Abstract
The interaction of papillomavirus E2 proteins with cellular Brd4 protein is important for transcriptional regulation of viral genes and partitioning of viral genomes. Bovine papillomavirus type 1 (BPV-1) E2 binds cellular chromatin in complex with Brd4 in both mitotic and interphase cells. To identify specific sites of E2 interaction on cellular chromatin, a genome-wide chromatin immunoprecipitation-on-chip analysis was carried out using human promoter sequences. Both E2 and Brd4 were found bound to most transcriptionally active promoters in C33A cells. These promoters were also bound by RNA polymerase II and were modified by histone H3 acetylation and K4 trimethylation, all indicators of active transcription. E2 binding strongly correlated with Brd4 and RNA polymerase II occupancy and H3K4me3 modification at all human promoters, indicating that E2 bound to active promoters. E2 binding did not correlate with the presence of consensus E2 binding sites in the promoters. Furthermore, the mRNA levels of E2-bound cellular genes were not significantly changed by E2 expression. Thus, the papillomavirus E2 proteins bind to transcriptionally active cellular genes but do not change their activity. We propose that this may be a way for the virus to ensure that the viral genome is retained in transcriptionally active regions of the nucleus to escape silencing. Therefore, E2-mediated tethering of viral genomes to host chromatin has multiple roles: to partition the viral genome to daughter cells, to ensure that the genomes are retained in the nucleus, and to make certain that the genomes are retained in functionally active nuclear domains.
Papillomavirus genomes replicate and are maintained as extrachromosomal elements that replicate in synchrony with cellular DNA in persistently infected cells. The viral E2 protein plays a central role in this process. E2 is a sequence-specific DNA binding protein that consists of an N-terminal transactivation domain linked to a C-terminal DNA binding/dimerization domain. Papillomavirus genomes contain a number of E2 binding sites important for viral transcription and replication. The E2 protein initiates viral DNA replication by loading the E1 helicase protein to the replication origin (14) and partitions the viral genomes to daughter cells by tethering them to cellular mitotic chromosomes (4, 8, 18). E2 is also the major transcriptional regulatory protein of the virus, and it can either activate or repress transcription depending on the relative position of its binding sites with respect to essential promoter elements.
Most activities of the E2 protein are mediated by interaction with cellular proteins in the nuclei of infected cells. E2 is localized in numerous nuclear speckles and has been demonstrated to interact with both chromatin and the nuclear matrix (3, 7, 12, 18, 24). Bovine papillomavirus type 1 (BPV-1) E2 interacts with chromatin in both mitotic and interphase cells, and this association requires the N-terminal transactivation domain of E2 (7, 12, 18). E2 binds to chromatin in association with the cellular bromodomain protein Brd4, and this interaction is important for the transcriptional function of all papillomaviruses (5, 11, 16, 17) and the genome partitioning function of some of them (11). McPhillips et al. demonstrated that the E2 protein stabilizes the interaction of Brd4 with both interphase and mitotic chromatin (11), and Kurg et al. demonstrated that a portion of BPV-1 E2 is bound to transcriptionally active chromatin (7).
To analyze whether the E2 proteins bind to specific chromosomal regions, we have studied the association of both BPV-1 and human papillomavirus type 1a (HPV-1a) E2 proteins with chromatin in the human cervical carcinoma-derived cell line C33A. We find that the E2 proteins bind to the chromatin of all active promoters, in association with the cellular Brd4 protein. However, this association does not affect the transcriptional activity of these promoters. We postulate that E2 associates with these regions to tether and maintain the viral genome in functionally active regions of the nucleus.
MATERIALS AND METHODS
Plasmids.
The pMEP4 expression vectors for FLAG-tagged E2 have previously been described (7). BPV-1 E2 proteins containing alanine substitutions in residues R37 and I73 were described previously (7). Standard mutagenesis procedures were used to mutate HPV-1a E2 residues R37 and I73 to alanines in pTZ18U/FLAG HPV-1a E2. FLAG-HPV-1a E2 (R37A/I73A) was subcloned into the Asp718 and blunted NheI sites of pMEP4. The FLAG-hemagglutinin tag was introduced into HindIII and blunted NotI sites of pMEP4 to generate the control plasmid pMEP-fh.
Antibodies.
Control mouse immunoglobulin G (IgG) and rabbit IgG were purchased from Jackson ImmunoResearch. Anti-FLAG M2 was from Sigma. Antibodies to RNA polymerase II (pol II; 8WG16, H5, H14) and histones (H3K4me3 and H3K9me3) were from Covance and Upstate, respectively. Antibodies to Cdk9 (C-20), TFIIB (C-18), TFIID (Sl-1), TFIIF (C-18), and RNA pol II (N-20) were from Santa Cruz. Anti-BPV-1 E2, B201 monoclonal antibody was a gift from Elliot Androphy. The polyclonal anti-Brd4 antiserum (directed against a C-terminal peptide) was described previously (1).
Establishment of E2-expressing cells.
Stable cell lines were generated by transfecting C33A cells with the pMEP-E2 expression plasmids using Fugene 6 (Roche). Cells containing the episomal plasmid were selected with 80 μg/ml of hygromycin B. Drug-resistant colonies were pooled after 2 weeks. E2 protein expression was induced by cadmium sulfate stimulation for 4 h before harvesting cells. The levels of E2 proteins were titrated and adjusted by differential cadmium sulfate concentration.
ChIP-chip analysis.
Chromatin immunoprecipitation-on-chip (ChIP-chip) experiments were carried out according to the protocol provided by NimbleGen. C33A cells were fixed in 1% formaldehyde for 10 min at room temperature, and the fixation was stopped by adding 125 mM glycine for 10 min. The cells were washed twice with phosphate-buffered saline and resuspended in lysis buffer 1 (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA [pH 8.0], 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, Complete protease inhibitor cocktail) for 10 min at 4°C, lysis buffer 2 (10 mM Tris [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, Complete protease inhibitor cocktail) for 10 min at room temperature, and then lysis buffer 3 (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, Complete protease inhibitor cocktail). Chromatin was sheared by sonication to a DNA fragment size of 500 bp, harvested by centrifugation, and used for ChIP analysis.
Chromatin samples were prepared from control cells and BPV-1 E2-expressing cells. Two milligrams of chromatin samples (as measured by A260) were incubated overnight with E2-specific B201 antibody prebound to Dynabeads conjugated to sheep anti-mouse IgG (Invitrogen). The immunoprecipitates were washed extensively with NimbleGen radioimmunoprecipitation assay buffer (50 mM HEPES [pH 8.0], 1 mM EDTA [pH 8.0], 1% NP-40, 0.7% deoxycholate, 0.5 M LiCl, Complete protease inhibitor cocktail) and once with Tris-EDTA buffer. Eluted chromatin DNA was reverse cross-linked, treated with proteinase K, and purified by phenol and phenol-chloroform extraction. After removal of RNA by RNase A treatment, purified chromatin DNA was blunted by incubation with T4 DNA pol, ligated with annealed linkers, and amplified by linker-mediated PCR. Amplified DNA was labeled and hybridized to HG18 build 36 promoter tiling array chips by NimbleGen (Madison, WI). These arrays contain 23,000 tiled regions and encompass 3,500 bp upstream and 750 bp downstream of known transcriptional start sites, with an interval spacing of 100 bp.
Each signal on the array has a corresponding scaled log2 ratio. This is the ratio of the input signals for the immunoprecipitated versus total DNA (not taken through immunoprecipitation steps) and is plotted against genomic position to identify regions where increased signal (i.e., DNA fragment enrichment) is observed relative to the control sample. The log2 ratio was computed and scaled to center the ratio data around zero. Scaling was performed by subtracting the bi-weight mean for the log2 ratio values for all features on the array from each log2 ratio value (Guide to ChIP-chip data, p. 3, 2007, Roche NimbleGen, Madison, WI). Peaks were generated from the scaled log2 ratio data files, and statistically significant binding/modification sites were identified and mapped to the transcription start site of each gene. Peaks with a false discovery rate (FDR) score of <0.05 were selected for analysis, as they represent the highest confidence protein binding site (see Table S2 in the supplemental data).
Statistical analysis of ChIP-chip data.
ChIP-chip data sets were obtained for BPV-1 E2, Brd4, RNA pol II, and H3K4me3. R software (http://www.r-project.org) was used to obtain pairwise correlations and scatter plots among the sets. The transcriptional start site information was obtained from the raw gene-finding format (GFF) files, 2006-07-18_HG18_PROMOTER_1of2.GFF and 2006-07-18_HG18_PROMOTER_2of2.GFF, provided by NimbleGen. Data were extracted from all promoters represented on the plus strand of chromosomes 1 to 10. This constituted 12,579 start sites; values representing the average ChIP signal obtained from a region spanning −750 bp to +750 bp of each transcriptional start site were calculated. These values were used to generate scatter plots and compute their pairwise correlations for the six pairs of data (BPV-1 E2 versus Brd4, BPV-1 E2 versus RNA pol II, BPV-1 E2 versus H3K4me3, Brd4 versus RNA pol II, Brd4 versus H3K4me3, and RNA pol II versus H3K4me3).
Conventional ChIP assay.
Chromatin samples were prepared from control and BPV-1 E2-expressing cells, as described above. Chromatin samples (0.5 mg) were incubated overnight with specific antibodies prebound to Dynabeads protein G (Invitrogen). After extensive washing, immunoprecipitated chromatin DNA was purified and analyzed by real-time PCR using specific primers.
Real-time QRT-PCR.
Total RNA was purified from C33A cells expressing pMEP4 control, BPV-1 E2, and HPV-1a E2 using Trizol (Invitrogen). One microgram of total RNA was reverse transcribed by Superscript III reverse transcriptase (Stratagene) using oligo(dT) primers. Real-time quantitative reverse transcription-PCR (QRT-PCR) was performed using an ABI Prism 7900HT sequence detector (Applied Biosystems) and SYBR Green PCR Master Mix (Applied Biosystems). Each reaction contained 12.5 μl of SYBR Green PCR Master Mix, cDNA from 0.5 ng of RNA, and a 0.3 μM concentration of each oligonucleotide primer in a total volume of 25 μl. In each run, a fourfold dilution series of pooled cDNA was used to generate a standard curve of the threshold cycle (CT) versus the log of quantity. PCR was performed at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 10 s and annealing and extension at 60°C for 60 s. The specificity of each primer pair was determined by dissociation curve analysis. The data were analyzed with SDS 2.1 software (Applied Biosystems). Specific signal for each mRNA transcript was normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal. See Table S1 in the supplemental data for a list of the primers used.
RESULTS
BPV-1 E2 binds to a subset of cellular promoter regions.
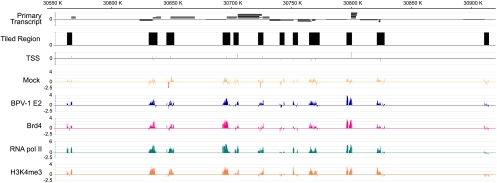
To identify E2 binding regions on cellular chromatin, we performed ChIP-chip analysis using NimbleGen microarrays containing 5-kb regions from 23,001 regions tiled around known human promoters with probes spaced, at most, 100 bp apart (HG18 5-kb human promoter arrays). C33A cell lines were established containing an episomal BPV-1 E2 expression plasmid. E2 expression was induced by Cd2+ treatment of the inducible metallothionein promoter, and care was taken to ensure that E2 expression was at low levels. Cellular chromatin, bound by E2, was collected by immunoprecipitation with the B201 E2-specific monoclonal antibody, and the extracted DNA was amplified by linker-mediated PCR. The amplified DNA was hybridized to the promoter arrays, and ChIP-chip data were analyzed using the SignalMap program (NimbleGen). From 59,557 transcriptional start sites in 23,001 tiled regions, there were 4,926 peaks of E2 binding with an FDR score of <0.05, which represent high-confidence protein binding sites. This is shown in the representative image in Fig. 1. For the data on the 4,926 E2 binding peaks, which are shared by 6,060 different genes, see Table S2 in the supplemental material. The binding peaks are often present in overlapping promoter elements adjacent to several different genes.
FIG. 1.
Representative ChIP-chip data from NimbleGen 5-kb promoter tiling arrays. Chromatin samples were isolated from BPV-1 E2-expressing cells using the antibodies against BPV-1 E2 (B201), Brd4, RNA pol II, and H3K4me3 and characterized by hybridization to promoter microarray chips by using NimbleGen. The chromosomal coordinates and position of primary transcripts are shown in the top two lines. The tiled regions of the chromosome surrounding the promoter regions and containing the oligonucleotide probes are represented by black blocks, and the transcriptional start sites are shown below these blocks. Note that only the promoter sequences from these blocked regions are represented on the arrays. The next two lines show the ChIP signals obtained from experimental samples from control and BPV-1 E2-expressing cells. The next three lines show ChIP signals for Brd4, RNA pol II, and H3K4me3, respectively. K, kilobase.
BPV-1 E2 binds transcriptionally active promoters.
The large number of target promoters indicated that E2 might be bound to all transcriptionally active promoters. To investigate this further, we performed additional ChIP-chip screenings using antibodies specific for Brd4, RNA pol II, and trimethylated lysine 4 of histone H3 (H3K4me3), as well as BPV-1 E2. RNA pol II and H3K4me3 are markers for active transcription of promoters, and Brd4 is a known chromatin binding partner of E2. Regions that showed BPV-1 E2 binding were also positive for binding of Brd4, RNA pol II, and H3K4me3. Therefore, we can conclude that most transcriptionally active promoters are occupied by both E2 and Brd4 proteins. Representative data from the ChIP-chip analysis for BPV-1 E2, Brd4, RNA pol II, and H3K4me3 are shown in Fig. 1. Shown are the tiled regions around 12 transcriptional start sites from a small region of chromosome six.
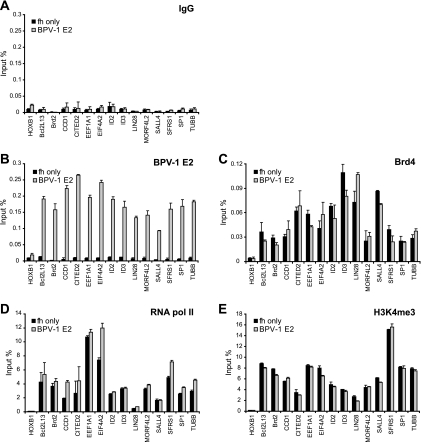
To confirm the ChIP-chip data, we performed conventional ChIP assays for E2 binding on selected cellular promoters. Fourteen positive E2 binding promoters were chosen from the top 100 ranked promoters along with one negative E2 binding promoter. The primers amplified regions that were mostly within 100 bp of the transcriptional start site. Only low levels of ChIP signal could be detected using control mouse IgG in both control and BPV-1 E2-expressing cells. However, all of the E2-positive promoters showed high signals of E2 binding in BPV-1 E2-expressing cells, but not in control cells (Fig. 2). The level of E2 binding to the negative promoter, HoxB1, was similar to the levels measured using control IgG. These data confirm that BPV-1 E2 binds selectively to different promoters.
FIG. 2.
BPV-1 E2 binds to transcriptionally active promoters. Chromatin samples from control (fh-only) and BPV-1 E2-expressing C33A cells were subjected to ChIP by IgG (A) and specific antibodies against BPV-1 E2 (B201) (B), Brd4 (C), RNA pol II (8WG16) (D), and H3K4me3 (E). ChIP DNA samples were analyzed by real-time PCR with specific primer sets for the target promoters indicated. The first promoter (HOXB1) was identified as negative for BPV-1 E2 binding by CHIP-chip analysis, and all of the others were positive for E2 binding. ChIP signals were expressed as percentages of chromatin DNA immunoprecipitated from the input amount of chromatin. TUBB, beta tubulin.
To further analyze the transcriptional status of the E2-bound promoters, the same promoters were analyzed for recruitment of RNA pol II and H3K4me3 modification by conventional ChIP. All of the E2-bound promoters showed high occupancy of RNA pol II and H3K4me3 and Brd4 proteins (Fig. 2) as well as acetylated H3, acetylated H4, and H3K4me2, which are other markers for active transcription of the promoters (data not shown). But H3K9me3 and H3K27me3, which are markers for inactive transcription, showed low signals on the selected promoters (data not shown). Therefore, E2 is recruited to transcriptionally active promoters but does not change the steady-state levels of RNA pol and Brd4 bound (Fig. 2C and D) or H3K4me3 histone modification (Fig. 2E).
BPV-1 E2 binding correlates with that of RNA pol II and H3K4me3.
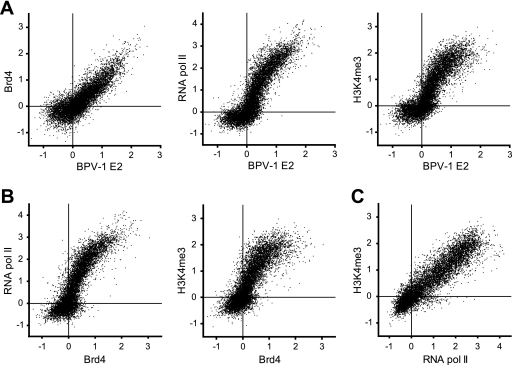
To further analyze the corelationship between BPV-1 E2 and Brd4, RNA pol II, and H3K4me3, ChIP-chip signals for 12,579 promoters (those from the plus strand of chromosomes 1 to 10) were compared in scatter plots (Fig. 3A). The mean binding signal for a region ±750 nucleotides from the transcriptional start site was plotted. The ChIP signals for BPV-1 E2 correlated well with those of Brd4 (R = 0.851), RNA pol II (R = 0.877), and H3K4me3 (R = 0.848). As shown in Fig. 3B, Brd4 binding also correlated with the binding of RNA pol II (R = 0.871) and H3K4me3 (R = 0.820). As expected for markers of active transcription, RNA pol II binding correlated with H3K4me3 binding on promoter regions (R = 0.810) (Fig. 3C). These data indicate that binding of both BPV-1 E2 and Brd4 to promoter regions of cellular genes correlates with the transcriptional activity of these promoters.
FIG. 3.
Binding of BPV-1 E2 to promoters correlates with binding of Brd4, RNA pol II, and H3K4me3. The mean values of binding signals (±750 bp from the transcriptional start site [TSS]) obtained from the ChIP-chip data for the 12,579 promoters on the plus strand of chromosomes 1 to 10 were plotted for BPV-1 E2 versus Brd4, RNA pol II, or H3K4me3 (A); Brd4 versus RNA pol II or H3K4me3 (B); and RNA pol II versus H3K4me3 (C).
BPV-1 E2 binding does not affect recruitment of cellular factors to the target promoters.
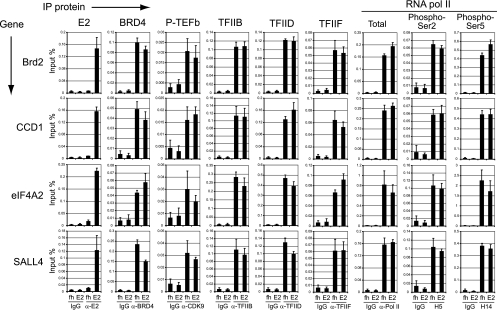
Further ChIP experiments were carried out to determine whether E2 binding increased the recruitment of cellular transcriptional factors to the promoter regions. ChIP chromatin samples were prepared from control and E2-expressing cells using control IgG- and E2-specific antibodies, B201, and FLAG. Antibodies to cellular factors, positive transcription elongation factor b (P-TEFb), TFIIB, TFIID, TFIIF, and RNA pol II were also used. Modification-specific antibodies directed against RNA pol II phosphorylated on serine 5, which is phosphorylated during initiation and recruits the capping enzyme, and RNA pol II phosphorylated on serine 2, which is phosphorylated during elongation by Cdk9/P-TEFb (15), were also used. The resulting chromatin DNA samples were analyzed by quantitative real-time PCR using primers specific for promoter regions of Brd2, Ccd1, eIF4A2, and Sall4 genes. BPV-1 E2 was specifically bound to these promoters in E2-expressing cells, but not in control cells (Fig. 4). A control immunoprecipitation with nonspecific IgG showed low background signals for all of the samples. Each cellular factor tested was strongly recruited to the promoter region of each gene in both control and E2-expressing cells. However, recruitment of the cellular factors to the promoters did not significantly change upon expression of BPV-1 E2. These data indicate that recruitment of BPV-1 E2 to cellular promoters does not change their transcriptional activity.
FIG. 4.
BPV-1 E2 does not alter the recruitment of cellular factors to active promoters. Chromatin samples from control (fh) and BPV-1 E2-expressing C33A cells (E2) were subjected to ChIP (IP) with control IgG or with specific antibodies against BPV-1 E2 (B201), Brd4, P-TEFb, TFIIB, TFIID, TFIIF, and different modification states of RNA pol II (total [N-20], Ser2-phosphorylated C-terminal domain [H5], and Ser5-phosphorylated C-terminal domain [H14]). ChIP DNA was analyzed by quantitative real-time PCR with primer sets for specific target promoters that were previously identified as positive for BPV-1 E2 binding. ChIP signals were expressed as the percentage of chromatin DNA immunoprecipitated from the input amount of chromatin. Phospho-Ser2 or -Ser5, RNA pol II phosphorylated on serine 2 or 5, respectively; α-, anti-.
The level of cellular mRNA expressed from cellular promoters is not affected by BPV-1 E2 binding.
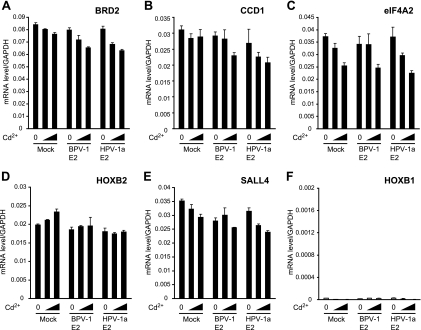
To further analyze the effect of E2 binding on the activity of the bound cellular genes, mRNA from several E2-bound promoters was reverse transcribed and the synthesized cDNA was analyzed by quantitative real-time PCR. RNA was prepared from control cells (expressing the tag only) and BPV-1 E2-expressing cells. Cells expressing HPV-1a E2 were also included in this analysis because HPV-1a has a Brd4 binding profile that is very similar to that of BPV-1 E2 (see Fig. 7A) (11). mRNA levels were assayed for five E2-bound promoters: Brd2, CCD1, eIF4A2, Sall4, and HoxB2. For all genes tested, there was no significant change in mRNA levels upon expression of either BPV-1 E2 or HPV-1a E2 (Fig. 5). This demonstrates that papillomavirus E2 proteins, although specifically bound to the regulatory regions, do not regulate the transcriptional activities of the target promoters. Consistent with our results, a recent microarray study showed that BPV-1 E2 expression had no effect on the expression of cellular genes (6).
FIG. 7.
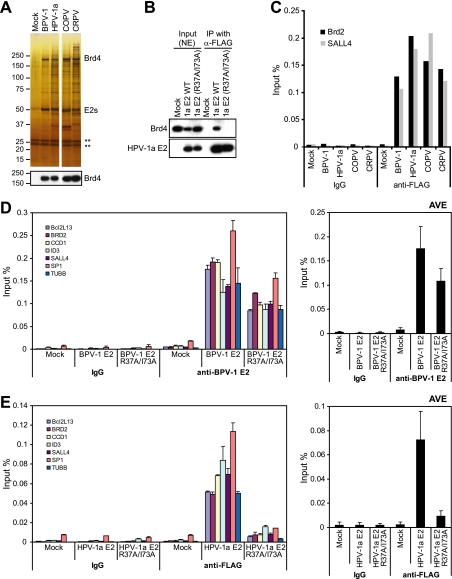
Brd4 association is important for E2 binding to cellular promoters. (A) E2 protein complexes were purified from C33A cells expressing tag only (Mock) and E2 proteins from BPV-1, HPV-1a, canine oral papillomavirus (COPV), and cottontail rabbit papillomavirus (CRPV) and visualized by silver staining and immunoblotting with anti-Brd4 antibody. ** represents Ig bands. (B) To confirm that HPV-1a E2 R37AI73A did not bind BRd4, a coimmunoprecipitation assay was carried out with C33 cells. (C) ChIP assays were performed with an anti-FLAG antibody, and signals for E2 binding on the Brd2 and SALL4 promoter regions were analyzed by real-time PCR with specific primers. (D) Chromatin samples were prepared from C33A cells expressing wild-type BPV-1 E2 or R37A/I73A mutated proteins. ChIP DNA was isolated using IgG and an anti-BPV-1 E2 antibody (B201). BPV-1 E2 binding on the promoter region was analyzed by real-time PCR with specific primers for the indicated target promoters. (E) ChIP assays were performed for C33A cells expressing FLAG-tagged wild-type HPV-1a E2 or the R37A/I73A mutated HPV-1a E2 protein using control IgG or anti-FLAG M2 antibody. HPV-1a E2 binding to the promoter regions was analyzed by real-time PCR with specific primers for the indicated target promoters. NE, nuclear abstract; IP, immunoprecipitation; α-, anti-; WT, wild type; TUBB, beta tubulin; AVE, average.
FIG. 5.
Expression of E2 proteins does not change the mRNA level of target genes. Expression of BPV-1 E2, HPV-1a E2, and cells expressing tag only (Mock) were induced by Cd2SO4 (0, 1, and 3 μM) in C33A cells for 4 h. Cellular mRNA was isolated from equivalent numbers of cells and used as a template to generate cDNA. Expression levels were assayed by QRT-PCR with specific primers for E2 binding promoters Brd2 (A), CCD1 (B), eIF4a2 (C), HoxB2 (D), and Sall4 (E) and for HoxB1 (F), a non-E2 binding promoter. All values were normalized to levels of GAPDH.
To further emphasize the association of E2 with active transcription, we used beta interferon to induce transcription of several interferon-responsive genes in U2OS cells expressing BPV1 and HPV-1a E2 proteins from the pMEP vectors. We hypothesized that the E2 proteins would be recruited to these induced promoters but might, in fact, repress induction of the interferon-responsive genes. However, consistent with the results presented above, E2 and Brd4 were rapidly recruited to these promoters, but E2 had no effect on the resulting induced levels of mRNA (M.K.J. and A.M., unpublished observations).
Binding of BPV-1 E2 is not dependent on consensus E2 binding sequences.
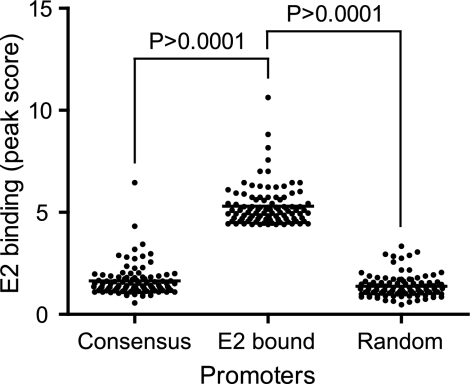
Promoters having consensus BPV-1 E2 binding sites (ACCGNNNNCGGT) between −1,000 and +200 were selected by searching the Database of Transcriptional Start Sites (DBTSS) (19). The level of E2 binding to 96 promoters containing consensus E2 binding sites was compared to the level of E2 binding to a similar number of promoters top ranked for E2 binding in the ChIP-chip analysis and to 99 randomly selected promoters. For the list of promoters analyzed, see Table S3 in the supplemental data. Only one promoter out of 96 with an E2 consensus site showed E2 protein binding in the ChIP-chip analysis. The level of BPV-1 E2 binding to promoters containing consensus E2 binding sites is very similar to that of the randomly selected group that does not contain E2 sites (Fig. 6). Therefore, consensus E2 binding sequences are not essential for E2 binding to active cellular promoters.
FIG. 6.
E2 promoter binding does not depend on E2 DNA binding motifs. Three groups of promoters were selected according to their BPV-1 E2 binding status. Promoters containing a consensus BPV-1 E2 binding motif (Consensus), promoters that were top ranked for BPV-1 E2 binding (E2 bound), and randomly selected promoters (Random). The E2 peak value from ChIP-chip signals for around 100 promoters from each of the three groups were plotted.
Several papillomavirus E2 proteins bind to active cellular promoters with Brd4.
C33A cell lines expressing 11 different papillomavirus E2 proteins were established, and E2-associated proteins were purified by tandem affinity purification (M.K.J., unpublished data). Four of the 11 E2 proteins, from BPV-1, HPV-1a, canine oral papillomavirus, and cottontail rabbit papillomavirus, showed strong interactions with Brd4 as shown by silver staining, mass spectrometric identification, and immunoblot analysis (Fig. 7A). We performed ChIP assays to check the binding of all four E2 proteins to the transcriptionally active Brd2 and Sall4 promoters. Each Brd4 binding E2 protein showed strong binding to the same promoter regions as BPV-1 E2 (Fig. 7C). Therefore, multiple papillomavirus E2 proteins can bind chromatin together with the Brd4 protein.
Brd4 is important for E2 binding to cellular promoter regions.
We have previously shown that the E2 proteins expressed from pMEP vectors in C33A cells are transcriptionally active and also that the double point mutation of R37A/I73A abrogates Brd4 binding and transcriptional activation in BPV-1 E2 (11). The R37A/I73A mutation was also introduced into the HPV-1a E2 protein and, as shown in the coimmunoprecipitation experiment in Fig. 7B, also abrogates Brd4 binding. To further investigate the role of Brd4 in the binding of E2 to cellular promoter regions, E2 binding was assayed by ChIP using BPV-1 and HPV-1a E2 proteins containing the R37A/I73 mutation. Chromatin samples from C33A cells expressing the control tag, the wild-type E2 proteins, and the R37A/I73A-mutated E2 proteins were subjected to ChIP assays with BPV-1 E2-specific B201 antibody or anti-FLAG M2 antibody for HPV-1a E2. Immunoprecipitated chromatin DNA was analyzed by real-time PCR using specific primers for selected active promoter regions. As shown in Fig. 7D and E, only low background binding signals could be detected in ChIP assays for IgG and from control cells (tag only). Both wild-type BPV-1 and HPV-1a E2s bound strongly to the promoter regions. However, E2 proteins defective for Brd4 binding showed decreased binding to the promoters. Notably, abrogation of Brd4 binding was more detrimental to HPV-1a E2 chromatin binding than it was for BPV-1 E2. Thus, the interaction of E2 with Brd4 is at least partially responsible for the interaction of E2 with transcriptionally active chromatin.
DISCUSSION
In this study, we have shown that the E2 proteins from several papillomaviruses associate with transcriptionally active regions of cellular chromatin. Using a ChIP-chip approach, we have demonstrated that the BPV-1 E2 protein binds to a subset of human promoters in concert with the bromodomain protein Brd4. This binding correlates with signatures of active transcription, such as binding of RNA pol II, acetylation of histones, and methylation of histone H3 K4. Therefore, the E2 proteins associate with regions of active chromatin.
Notably, binding of the E2 proteins to cellular chromatin does not correlate with the presence of consensus E2 binding motifs in the cellular DNA. This is consistent with previous findings showing that the DNA binding function of E2 was not required for the association of E2 with cellular chromatin and that the chromatin binding determinants resided in the E2 transactivation domain (7, 12). Furthermore, it is unlikely that the binding sites for a viral protein would be conserved in host genes unless, by chance, they overlapped the binding site for an essential cellular factor or provided some other advantage to the host organism. Therefore, these interactions are most likely mediated by interaction of E2 with cellular host factors.
Although the E2 proteins bind to active cellular genes, there is no change in the occupancy of transcription factors bound to these sites and no change in the activity of these promoters, as measured by mRNA levels. This finding is actually not surprising. Papillomaviruses are persistent viruses that develop a long-term relationship with the host cell, and it is unlikely that they would cause global changes in cellular gene expression in the persistent phase of their life cycle. This is completely consistent with a recent microarray study that showed that when expressed in HeLa cells, BPV-1 E2 had no effect on the expression of cellular genes (6). Rather than cause global changes in cellular gene expression, persistent viruses are much more likely to encode functions that specifically evade the host defense mechanisms and allow the virus to escape detection. For example, the papillomavirus E6 and E7 proteins can abrogate the cellular interferon response (reviewed in reference 20).
We find that the cellular bromodomain protein, Brd4, also binds to all active cellular promoters. Brd4 is a ubiquitous protein that is a component of both the transcriptional elongation complex pTEFb (23) and the Mediator complex (10) and functions as a chromatin adaptor (21). Brd4 is an essential gene (2) that is important for G1 transcription and progression into S phase (13). Based on the results presented here, it is likely that Brd4 is involved in the transcription of most, if not all, cellular genes. Binding of BPV-1 E2 to these promoters does not change the occupancy of Brd4 or other cellular factors. The effect of E2 and Brd4 on viral promoters, which are regulated by E2-specific binding sites, is very different. Under these circumstances, E2 can bind tightly to the promoters through its cognate DNA binding site and recruit Brd4 and other transcription factors. Notably, Brd4 seems to be important for the E2-dependent transcriptional regulation of all papillomavirus E2 proteins tested to date (11, 16, 17, 22).
Host cells have innate antiviral defenses to limit the expression of incoming foreign viral DNA, and in many cases, viral DNA is transcriptionally repressed and heterochromatinized (reviewed in reference 9). However, viruses are equipped to counteract this response and maintain their genomes in a functional state. Furthermore, small DNA viruses, such as the papillomaviruses, must use all of the host transcriptional and replication machinery to express and replicate their own genome. Thus, we propose that the interaction of the E2 proteins with transcriptionally active promoters is an additional mechanism to localize E2, and the associated viral genomes, in subregions of the nucleus that are permissive for transcription and replication. Our results are very consistent with those of Kurg et al., who demonstrated biochemically that a fraction of E2, and associated viral genomes, was associated with fractions of transcriptionally active cellular chromatin (7).
Studies are in progress to characterize the E2-Brd4 complexes that bind to cellular chromatin in mitosis, when cellular transcription is turned off. E2 tethers the viral genome to the host chromosomes in mitosis, and it has been proposed that this is important to partition the viral genome in approximately equal numbers to daughter cells and also to ensure that the viral genome is retained in the nucleus after cell division. We can now add a third reason for E2 to tether the viral genomes to cellular chromatin: to provide epigenetic memory and ensure that the genomes are retained in euchromatic regions of the nucleus to allow continued expression of the viral proteins important for its survival.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH.
We are grateful to Jonathan Spindler for mutagenesis of the HPV-1a E2 gene, to Elliot Androphy for the B201 antibody, and to Tom Kristie for comments on the manuscript.
Footnotes
Published ahead of print on 7 January 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 206537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 223794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbert, N. L., J. T. Schiller, D. R. Lowy, and E. J. Androphy. 1988. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc. Natl. Acad. Sci. USA 855864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 734404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 803660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johung, K., E. C. Goodwin, and D. DiMaio. 2007. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 812102-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurg, R., K. Sild, A. Ilves, M. Sepp, and M. Ustav. 2005. Association of bovine papillomavirus E2 protein with nuclear structures in vivo. J. Virol. 7910528-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 954338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman, P. M. 2008. Chromatin organization and virus gene expression. J. Cell. Physiol. 216295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25277-283. [DOI] [PubMed] [Google Scholar]
- 11.McPhillips, M. G., J. G. Oliveira, J. E. Spindler, R. Mitra, and A. A. McBride. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 809530-9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 798920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mochizuki, K., A. Nishiyama, M. K. Jang, A. Dey, A. Ghosh, T. Tamura, H. Natsume, H. J. Yao, and K. Ozato. 2008. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2839040-9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 2501694-1699. [DOI] [PubMed] [Google Scholar]
- 15.Morris, H., and S. Price. 1986. Langerhans' cells, papillomaviruses and oesophageal carcinoma: a hypothesis. S. Afr. Med. J. 69413-417. [PubMed] [Google Scholar]
- 16.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 804276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senechal, H., G. G. Poirier, B. Coulombe, L. A. Laimins, and J. Archambault. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 35810-17. [DOI] [PubMed] [Google Scholar]
- 18.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 722079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakaguri, H., R. Yamashita, Y. Suzuki, S. Sugano, and K. Nakai. 2008. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 36D97-D101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodworth, C. D. 2002. HPV innate immunity. Front. Biosci. 7d2058-d2071. [DOI] [PubMed] [Google Scholar]
- 21.Wu, S. Y., and C. M. Chiang. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 28213141-13145. [DOI] [PubMed] [Google Scholar]
- 22.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 202383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19535-545. [DOI] [PubMed] [Google Scholar]
- 24.Zou, N., B. Y. Lin, F. Duan, K. Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 743761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.