1 sequence distances. We used HIV clade B and C sequences in the Los Alamos HIV Sequence Database to generate our estimates of HIV-1 interclade and intraclade diversity. We used one sequence per person for these alignments. We analyzed 11,484 pairs of sequences for gag, 21,177 pairs of sequences for env, 32,465 pairs of sequences for nef, and 7,140 pairs of sequences for pol. Figure 1 shows the distribution of normalized frequencies for percent similarity of intraclade and interclade pairwise comparisons. The calculated distances between SIVsmE660 and SIVmac251 at gag, pol, env, and nef are plotted in each panel. As shown in Fig. 1A and B, the distance between gag and env of the two SIV strains is similar to HIV-1 clade B and C intraclade distances, with distances of 0.91 and 0.83, respectively. In contrast, the distances between the two SIV isolates in pol and nef are of the magnitude seen in interclade differences in HIV-1 (Fig. 1C and D). Therefore, these two pathogenic SIV isolates are well suited for use in an SIV model of superinfection because their two key foci, env and gag, have differences that reflect a degree of sequence heterogeneity comparable to different circulating HIV-1 isolates within the same clade.
FIG. 1.
Genetic distances between SIVmac251 and SIVsmE660 in relation to HIV-1 clade B and C intraclade and interclade distances. We performed pairwise comparisons of 11,484 gag (A), 21,177 env (B), 7,140 pol (C), and 32,465 nef (D) sequences from individuals infected with HIV-1. The genetic distance for each of these comparisons was graphed as fractional similarity between a given pair (x axis). The amplitude of the bar graph reflects the percentage of pairwise comparisons exhibiting a given similarity (y axis). Comparisons between pairs of sequences within each clade and pairs of sequences from different clades are distinguished by shading: intraclade B (light hatched bars), intraclade C (gray bars), and interclade B versus C (dark hatched bars). Genetic distances between SIVmac251 and SIVsmE660 sequences are plotted simultaneously at each genetic locus as black diamonds.
Plasma SIV RNA levels following primary infection.
We then established cohorts of rhesus monkeys that were infected with one or the other of these two strains of SIV. The viruses and routes of administration used to initiate these infections are summarized in Table 2. Eight animals were initially infected with SIVmac251 (Fig. 2A), and six animals were initially infected with SIVsmE660 (Fig. 2B). Infection was successfully established in 9 of these 14 monkeys via the intrarectal route. However, 5 of 14 monkeys did not exhibit detectable viremia after 18 sequential intrarectal inoculations and had to be inoculated intravenously to initiate the primary infection (CR53, AV74, and CG5G with SIVmac251 and CR54 and CP37 with SIVsmE660).
TABLE 2.
Viruses, routes of infection, viral load, and time of superinfection
| Monkey | Primary virus | MHC class I allele(s)a | Route of infection by primary virusb | No. of days of infection | Viral RNA set point (copies/ml)c | Second virusd | Superinfection |
|---|---|---|---|---|---|---|---|
| CP3C | SIVsmE660 | Mamu-A*01, -B*17 | i.r. | 768 | 1.00 × 102 | SIVmac251 | Yes |
| CG7G | SIVsmE660 | Mamu-A*01, -B*17 | i.r. | 775 | 5.02 × 103 | SIVmac251 | Yes |
| AK9F | SIVsmE660 | Mamu-B*17 | i.r. | 748 | 1.00 × 102 | SIVmac251 | Yes |
| CP23 | SIVsmE660 | Mamu-A*01 | i.r. | 768 | 8.70 × 104 | SIVmac251 | Yes |
| CR54 | SIVsmE660 | Mamu-A*01 | i.v. | 105 | 2.21 × 102 | SIVmac251 | Yes |
| CP37 | SIVsmE660 | Mamu-A*01 | i.v. | 105 | 3.77 × 106 | SIVmac251 | Yes |
| CP1W | SIVmac251 | Mamu-A*01, -B*08 | i.r. | 775 | 6.98 × 104 | SIVsmE660 | Yes |
| CT76 | SIVmac251 | Mamu-A*01 | i.r. | 775 | 2.45 × 102 | SIVsmE660 | Yes |
| PBE | SIVmac251 | Mamu-A*02 | i.r. | 748 | 4.85 × 105 | SIVsmE660 | Yes |
| CG71 | SIVmac251 | Mamu-A*01, -B*17 | i.r. | 105 | 1.12 × 105 | SIVsmE660 | Yes |
| AH4X | SIVmac251 | Negative | i.r. | 365 | 6.23 × 104 | SIVsmE660 | Yes |
| CR53 | SIVmac251 | Mamu-A*01, -B*17 | i.v. | 762 | 1.17 × 104 | SIVsmE660 | Yes |
| AV74e | SIVmac251 | Negative | i.v. | 105 | 2.73 × 105 | SIVsmE660 | No |
| CG5Ge | SIVmac251 | Mamu-A*01 | i.v. | 105 | 1.37 × 104 | SIVsmE660 | No |
Alleles that are present in each monkey are indicated. “Negative” indicates all four alleles were not detected.
i.r., intrarectal; i.v., intravenous.
Set point plasma viral RNA in copies/ml of primary virus at time of exposure to the second virus.
All monkeys were exposed to the second virus via intrarectal inoculation.
Results for monkeys who resisted superinfection are highlighted in boldface.
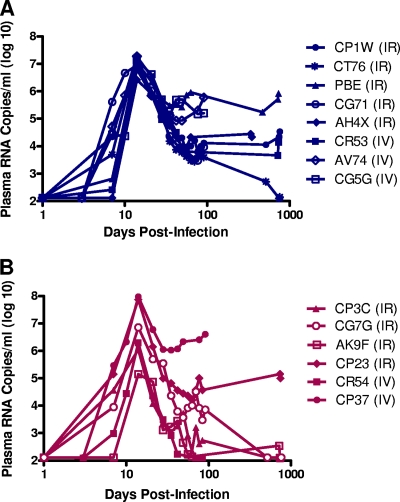
FIG. 2.
Plasma viral RNA levels following primary infection with either SIVmac251 or SIVsmE660. (A) Six rhesus monkeys were infected with SIVmac251, and (B) eight were infected with SIVsmE660 via either intrarectal (IR) or intravenous (IV) inoculations. Although the animals were infected after different numbers of intrarectal exposures or a single intravenous inoculation, the viral RNA levels are displayed synchronously as days postinfection. Viral RNA levels are shown as log10 copies of plasma viral RNA/ml of plasma for individual monkeys at each time point.
Viral replication during primary infection occurred with kinetics typical of SIV replication in naïve rhesus monkeys. Moreover, SIV replication kinetics did not differ significantly between animals that became infected by mucosal or intravenous routes. Monkeys that were infected with SIVmac251 all developed uniform peak plasma viral RNA levels of 6 to 7 logs at 14 days after virus inoculation followed by a sustained viremia of 4 to 6 logs of plasma viral RNA, with the exception of one monkey (CT76) who had undetectable viremia by 700 days postinfection.
In the cohort of monkeys infected by SIVsmE660, monkeys had peak plasma viral RNA levels of 5 to 8 logs at 14 days after virus inoculation, followed by sustained viremia of 5 to 7 logs of plasma viral RNA in animals CP37 and CP23. However, three of the monkeys infected with SIVsmE660 (CP3C, CG7G, and AK9F) had undetectable plasma viral RNA levels by 700 days postinfection, while monkey CR54 had an undetectable plasma viral RNA levels by 85 days postinfection. This wide range in peak and set point viremias in monkeys infected with SIVsmE660 has been previously described (7, 19, 35). Since plasma viral RNA levels at peak and set point in some of the SIVsmE660-infected monkeys (CP37, CP23, and CG7G) were of a magnitude comparable to that seen in monkeys following SIVmac251 infection, the variability in SIVsmE660 replication levels in monkeys likely reflects a host factor effect rather than an intrinsic lack of replicative capacity of the SIVsmE660 strain.
Plasma SIV RNA levels following superinfection.
Once set point plasma virus RNA levels were reached, all monkeys were exposed to the heterologous virus by 6 weekly intrarectal inoculations. The duration of primary infection and plasma virus RNA levels at time of exposure to the second virus are summarized in Table 2. The eight SIVmac251-infected monkeys and six SIVsmE660-infected monkeys were then monitored for evidence of superinfection by assessing plasma SIVmac251 and SIVsmE660 RNA weekly for 20 weeks.
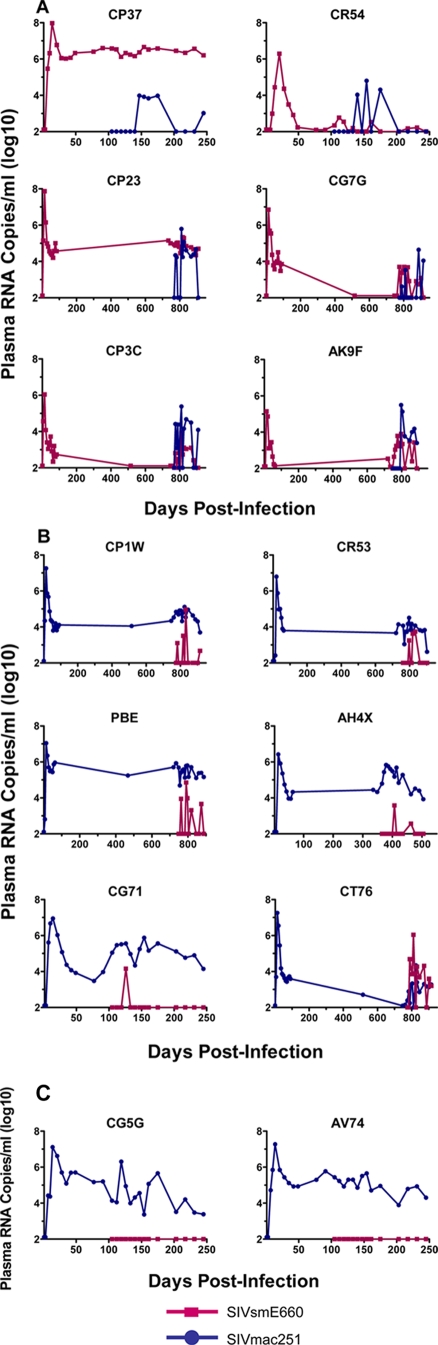
To monitor the viral replication dynamics for each SIV strain in the dually infected monkeys, we developed a qRT-PCR assay using strain-specific probes. Figure 3 shows the replication kinetics of the two strains of SIV following the first and second infections. As depicted in Fig. 3A, six of six monkeys that were initially infected with SIVsmE660 became superinfected with SIVmac251. Of the eight monkeys that were initially infected with SIVmac251, six became superinfected with SIVsmE660 (Fig. 3B). Viral RNA of the heterologous SIV strain was detected by 14 to 21 days after challenge. In 11 of 12 superinfected animals, with the exception of AK9F, the levels of plasma viral RNA of the second virus at peak viremia were 1 to 4 logs lower than the peak viremia of the first virus. In addition, the levels of plasma viral RNA of the second virus also declined rapidly to undetectable levels in six animals (CR54, CP23, CR53, PBE, AH4X, and CG71), while the viral load persisted at low levels in the remaining six animals (CP37, CG7G, CP3C, AK9F, CP1W, and CT76). The presence of the superinfecting virus at multiple time points was confirmed in each animal by direct sequencing.
FIG. 3.
Plasma viral RNA levels of both SIV strains following the primary infection and superinfection in each monkey. Monkeys were either first infected with SIVsmE660 and then with SIVmac251 (A) or first infected with SIVmac251 followed by SIVsmE660 (B). Only two monkeys that were initially infected with SIVmac251 resisted superinfection with SIVsmE660 after six intrarectal challenges (C). The red lines and symbols represent RNA levels of SIVsmE660, while the blue lines and symbols represent plasma RNA levels of SIVmac251.
Of the 14 infected animals that were exposed to a heterologous virus, only 2 (AV74 and CG5G) that were initially infected with SIVmac251 resisted superinfection with the heterologous virus (Fig. 3C). There was no detectable SIVsmE660 viral RNA in these animals for 20 weeks after exposure. The absence of replication by the second virus was verified by direct sequencing (data not shown).
No apparent acceleration in disease progression after superinfection.
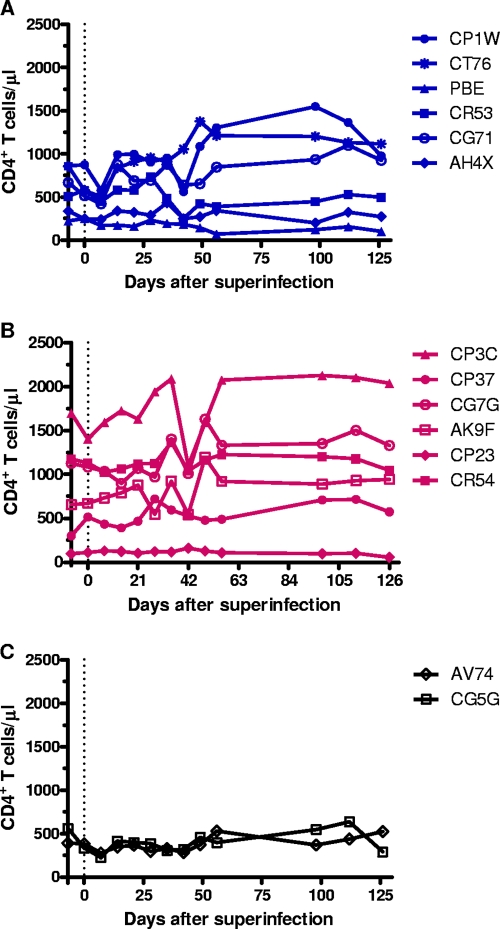
Interestingly, we observed an increase in plasma viral RNA levels of the primary virus (Fig. 3A and B) and a transient decline in CD4+ T cells following superinfection in all of the animals, except AH4X (Fig. 4A), CP3C, and AK9F (Fig. 4B). This finding is consistent with case reports of HIV superinfection in which superinfected individuals developed a transient perturbation in total plasma viral RNA levels in association with a clinical prodrome that aroused suspicion that an intervening event might have caused a sudden rise in viral load (2, 26, 27, 42, 60, 67). The CD4+ T-cell counts re-equilibrated 2 to 6 weeks after superinfection, and a small increase in the CD4+ T-cell counts in some of the animals was observed from 42 to 126 days after superinfection (CT76, CP1W, CG71, CP3C, AK9F, and CG7G). We did not perform statistical analyses on the differences in the CD4+ T-cell decline between superinfected and nonsuperinfected animals due to the small sample size of animals that resisted superinfection, but the trends in changes of CD4+ T-cell counts were indistinguishable between all animals. Therefore, there appeared to be no acceleration in disease progress in the superinfected monkeys as a consequence of superinfection.
FIG. 4.
Absolute CD4+ T-cell counts for 126 days after superinfection. The CD4+ T-cell counts in the peripheral blood are shown in blue for the six animals that were initially infected with SIVmac251 and then superinfected with SIVsmE660 (A), in red for the six animals that were first infected with SIVsmE660 and then superinfected with SIVmac251 (B), and in black for the two animals that resisted superinfection (C). The dotted line indicates day 0 prior to superinfection. The presuperinfection CD4+ T-cell counts were obtained 7 days prior to superinfection.
Peak viral replication following the second infection was lower than peak viral replication following the first infection.
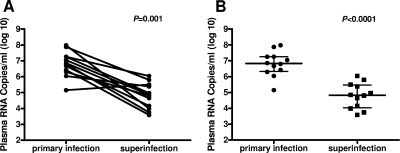
Of the 12 monkeys that became superinfected, 11 animals efficiently controlled the second virus at peak viremia, with the exception of AK9F. Peak replication following the second virus infection was lower than peak replication after the first infection in each monkey (Fig. 5A). The decrease in peak viremia was statistically significant as determined by the paired Wilcoxon rank sum test (P = 0.001). Furthermore, when considered as a cohort, the median peak viral load value following the second infection was lower than that observed following the first infection (Fig. 5B). The difference in the median values and interquartile ranges of peak viremia between the first and second infections was statistically significant as determined by the unpaired Mann-Whitney U test (P < 0.0001).
FIG. 5.
Peak plasma viral RNA levels were higher following the first infection than after the second infection. (A) Peak plasma viral RNA levels for each monkey following primary infection and superinfection are indicated by individual filled circles and are connected by lines. In 11 of 12 superinfected animals, there was a lower peak plasma viral RNA level following the superinfection than following the primary infection. These comparisons were done using the two-tailed paired Wilcoxon's rank sum test (P = 0.001). (B) Peak plasma viral RNA levels are depicted as separate points following primary infection and following superinfection. Bars representing the median value and interquartile ranges are shown for each group. The two-tailed unpaired Mann-Whitney U test (P < 0.0001) was used to evaluate the statistical significance of the differences between the peak viremias at the two time points.
Susceptibility to superinfection was not associated with time after the first infection or persistence of the primary virus.
In these two cohorts of monkeys, superinfection was initiated between 3 and 20 months after the primary infection (Table 2). This large window of susceptibility suggests that infected individuals are likely susceptible to superinfection regardless of the state of immune competence of the host or the maturity of the immune response to the initial virus. Superinfection can occur after the immune response against the initial infection has had time to develop and mature. In addition, since 10 of 12 superinfected animals harbored the Mamu-A*01, -B*08, and -B*17 alleles (Table 2), susceptibility to superinfection appears not to be a consequence of major histocompatibility complex alleles that are associated with relatively efficient viral control.
Furthermore, the likelihood of acquiring a second virus appears not to be correlated with the persistence of replication of the primary virus at the time of exposure to the heterologous virus (Table 2). Some animals became superinfected despite relatively high levels of replication of the primary virus, ranging from 104 to 106 RNA copies/ml in the plasma (CP23, CP37, CP1W, PBE, CG71, AH4X, and CR53), while others became superinfected in the setting of undetectable or low-level replication of the primary virus, ranging from 102 to 103 RNA copies/ml in the plasma (CP3C, CG7G, AK9F, CR54, and CT76).
Interestingly, in animals that had a high-set-point viremia following exposure to the first virus, either SIVmac251 (CP1W, CR53, PBE, AH4X, and CG71) or SIVsmE660 (CP37 and CP23), the second virus was efficiently controlled after superinfection while the first infecting virus remained the predominant viral quasispecies in the plasma. In contrast, in animals that had undetectable plasma viral RNA levels following exposure to SIVsmE660 (CG7G, CP3C, and AK9F) or SIVmac251 (CT76) prior to superinfection, the heterologous virus replaced the first viral strain after superinfection even in monkeys with blunted peak replication of the second virus. Only one monkey in the cohort, CR54, was able to control both viruses to undetectable levels. These data suggest that, although direct viral interference did not contribute to susceptibility to superinfection, it may have influenced the viral replication dynamics of the second virus relative to the primary virus after superinfection.
Susceptibility to superinfection was not associated with absolute CD4+ T-cell counts or percent central memory CD4+ T cells.
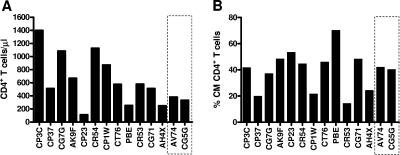
To determine if there were any clinical parameters associated with relative susceptibility to superinfection in these cohorts of monkeys, we assessed the absolute CD4+ T-cell counts and the percentage of CD4+ T lymphocytes that were central memory cells immediately prior to the exposure of these animals to the heterologous virus. There was no difference between absolute CD4+ T-cell counts or the percentage of CD4+ central memory T cells in the animals that became superinfected and those that resisted superinfection (Fig. 6A and B). Although a statistical analysis could not be performed to validate this observation due to the small sample size of animals that resisted superinfection, the absolute CD4+ T-cell counts and the percentage of central memory CD4+ T cells of animals that resisted superinfection were within the range of the corresponding parameters in animals that became superinfected. In addition, we also analyzed the percentages of effector and naïve memory CD4+ T cells and found that there were no differences in these values between the two groups of monkeys (data not shown). Together, these data indicate that animals with immune systems that are more damaged by a prior SIV infection appeared not to have an increased susceptibility to superinfection.
FIG. 6.
Resistance to SIV superinfection was not associated with peripheral blood absolute CD4+ counts or central memory CD4+ T cells at the time of exposure to the superinfecting virus. (A) CD4+ T-lymphocyte counts on the day of challenge with the heterologous SIV isolate did not differ between the monkeys that became superinfected and those that resisted superinfection. (B) There was also no significant difference in these groups of monkeys in the percentage of central memory (CM) CD4+ T lymphocytes as identified by their expression of CD28 and CD95. The dashed boxes highlight the animals that resisted superinfection.
Susceptibility to superinfection was not associated with virus-specific cellular immune responses.
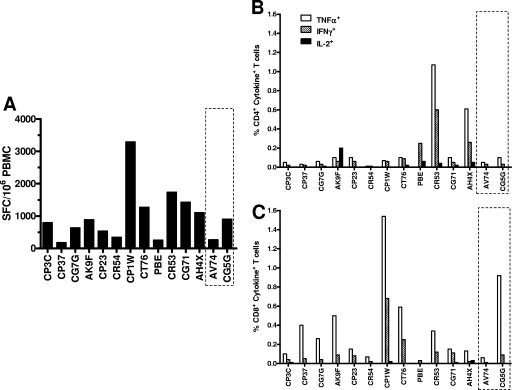
To determine whether systemic virus-specific cellular immune responses conferred protection against heterologous virus in the monkeys that resisted superinfection, all rhesus monkeys were evaluated for SIV-specific cellular immunity immediately prior to exposure to the heterologous virus. Cellular immunity to SIV was first evaluated using an ELISPOT assay to assess PBMC IFN-γ responses following exposure to a pool of SIV Gag peptides (Fig. 7A). SIV-specific T-cell responses were indistinguishable between the animals that became superinfected and those that resisted superinfection.
FIG. 7.
Resistance to superinfection was not associated with SIV Gag-specific CD4+ and CD8+ T-lymphocyte responses at the time of exposure to the superinfecting virus. Peripheral blood lymphocytes obtained from the monkeys prior to challenge with the superinfecting virus were exposed to a pool of overlapping SIV Gag peptides, and their responses were assessed in IFN-γ ELISPOT assays (A). SFC, spot-forming cells. By gating on CD4+ (B) or CD8+ (C) T lymphocytes, the cells were assessed for production of TNF-α, IFN-γ, and IL-2 in intracellular cytokine staining assays. The dashed boxes highlight the animals that resisted superinfection.
SIV-specific CD4+ and CD8+ T-lymphocyte function was further evaluated by intracellular cytokine staining. Immediately prior to exposure to the heterologous virus, PBMC production of IFN-γ, TNF-α, and IL-2 was assessed after stimulation with SIV Gag peptide pools. We were able to detect virus-specific CD4+ (Fig. 7B) and CD8+ (Fig. 7C) T-lymphocyte responses in PBMCs of all monkeys. We did not perform statistical analyses on the differences in cytokine secretion between the two groups of monkeys due to the small sample size of animals that resisted superinfection. However, the cytokine responses of the two animals that resisted superinfection were within the range of the corresponding parameters in animals that became superinfected. Therefore, the qualitative and quantitative cell-mediated SIV-specific immune responses of monkeys that became superinfected and those that resisted superinfection appeared to be indistinguishable. These findings suggest that SIV-specific cellular immune responses likely did not account for the variability in the susceptibility of these monkeys to superinfection.
Antibody responses did not protect against superinfection.
The role of neutralizing antibody responses in protecting against HIV superinfection is not clear (5, 49, 50). To assess whether SIV-specific antibodies played a role in the resistance to superinfection in these cohorts of animals, plasma samples harvested just prior to the heterologous viral challenge were assayed for neutralizing antibody responses elicited by the primary SIV infection. The ability of plasma antibody to neutralize SIVsmE660 and SIVmac251 was measured in luciferase reporter gene neutralizing antibody assays using uncloned SIVsmE660 and pseudoviruses expressing viral Envelope cloned from SIVmac251CS.41 (33). The serum ID50 neutralizing titers against both viruses are shown in Table 3, Plasma from five of six monkeys (except CR54) that were first infected with SIVsmE660 neutralized the homologous SIVsmE660 (1:62 to 1:508), while plasma from five of eight SIVmac251-infected monkeys neutralized homologous SIVmac251 (1:33 to 1:215).
TABLE 3.
Neutralizing antibodies in rhesus animals after primary infection prior to superinfection
| Animal | Primary virus | No. of days of infection | ID50 ina:
|
|
|---|---|---|---|---|
| TZM-bl cells infected with SIVmac251/CS.41b | 5.25.EGFP.Luc.M7 cells infected with SIVsmE660c | |||
| CP3C | SIVsmE660 | 768 | <20 | 92 |
| CG7G | SIVsmE660 | 775 | <20 | 508 |
| AK9F | SIVsmE660 | 748 | 20 | 136 |
| CP23 | SIVsmE660 | 768 | 37 | 62 |
| CR54 | SIVsmE660 | 105 | <20 | <20 |
| CP37 | SIVsmE660 | 105 | 41 | 79 |
| CP1W | SIVmac251 | 775 | 43 | 245 |
| CT76 | SIVmac251 | 775 | 50 | 110 |
| PBE | SIVmac251 | 748 | 50 | <20 |
| CG71 | SIVmac251 | 105 | <20 | 188 |
| AH4X | SIVmac251 | 365 | 33 | 120 |
| CR53 | SIVmac251 | 762 | 215 | <20 |
| AV74d | SIVmac251 | 105 | <20 | 73 |
| CG5Gd | SIVmac251 | 105 | <20 | 124 |
Values are the sample serum dilution at which relative luminescence units (RLU) were reduced 50% compared to virus control wells (no serum sample).
Pseudoviruses containing Env cloned from single expansion of uncloned SIVmac251 challenge stock were generated in 293T cells.
Uncloned SIVsmE660 virus stock was generated in CEMx174 cells.
Results for monkeys who resisted superinfection are highlighted in boldface.
To investigate whether the antibodies generated by these animals following primary infection have the ability to neutralize the heterologous virus, we assayed the plasma of the monkeys for neutralization activity against the second virus before their exposure to that virus. As shown in Table 3, animals initially infected with SIVsmE660 generated undetectable or low titers of neutralizing antibodies to SIVmac251 (ranging from undetectable to 1:41). We also detected neutralizing antibodies against SIVsmE660 in six of eight animals that were initially infected with SIVmac251 (ranging from 1:73 to 1:245). The titers against the heterologous SIVsmE660 in the SIVmac251-infected animals were not significantly lower than the titers against the homologous SIVsmE660 in SIVsmE660-infected animals (P = 0.95, Mann-Whitney test).
Interestingly, animals AV74 and CG5G, who were initially infected with SIVmac251 and subsequently resisted superinfection with SIVsmE660, had neutralizing antibodies against SIVsmE660 prior to exposure to this heterologous virus. However, the titers of these antibodies were within the range of antibody titers against SIVsmE660 that were generated by other SIVmac251-infected animals that became superinfected following exposure to SIVsmE660. We did not perform statistical analyses of the differences in antibody titers against SIVsmE660 between the SIVmac251-infected monkeys that resisted superinfection and the SIVmac251- infected monkeys that became superinfected because of the small number of animals that resisted superinfection. Nevertheless, the titers of neutralizing antibodies specific for the heterologous viruses that were elicited during primary infection appear to not have influenced the susceptibility of monkeys to superinfection.
DISCUSSION
HIV superinfection has important implications for vaccine prevention of HIV infection and the global genetic diversity of HIV. In this study, we used intrarectal inoculations of two replication-competent strains of SIV to simulate HIV-1 superinfection and employed quantitative analyses of viral RNA using strain-specific primers to define the replication dynamics of each virus over time. We demonstrated that immune responses generated during primary infection that are capable of controlling one strain of SIV do not preclude subsequent infection with a second strain of SIV. Superinfection occurred as early as 3 months and as late as 2 years following primary infection, and susceptibilities to superinfection appeared to be independent of classical adaptive immune responses or the level of replication of the primary virus, even though we were not able to evaluate the statistical significance of these parameters because of the small number of animals that resisted superinfection in this study. Importantly, the replication of the superinfecting virus during the first days following exposure was attenuated compared with the replication of the primary virus. The relative susceptibility of monkeys to superinfection in the present study could not be attributed to a difference in the replication capacities of these two strains of SIV, since superinfection occurred in both cohorts of animals regardless of which virus was used to establish the first infection. Furthermore, the ability of both SIVmac251 and SIVsmE660 to maintain dominance in superinfected monkeys suggests that these two SIV strains are comparable in their fitness.
Previous nonhuman primate studies using a live attenuated immunodeficiency virus to generate protection against a pathogenic immunodeficiency virus challenge provide an important context for the present findings. Although such live attenuated viruses can confer protection against a homologous virus challenge (11, 14, 25, 36, 57, 64), they provide only partial protection against a heterologous virus infection (16, 34, 44, 63). The results of the present study are consistent with those findings in that prior infection did not prevent superinfection with a heterologous virus, but did damp replication of the second virus at peak and in the post-acute phase of superinfection. Interestingly, the two animals that resisted superinfection had also resisted 18 attempts at the first infection by the intrarectal route and required intravenous inoculation to establish primary infection. This finding raises the possibility that variations in the mucosal barrier rather than specific immunological mechanisms may have contributed to differences in susceptibility to mucosal infection in this cohort of animals (29).
Just as the correlates of protective immunity have not yet been defined for the protection observed in monkeys that have received a live attenuated SIV vaccine (1, 3, 11, 44, 45, 54, 55), the mechanisms accounting for the partial protection observed against superinfection in our study are not clear. We used pooled peptides corresponding to SIVmac239 Gag to evaluate virus-specific cellular immune responses because the cross-reactive responses are likely the most germane to controlling the replication of the heterologous virus. Nevertheless, there may be additional T-cell responses that contribute to controlling the second virus that are not detected using SIVmac239 peptides. A recent study by Reynolds et al. examining the ability of live-attenuated SIV to protect macaques against heterologous virus challenge implicated major histocompatibility complex (MHC) class I-restricted CD8+ cellular responses in reducing heterologous viral replication during the chronic phase of infection (44). However, further studies are needed to elucidate the relative contributions of CD8+ T cells and other factors, including CD4+ T cells, antibodies, and NK cells, in the acute phase of replication of the second virus. A decrease in the number of potential target cells as a result of depletion of memory CD4+ T cells in the lamina propria in the gut and lymph nodes following the first infection may have contributed to the reduction and magnitude of peak viremia observed following the second infection. Further detailed characterization of CCR5+ transitional and effector memory T cells in mucosal effector sites is needed to determine the availability of target cells. Other factors, such as innate immune responses or viral interference, may have also contributed to the relative protection observed against the superinfecting virus.
The present study of superinfection in the SIV/rhesus monkey model has important implications for HIV pathogenesis and vaccine development. Although this SIV model of superinfection utilized a higher-dose mucosal challenge to establish superinfection than likely occurs in human cases of HIV superinfection, the findings in the present study suggest that HIV superinfection can occur readily throughout the course of infection. Therefore, the prevalence of HIV superinfection is likely underestimated, especially in cases whose only clinical manifestation is transient low-level replication of the second virus. Similar to human cases of HIV superinfection described by Casado and Piantadosi et al. (8, 40), SIV superinfection also did not necessarily lead to increases in viral load and clinical deterioration. This could be because both SIV strains that were used in this study are comparably fit, and therefore the persistence of either one or both may not dramatically affect disease progression. In contrast, the clinical sequelae in HIV superinfection may have more variable outcomes than what we observed in this study, since the relative dynamics of the two viruses may be markedly different as a consequence of their relative replication fitness. Interestingly, although superinfection is likely a common phenomenon in HIV-1 infections, it may not have clinical consequences if the two viruses are equivalent in their fitness or if the superinfecting virus transiently replicates at a low level. In contrast to this, superinfection likely has a more profound impact on the sensitivity of circulating viruses to antiretroviral therapy and global HIV genetic diversity as a consequence of viral recombination.
Creating a vaccine that can protect against infection by a virus with the genetic heterogeneity of HIV is a daunting challenge, given that immune responses generated after live SIV infection do not prevent infection of macaques by a heterologous SIV isolate in the nonhuman primate model. Nevertheless, the phenomenon of HIV/SIV superinfection should not discourage the pursuit of an AIDS vaccine, since effective vaccines for viruses such as the mumps and measles viruses also do not prevent entry of virus into the body. While the immune system does not prevent new strains of virus from establishing infections, it can limit the spread of those viruses and attenuate the pathogenic sequelae of infection. Further dissection of the virologic and immune correlates of protection against superinfection in monkeys may provide important insights into the nature of immune responses that are required to provide protective immunity against an immunodeficiency virus infection.
Acknowledgments
We thank Vanessa Hirsch for providing virus stock SIVsmE660.
This work was supported by NIH NIAID PHS grants K08-AI069995 (W.W.Y.) and AI-067854 (W.W.Y. and N.L.L.) and the Center for HIV/AIDS Vaccine Immunology.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Almond, N., T. Corcoran, R. Hull, B. Walker, J. Rose, R. Sangster, K. Silvera, P. Silvera, M. Cranage, E. Rud, and E. J. Stott. 1997. Mechanisms of protection induced by attenuated simian immunodeficiency virus. IV. Protection against challenge with virus grown in autologous simian cells. J. Med. Primatol. 2634-43. [DOI] [PubMed] [Google Scholar]
- 2.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420434-439. [DOI] [PubMed] [Google Scholar]
- 3.Berry, N., R. Stebbings, D. Ferguson, C. Ham, J. Alden, S. Brown, A. Jenkins, J. Lines, L. Duffy, L. Davis, W. Elsley, M. Page, R. Hull, J. Stott, and N. Almond. 2008. Resistance to superinfection by a vigorously replicating, uncloned stock of simian immunodeficiency virus (SIVmac251) stimulates replication of a live attenuated virus vaccine (SIVmacC8). J. Gen. Virol. 892240-2251. [DOI] [PubMed] [Google Scholar]
- 4.Blick, G., R. M. Kagan, E. Coakley, C. Petropoulos, L. Maroldo, P. Greiger-Zanlungo, S. Gretz, and T. Garton. 2007. The probable source of both the primary multidrug-resistant (MDR) HIV-1 strain found in a patient with rapid progression to AIDS and a second recombinant MDR strain found in a chronically HIV-1-infected patient. J. Infect. Dis. 1951250-1259. [DOI] [PubMed] [Google Scholar]
- 5.Blish, C. A., O. C. Dogan, N. R. Derby, M.-A. Nguyen, B. Chohan, B. A. Richardson, and J. Overbaugh. 2008. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J. Virol. 8212094-12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner, B., J. P. Routy, Y. Quan, D. Moisi, M. Oliveira, D. Turner, and M. A. Wainberg. 2004. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection. AIDS 181653-1660. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. R., M. Czapiga, J. Kabat, Q. Dang, I. Ourmanov, Y. Nishimura, M. A. Martin, and V. M. Hirsch. 2007. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. J. Virol. 815594-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casado, C., M. Pernas, T. Alvaro, V. Sandonis, S. Garcia, C. Rodriguez, J. del Romero, E. Grau, L. Ruiz, and C. Lopez-Galindez. 2007. Coinfection and superinfection in patients with long-term, nonprogressive HIV-1 disease. J. Infect. Dis. 196895-899. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty, B., L. Valer, C. De Mendoza, V. Soriano, and M. E. Quinones-Mateu. 2004. Failure to detect human immunodeficiency virus type 1 superinfection in 28 HIV-seroconcordant individuals with high risk of reexposure to the virus. AIDS Res. Hum Retrovir. 201026-1031. [DOI] [PubMed] [Google Scholar]
- 10.Chohan, B., L. Lavreys, S. M. J. Rainwater, and J. Overbaugh. 2005. Evidence for frequent reinfection with human immunodeficiency virus type 1 of a different subtype. J. Virol. 7910701-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connor, R. I., D. C. Montefiori, J. M. Binley, J. P. Moore, S. Bonhoeffer, A. Gettie, E. A. Fenamore, K. E. Sheridan, D. D. Ho, P. J. Dailey, and P. A. Marx. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 727501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cranage, M. P., S. A. Sharpe, A. M. Whatmore, N. Polyanskaya, S. Norley, N. Cook, S. Leech, M. J. Dennis, and G. A. Hall. 1998. In vivo resistance to simian immunodeficiency virus superinfection depends on attenuated virus dose. J. Gen. Virol. 791935-1944. [DOI] [PubMed] [Google Scholar]
- 13.Cranage, M. P., A. M. Whatmore, S. A. Sharpe, N. Cook, N. Polyanskaya, S. Leech, J. D. Smith, E. W. Rud, M. J. Dennis, and G. A. Hall. 1997. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology 229143-154. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 2581938-1941. [DOI] [PubMed] [Google Scholar]
- 15.Diaz, R. S., R. Pardini, M. Catroxo, E. A. Operskalski, J. W. Mosley, and M. P. Busch. 2005. HIV-1 superinfection is not a common event. J. Clin. Virol. 33328-330. [DOI] [PubMed] [Google Scholar]
- 16.Evans, D. T., J. E. Bricker, H. B. Sanford, S. Lang, A. Carville, B. A. Richardson, M. Piatak, Jr., J. D. Lifson, K. G. Mansfield, and R. C. Desrosiers. 2005. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J. Virol. 797707-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, G., B. Weiser, C. Kuiken, S. M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, O. Anzala, and H. Burger. 2004. Recombination following superinfection by HIV-1. AIDS 18153-159. [DOI] [PubMed] [Google Scholar]
- 18.Fultz, P. N., A. Srinivasan, C. R. Greene, D. Butler, R. B. Swenson, and H. M. McClure. 1987. Superinfection of a chimpanzee with a second strain of human immunodeficiency virus. J. Virol. 614026-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, S., C. R. Brown, H. Dehghani, J. D. Lifson, and V. M. Hirsch. 2000. Intrinsic susceptibility of rhesus macaque peripheral CD4+ T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J. Virol. 749388-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzales, M. J., E. Delwart, S. Y. Rhee, R. Tsui, A. R. Zolopa, J. Taylor, and R. W. Shafer. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J. Infect. Dis. 188397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb, G. S., D. C. Nickle, M. A. Jensen, K. G. Wong, J. Grobler, F. Li, S. L. Liu, C. Rademeyer, G. H. Learn, S. S. Karim, C. Williamson, L. Corey, J. B. Margolick, and J. I. Mullins. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363619-622. [DOI] [PubMed] [Google Scholar]
- 22.Gottlieb, G. S., D. C. Nickle, M. A. Jensen, K. G. Wong, R. A. Kaslow, J. C. Shepherd, J. B. Margolick, and J. I. Mullins. 2007. HIV type 1 superinfection with a dual-tropic virus and rapid progression to AIDS: a case report. Clin. Infect. Dis. 45501-509. [DOI] [PubMed] [Google Scholar]
- 23.Grobler, J., C. M. Gray, C. Rademeyer, C. Seoighe, G. Ramjee, S. A. Karim, L. Morris, and C. Williamson. 2004. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J. Infect. Dis. 1901355-1359. [DOI] [PubMed] [Google Scholar]
- 24.Hu, D. J., S. Subbarao, S. Vanichseni, P. A. Mock, A. Ramos, L. Nguyen, T. Chaowanachan, F. Griensven, K. Choopanya, T. D. Mastro, and J. W. Tappero. 2005. Frequency of HIV-1 dual subtype infections, including intersubtype superinfections, among injection drug users in Bangkok, Thailand. AIDS 19303-308. [PubMed] [Google Scholar]
- 25.Johnson, R. P., J. D. Lifson, S. C. Czajak, K. S. Cole, K. H. Manson, R. Glickman, J. Yang, D. C. Montefiori, R. Montelaro, M. S. Wyand, and R. C. Desrosiers. 1999. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J. Virol. 734952-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jost, S., M. C. Bernard, L. Kaiser, S. Yerly, B. Hirschel, A. Samri, B. Autran, L. E. Goh, and L. Perrin. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347731-736. [DOI] [PubMed] [Google Scholar]
- 27.Jurriaans, S., K. Kozaczynska, F. Zorgdrager, R. Steingrover, J. M. Prins, A. C. van der Kuyl, and M. Cornelissen. 2008. A sudden rise in viral load is infrequently associated with HIV-1 superinfection. J. Acquir. Immune Defic. Syndr. 4769-73. [DOI] [PubMed] [Google Scholar]
- 28.Koelsch, K. K., D. M. Smith, S. J. Little, C. C. Ignacio, T. R. Macaranas, A. J. Brown, C. J. Petropoulos, D. D. Richman, and J. K. Wong. 2003. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS 17F11-F16. [DOI] [PubMed] [Google Scholar]
- 29.Letvin, N. L., S. S. Rao, V. Dang, A. P. Buzby, B. Korioth-Schmitz, D. Dombagoda, J. G. Parvani, R. H. Clarke, L. Bar, K. R. Carlson, P. A. Kozlowski, V. M. Hirsch, J. R. Mascola, and G. J. Nabel. 2007. No evidence for consistent virus-specific immunity in simian immunodeficiency virus-exposed, uninfected rhesus monkeys. J. Virol. 8112368-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, S.-L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 7610674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manigart, O., V. Courgnaud, O. Sanou, D. Valea, N. Nagot, N. Meda, E. Delaporte, M. Peeters, and P. Van de Perre. 2004. HIV-1 superinfections in a cohort of commercial sex workers in Burkina Faso as assessed by an autologous heteroduplex mobility procedure. AIDS 181645-1651. [DOI] [PubMed] [Google Scholar]
- 32.McCutchan, F. E., M. Hoelscher, S. Tovanabutra, S. Piyasirisilp, E. Sanders-Buell, G. Ramos, L. Jagodzinski, V. Polonis, L. Maboko, D. Mmbando, O. Hoffmann, G. Riedner, F. von Sonnenburg, M. Robb, and D. L. Birx. 2005. In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J. Virol. 7911693-11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montefiori, D. C. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Chapter 12, unit 12.11. In R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, Inc., Hoboken, NJ. [DOI] [PubMed]
- 34.Nilsson, C., B. Makitalo, R. Thorstensson, S. Norley, D. Binninger-Schinzel, M. Cranage, E. Rud, G. Biberfeld, and P. Putkonen. 1998. Live attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS 122261-2270. [DOI] [PubMed] [Google Scholar]
- 35.Nishimura, Y., T. Igarashi, A. Buckler-White, C. Buckler, H. Imamichi, R. M. Goeken, W. R. Lee, B. A. P. Lafont, R. Byrum, H. C. Lane, V. M. Hirsch, and M. A. Martin. 2007. Loss of naive cells accompanies memory CD4+ T-cell depletion during long-term progression to AIDS in simian immunodeficiency virus-infected macaques. J. Virol. 81893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norley, S., B. Beer, D. Binninger-Schinzel, C. Cosma, and R. Kurth. 1996. Protection from pathogenic SIVmac challenge following short-term infection with a nef-deficient attenuated virus. Virology 219195-205. [DOI] [PubMed] [Google Scholar]
- 37.Novembre, F. J., J. de Rosayro, S. Nidtha, S. P. O'Neil, T. R. Gibson, T. Evans-Strickfaden, C. E. Hart, and H. M. McClure. 2001. Rapid CD4+ T-cell loss induced by human immunodeficiency virus type 1NC in uninfected and previously infected chimpanzees. J. Virol. 751533-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otten, R. A., D. L. Ellenberger, D. R. Adams, C. A. Fridlund, E. Jackson, D. Pieniazek, and M. A. Rayfield. 1999. Identification of a window period for susceptibility to dual infection with two distinct human immunodeficiency virus type 2 isolates in a Macaca nemestrina (pig-tailed macaque) model. J. Infect. Dis. 180673-684. [DOI] [PubMed] [Google Scholar]
- 39.Pernas, M., C. Casado, R. Fuentes, M. J. Perez-Elias, and C. Lopez-Galindez. 2006. A dual superinfection and recombination within HIV-1 subtype B 12 years after primoinfection. J. Acquir. Immune Defic. Syndr. 4212-18. [DOI] [PubMed] [Google Scholar]
- 40.Piantadosi, A., B. Chohan, V. Chohan, R. S. McClelland, and J. Overbaugh. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piantadosi, A., M. O. Ngayo, B. Chohan, and J. Overbaugh. 2008. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res. Hum. Retrovir. 241221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plantier, J. C., V. Lemee, I. Dorval, M. Gueudin, J. Braun, P. Hutin, A. Ruffault, and F. Simon. 2004. HIV-1 group M superinfection in an HIV-1 group O-infected patient. AIDS 182444-2446. [PubMed] [Google Scholar]
- 43.Ramos, A., D. J. Hu, L. Nguyen, K.-O. Phan, S. Vanichseni, N. Promadej, K. Choopanya, M. Callahan, N. L. Young, J. McNicholl, T. D. Mastro, T. M. Folks, and S. Subbarao. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 767444-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. Leon, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 2052537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz, J. E., R. P. Johnson, H. M. McClure, K. H. Manson, M. S. Wyand, M. J. Kuroda, M. A. Lifton, R. S. Khunkhun, K. J. McEvers, J. Gillis, M. Piatak, J. D. Lifson, G. Grosschupff, P. Racz, K. Tenner-Racz, E. P. Rieber, K. Kuus-Reichel, R. S. Gelman, N. L. Letvin, D. C. Montefiori, R. M. Ruprecht, R. C. Desrosiers, and K. A. Reimann. 2005. Effect of CD8+ lymphocyte depletion on virus containment after simian immunodeficiency virus SIVmac251 challenge of live attenuated SIVmac239Δ3-vaccinated rhesus macaques. J. Virol. 798131-8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sernicola, L., F. Corrias, M. L. Koanga-Mogtomo, S. Baroncelli, S. Di Fabio, M. T. Maggiorella, R. Belli, Z. Michelini, I. Macchia, A. Cesolini, L. Cioe, P. Verani, and F. Titti. 1999. Long-lasting protection by live attenuated simian immunodeficiency virus in cynomolgus monkeys: no detection of reactivation after stimulation with a recall antigen. Virology 256291-302. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe, S. A., A. Cope, S. Dowall, N. Berry, C. Ham, J. L. Heeney, D. Hopkins, L. Easterbrook, M. Dennis, N. Almond, and M. Cranage. 2004. Macaques infected long-term with attenuated simian immunodeficiency virus (SIVmac) remain resistant to wild-type challenge, despite declining cytotoxic T lymphocyte responses to an immunodominant epitope. J. Gen. Virol. 852591-2602. [DOI] [PubMed] [Google Scholar]
- 48.Sharpe, S. A., A. M. Whatmore, G. A. Hall, and M. P. Cranage. 1997. Macaques infected with attenuated simian immunodeficiency virus resist superinfection with virulence-revertant virus. J. Gen. Virol. 781923-1927. [DOI] [PubMed] [Google Scholar]
- 49.Shibata, R., T. Igarashi, N. Haigwood, A. Buckler-White, R. Ogert, W. Ross, R. Willey, M. W. Cho, and M. A. Martin. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5204-210. [DOI] [PubMed] [Google Scholar]
- 50.Smith, D. M., M. C. Strain, S. D. Frost, S. K. Pillai, J. K. Wong, T. Wrin, Y. Liu, C. J. Petropolous, E. S. Daar, S. J. Little, and D. D. Richman. 2006. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 3551-5. [DOI] [PubMed] [Google Scholar]
- 51.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, E. S. Daar, D. D. Richman, and S. J. Little. 2004. Incidence of HIV superinfection following primary infection. JAMA 2921177-1178. [DOI] [PubMed] [Google Scholar]
- 52.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, C. J. Petropoulos, D. D. Richman, and S. J. Little. 2005. HIV drug resistance acquired through superinfection. AIDS 191251-1256. [DOI] [PubMed] [Google Scholar]
- 53.Stebbings, R., N. Berry, J. Stott, R. Hull, B. Walker, J. Lines, W. Elsley, S. Brown, A. Wade-Evans, G. Davis, J. Cowie, M. Sethi, and N. Almond. 2004. Vaccination with live attenuated simian immunodeficiency virus for 21 days protects against superinfection. Virology 330249-260. [DOI] [PubMed] [Google Scholar]
- 54.Stebbings, R., N. Berry, H. Waldmann, P. Bird, G. Hale, J. Stott, D. North, R. Hull, J. Hall, J. Lines, S. Brown, N. D'Arcy, L. Davis, W. Elsley, C. Edwards, D. Ferguson, J. Allen, and N. Almond. 2005. CD8+ lymphocytes do not mediate protection against acute superinfection 20 days after vaccination with a live attenuated simian immunodeficiency virus. J. Virol. 7912264-12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stebbings, R. J., N. M. Almond, E. J. Stott, N. Berry, A. M. Wade-Evans, R. Hull, J. Lines, P. Silvera, R. Sangster, T. Corcoran, J. Rose, and K. B. Walker. 2002. Mechanisms of protection induced by attenuated simian immunodeficiency virus. Virology 296338-353. [DOI] [PubMed] [Google Scholar]
- 56.Stephens, E. B., S. V. Joag, B. Atkinson, M. Sahni, Z. Li, L. Foresman, I. Adany, and O. Narayan. 1997. Infected macaques that controlled replication of SIVmac or nonpathogenic SHIV developed sterilizing resistance against pathogenic SHIV(KU-1). Virology 234328-339. [DOI] [PubMed] [Google Scholar]
- 57.Tenner-Racz, K., C. Stahl Hennig, K. Uberla, H. Stoiber, R. Ignatius, J. Heeney, R. M. Steinman, and P. Racz. 2004. Early protection against pathogenic virus infection at a mucosal challenge site after vaccination with attenuated simian immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1013017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Titti, F., L. Sernicola, A. Geraci, G. Panzini, S. Di Fabio, R. Belli, F. Monardo, A. Borsetti, M. T. Maggiorella, M. Koanga-Mogtomo, F. Corrias, R. Zamarchi, A. Amadori, L. Chieco-Bianchi, and P. Verani. 1997. Live attenuated simian immunodeficiency virus prevents super-infection by cloned SIVmac251 in cynomolgus monkeys. J. Gen. Virol. 782529-2539. [DOI] [PubMed] [Google Scholar]
- 59.Tsui, R., B. L. Herring, J. D. Barbour, R. M. Grant, P. Bacchetti, A. Kral, B. R. Edlin, and E. L. Delwart. 2004. Human immunodeficiency virus type 1 superinfection was not detected following 215 years of injection drug user exposure. J. Virol. 7894-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Kuyl, A. C., K. Kozaczynska, R. van den Burg, F. Zorgdrager, N. Back, S. Jurriaans, B. Berkhout, P. Reiss, and M. Cornelissen. 2005. Triple HIV-1 infection. N. Engl. J. Med. 3522557-2559. [DOI] [PubMed] [Google Scholar]
- 61.Wakrim, L., R. Le Grand, B. Vaslin, A. Cheret, F. Matheux, F. Theodoro, P. Roques, I. Nicol-Jourdain, and D. Dormont. 1996. Superinfection of HIV-2-preinfected macaques after rectal exposure to a primary isolate of SIVmac251. Virology 221260-270. [DOI] [PubMed] [Google Scholar]
- 62.Walther-Jallow, L., C. Nilsson, J. Soderlund, P. ten Haaft, B. Makitalo, P. Biberfeld, P. Bottiger, J. Heeney, G. Biberfeld, and R. Thorstensson. 2001. Cross-protection against mucosal simian immunodeficiency virus (SIVsm) challenge in human immunodeficiency virus type 2-vaccinated cynomolgus monkeys. J. Gen. Virol. 821601-1612. [DOI] [PubMed] [Google Scholar]
- 63.Wyand, M. S., K. Manson, D. C. Montefiori, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 738356-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 703724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiridou, M., F. van Griensven, J. W. Tappero, M. Martin, M. Gurwith, S. Vanichseni, W. Kittikraisak, R. Coutinho, and K. Choopanya. 2007. The spread of HIV-1 subtypes B and CRF01_AE among injecting drug users in Bangkok, Thailand. J. Acquir. Immune Defic. Syndr. 45468-475. [DOI] [PubMed] [Google Scholar]
- 66.Yang, O. O., E. S. Daar, B. D. Jamieson, A. Balamurugan, D. M. Smith, J. A. Pitt, C. J. Petropoulos, D. D. Richman, S. J. Little, and A. J. L. Brown. 2005. Human immunodeficiency virus type 1 clade B superinfection: evidence for differential immune containment of distinct clade B strains. J. Virol. 79860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yerly, S., S. Jost, M. Monnat, A. Telenti, M. Cavassini, J. P. Chave, L. Kaiser, P. Burgisser, and L. Perrin. 2004. HIV-1 co/super-infection in intravenous drug users. AIDS 181413-1421. [DOI] [PubMed] [Google Scholar]