Abstract
High-risk types of human papillomavirus (HPV) are considered the major causative agents of cervical carcinoma. The transforming ability of HPV resides in the E6 and E7 oncogenes, yet the pathway to transformation is not well understood. Cells expressing the oncogene E7 from high-risk HPVs have a high incidence of polyploidy, which has been shown to occur as an early event in cervical carcinogenesis and predisposes the cells to aneuploidy. The mechanism through which E7 contributes to polyploidy is not known. It has been hypothesized that E7 induces polyploidy in response to mitotic stress by abrogating the mitotic spindle assembly checkpoint. It was also proposed that E7 may stimulate rereplication to induce polyploidy. We have tested these hypotheses by using human epithelial cells in which E7 expression induces a significant amount of polyploidy. We find that E7-expressing cells undergo normal mitoses with an intact spindle assembly checkpoint and that they are able to complete cytokinesis. Our results also exclude DNA rereplication as a major mechanism of polyploidization in E7-expressing cells upon microtubule disruption. Instead, we have shown that while normal cells arrest at the postmitotic checkpoint after adaptation to the spindle assembly checkpoint, E7-expressing cells replicate their DNA and propagate as polyploid cells. Thus, abrogation of the postmitotic checkpoint leads to polyploidy formation in E7-expressing human epithelial cells. Our results suggest that downregulation of pRb is important for E7 to induce polyploidy and abrogation of the postmitotic checkpoint.
An important hallmark of human cancers is aneuploidy, the state in which a cell has extra or missing chromosomes (12, 25). Polyploidy is the state in which cells have more than two equal sets of chromosomes and is thought to be an early event in multistep carcinogenesis that can lead to aneuploidy (1, 24), as exemplified in Barrett's esophagus (11). Polyploidy has recently been shown to occur as an early event in cervical carcinogenesis and to predispose the cells to aneuploidy (26). Other recent studies have shown that tetraploid but not diploid mouse or human cells induce tumor formation in mice (3, 9). These studies highlight the potential importance of polyploidy in carcinogenesis.
The cellular mechanisms responsible for this polyploidy formation are as of yet undetermined, but several models have been proposed. First, abrogation of the spindle assembly checkpoint followed by cleavage failure may lead to polyploidy formation (36, 40). A second proposed model is rereplication, a process of multiple rounds of DNA replication without an intervening mitosis. Third, cells that adapt to the mitotic spindle checkpoint halt in a G1-like state with 4C DNA content. Abrogation of this postmitotic checkpoint allows the cells to replicate their 4C DNA content, leading to polyploidy formation. This has been shown in cells that express the human papillomavirus type 16 (HPV-16) E6 oncogene that degrades p53 (21). Finally, cleavage failure, which yields binucleate cells with 4C DNA content, is also a potential mechanism for polyploidy formation (31).
The postmitotic checkpoint becomes activated when cells with an intact spindle assembly checkpoint become arrested during mitosis for a prolonged period of time and eventually adapt to the checkpoint, exit mitosis without cleavage, and progress into a G1-like state with 4C DNA content (19, 22). The cells are prevented from continuing through the cell cycle and replicating their DNA by a proposed p53- and pRb-dependent postmitotic checkpoint (18, 19).
High-risk types of HPV (of which HPV-16 is the most prevalent) are commonly associated with lesions that can progress to cervical carcinoma, which is one of the leading causes of cancer death in women worldwide (42). The transforming properties of high-risk HPVs primarily reside in the E6 and E7 oncogenes (reviewed in reference 7). The ability of high-risk HPV E6 and E7 proteins to promote the degradation of p53 and pRb, respectively, has been suggested as a mechanism by which HPV induces cellular transformation (6, 30). Expression of the high-risk HPV E6 and E7 oncogenes in human keratinocytes leads to polyploidy, which is enhanced by DNA damage and by activation of the spindle checkpoint through microtubule disruption (15, 27, 37, 38).
Previously, it was thought but not directly shown that high-risk E6 and E7 induce polyploidy in response to microtubule disruption by abrogating the spindle checkpoint and that degradation of the tumor suppressor p53 by E6 is the mechanism by which E6 accomplishes this polyploidy formation (27, 37, 38). Others have proposed that E7 may play a role in stimulating DNA rereplication that occurs prior to mitosis initiation and polyploidy formation (20). Our recent studies demonstrate that E6 does not affect the mitotic spindle checkpoint (21). Instead, E6 abrogates the postmitotic checkpoint to induce polyploidy after microtubule disruption. Interestingly, E6 mutant proteins defective in inducing p53 degradation also induce polyploidy (21). The mechanism by which HPV E7 induces polyploidy remains to be determined. In this study, we investigate these possible mechanisms through which HPV-16 E7 induces polyploidy formation.
MATERIALS AND METHODS
Cell culture.
Primary human keratinocytes (PHKs) were derived from neonatal human foreskin epithelium obtained from the University of Massachusetts Hospital as described previously (21). These cells were maintained on mitomycin C-treated J2-3T3 feeder cells in F medium composed of three parts Ham's F12 medium and one part Dulbecco's modified Eagle medium (DMEM) plus 5% fetal bovine serum (FBS), with all supplements as previously described (8). Retinal pigment epithelial (RPE1) cells (39) were maintained in a 1:1 dilution of DMEM and Ham's F12 medium plus 10% FBS. Mouse embryonic fibroblasts (MEFs) (provided by Stephen Jones, University of Massachusetts Medical School) were maintained in DMEM plus 15% FBS.
Retroviral infections.
PHKs and RPE1 cells expressing vector or E7 were established by retrovirus-mediated infection using the pBabe-puro-based retroviral construct. After puromycin selection, populations of infected cells were pooled. PHKs and RPE1 cells were maintained in puromycin and used within eight passages. E7 expression was confirmed by reverse transcription-PCR (RT-PCR) using previously described oligonucleotides (21) and by Western blotting.
Immunoblotting.
Protein extraction was performed in radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris, 5 mM EDTA, protease inhibitors [Complete mini EDTA-free, Roche]). Protein concentrations were determined by bicinchoninic acid analysis (Pierce). Equal amounts of protein from each cell lysate were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride membrane. The membranes were blotted with antibodies against E7 (Santa Cruz), pRb (BD Biosciences), cyclin B1 (BD Biosciences), cyclin D1 (Santa Cruz), and β-tubulin (Sigma). Immunoreactive proteins were visualized with SuperSignal West Femto maximum sensitivity substrate (Pierce). The membranes were scanned with an LAS-1000 image reader (Fuji Photo Film Inc.).
Flow cytometry.
Asynchronous cultures of cells were treated with dimethyl sulfoxide (DMSO; Sigma), nocodazole (50 ng/ml; Sigma), or monastrol (100 μM; Sigma). For the polyploidy experiments, the cells were harvested, fixed in 70% ethanol, and resuspended in a phosphate-buffered saline (PBS)-propidium iodide (PI) (50 μg/ml; Sigma)-RNase A (70 μg/ml; Sigma) solution. For the mitotic index and mitotic shake-off experiments, the cells were harvested at various time points, fixed in 70% ethanol, permeabilized in 0.25% Triton X-100, stained with 0.5 μg of the rat anti-phospho-histone H3 (anti-P-His H3) immunoglobulin G2a (IgG2a) (Sigma) and then with fluorescein isothiocyanate (FITC)-conjugated anti-rat IgG2a (BD Biosciences), and counterstained with PBS-PI-RNase A. For the bromodeoxyuridine (BrdU)-labeling experiments, the plated cells were incubated with BrdU (final 10 μM) for 2 h before being harvested and fixed in 70% ethanol. After fixation, the cells were permeabilized with 2 N HCl-0.5% Triton X-100, neutralized with 0.1 M sodium tetraborate, stained with anti-BrdU-FITC (BD Biosciences), stained with anti-mouse IgG F(ab′)2-FITC (Sigma), and counterstained with PBS-PI-RNase A. Cell cycle analysis was performed using FlowJo software (Becton Dickinson).
Time-lapse videomicroscopy.
Live-cell imaging was performed as previously described (32). Briefly, RPE1 cells were grown on 22- by 22-mm glass coverslips in low-HEPES (12.5 mM) DMEM for time-lapse videomicroscopy. Phase contrast images were acquired with a Leica DMIRE2 inverted microscope with a 10× objective lens, and images were acquired using Openlab 3.5.2 (Improvision Inc., Lexington, MA) and recorded using a QImaging Retiga EXi camera. Images were taken every 3 min with a 3-s exposure and later compressed to view as a movie using QuickTime 6.5 (Apple Computer, Inc., Cupertino, CA).
Statistical analysis.
Data were expressed as means ± standard deviations. Differences between means were assessed by Student's t test. P values of ≤0.05 were considered significant.
RESULTS
Expression of HPV E7 results in polyploidy.
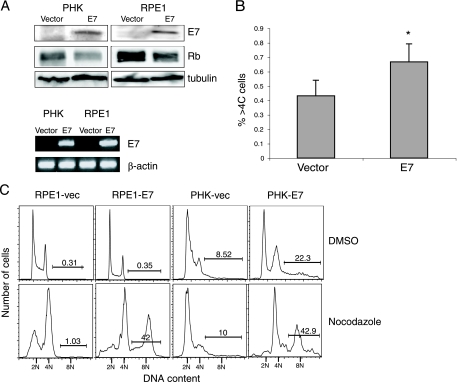
Stable cell lines were created by infection of both PHKs and hTERT-immortalized human RPE1 cells with retroviruses containing HPV-16 E7 or an empty vector. E7 expression in the stable cell lines was confirmed by Western blotting and by RT-PCR (Fig. 1A). As expected, the steady-state level of pRb is reduced in E7-expressing PHKs and RPE1 cells (Fig. 1A). To examine the incidence of polyploidization over an extended period of time, wild-type and E7-expressing RPE1 cells were serially passaged and stained with PI and analyzed by a fluorescence-activated cell sorter for DNA content. Because PHKs have a very limited life span in culture, the RPE1 cells were used for this experiment. Figure 1B shows that there was a small but statistically significant increase (P < 0.001) in the percentage of polyploid cells in RPE1-E7 cells (0.67% ± 0.13%) compared to that of the RPE1-vector control cells (0.43% ± 0.11%).
FIG. 1.
Expression of HPV E7 results in polyploidy formation. (A) Total protein extracts from PHKs and RPE1 cells expressing vector or E7 were resolved by SDS-PAGE and blotted with antibodies against E7, pRb, and tubulin (top). Total RNA isolated from PHKs and RPE1 cells expressing vector or E7 were subjected to RT-PCR using specific primers. The products were resolved on a 1% agarose gel. β-Actin was used as a control (bottom). (B) Untreated RPE1 cells expressing E7 or vector were harvested, fixed, and stained with PI. The percentages of polyploidy (>4C DNA content) (%>4C cells) for each were measured and averaged. *, P < 0.001. (C) Asynchronous cultures of RPE1 cells and PHKs expressing vector (vec) or E7 were treated with DMSO or nocodazole (50 ng/ml). Seventy-two or forty-eight hours later, the RPE1 cells and PHKs, respectively, were collected, fixed, stained with PI, and analyzed by flow cytometry. Data from a representative experiment (of three) are shown. The percentages of cells with more than 4C DNA content are indicated.
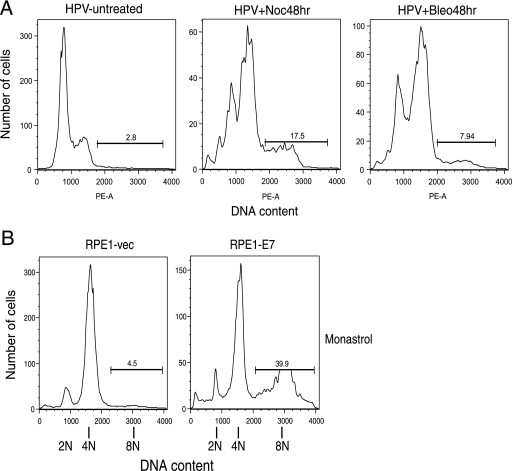
Because the level of spontaneous polyploidization was low in E7-expressing cells, we used the drug nocodazole to trigger the spindle assembly checkpoint instead of waiting for its activation through acquired mutations or long-term in vitro culture. This is biologically relevant since in HPV-infected cells, E6 could potentially trigger the spindle checkpoint as it induces a high percentage of metaphase-lagging (misaligned) chromosomes (5, 28). After treatment with nocodazole, E7-expressing PHKs and RPE1 cells showed a significant increase in polyploidy compared to vector controls (42.9% versus 10.0% in PHKs and 42.0% versus 1.0% in RPE1 cells; Fig. 1C). To examine the effect of E7 on polyploidy in conjunction with other HPV genes, we also used PHKs containing the HPV genome. Our results indicate that in response to nocodazole or bleomycin, PHKs containing the HPV genome become polyploid (Fig. 2A). To investigate to what extent this result is specific to nocodazole, we used the drug monastrol, a small-molecule inhibitor of the mitotic kinesin motor protein Eg5, which triggers the spindle assembly checkpoint through the formation of monopolar spindles. The RPE1-E7 cells also showed an increase in polyploidy (39.9% versus 4.5%) in response to monastrol (Fig. 2B).
FIG. 2.
E7 promotes polyploidy in the context of the HPV genome and upon monastrol treatment. (A) Asynchronous cultures of PHKs containing the HPV genome were either left untreated or treated with nocodazole (Noc; 50 ng/ml) or bleomycin (Bleo; 100 ng/ml). Forty-eight hours later, the cells were collected, fixed, stained with PI, and analyzed by flow cytometry. PE-A, phycoerythrin A. (B) Asynchronous cultures of RPE1 cells expressing vector (vec) or E7 were treated with DMSO or monastrol (100 μM). Seventy-two hours later, the RPE1 cells were collected, fixed, stained with PI, and analyzed by flow cytometry.
HPV E7 expression does not affect the spindle assembly checkpoint or mitosis.
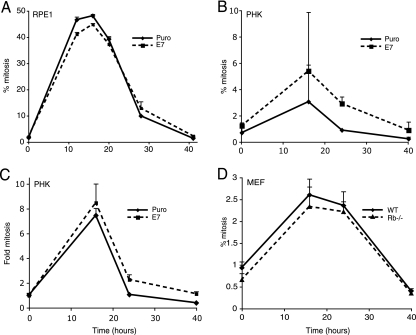
Previously, it was thought but not directly shown that high-risk E7 induces polyploidy in response to microtubule disruption by abrogating the spindle checkpoint (27, 37). Here, we examined the effect of HPV E7 expression on the spindle checkpoint by using an antibody against the mitotic marker P-His H3 that stains mitotic cells from prophase to late telophase (13). Concurrent staining with both the P-His H3 antibody and PI allows us to differentiate between mitotic cells and interphasic cells with 4C DNA content. We constructed a mitotic index curve by treating E7-expressing and vector control cells with nocodazole and examining the percentage of mitotic cells with 4C DNA content. By staining the cells at various time points, we determine the percentage of cells in mitosis over time. This allows us to determine whether the spindle assembly checkpoint becomes activated or is abrogated. When the cells halt in mitosis, the percentage of cells positive for P-His H3 increases. As the cells adapt to the spindle assembly checkpoint, the percentage of cells positive for P-His H3 decreases. Cells defective for the spindle assembly checkpoint should fail to arrest in mitosis. Therefore, there would be no accumulation of P-His H3-positive cells, and the percentage of E7-expressing cells in mitosis would be significantly lower than that in the control cells.
Figure 3A shows that the mitotic index curves for HPV-16 E7-expressing RPE1 cells are not significantly different from those for the vector control cells. In PHKs, the percentage of E7-expressing cells in mitosis was even higher than that in the control cells, though the difference is not statistically significant (Fig. 3B). We believe this is due to the fact that control PHKs do not proliferate as efficiently as E7-expressing PHKs. When the curves of mitosis are compared, there is essentially no difference between the mitotic index of E7 and that of control PHKs (Fig. 3C). Therefore, the spindle assembly checkpoint is functional in E7-expressing cells, which suggests that an alternate mechanism leads to polyploidization in E7-expressing cells.
FIG. 3.
E7 expression does not affect the spindle assembly checkpoint. Mitotic index of RPE1 cells (A), PHKs containing vector or expressing E7 (B and C), and wild-type (WT) and Rb−/− MEFs (D). Cells under nocodazole treatment (50 ng/ml) were harvested at various time points, fixed, stained with rat anti-P-His H3 IgG2a and FITC-conjugated anti-rat IgG2a, stained with PI, and analyzed by flow cytometry. The percentage of cells positive for P-His H3 with 4C DNA content was plotted as a function of time (A, B, and D). The increase (relative to interphase cells) of cells positive for P-His H3, with DNA content around 4C, was plotted as a function of time (C). Puro, puromycin. Data represent the means of two to three determinants from a representative experiment of three.
We also used time-lapse videomicroscopy to compare the lengths of normal mitosis and cytokinesis for RPE1-vector versus those for RPE1-E7 cells. The duration of mitosis was measured as the time between nuclear envelope breakdown and nuclear envelope reformation. Table 1 shows that the time spent in mitosis was not significantly different between RPE1-vector and RPE1-E7 cells (34 min ± 7 min versus 37 min ± 6 min, respectively). These data are consistent with the mitotic index curves in Fig. 3, supporting the hypothesis that the expression of E7 does not have a significant effect on the duration of mitosis.
TABLE 1.
HPV E7 expression does not affect mitosis as determined by time-lapse videomicroscopy
| RPE1 cells or MEFsa | Number of cells | Length of mitosis (min) | Length of cytokinesis (min) | Length of mitosis + cytokinesis (min) |
|---|---|---|---|---|
| RPE1 cells | ||||
| RPE1-vector | 56 | 34 ± 7 | 96 ± 28 | 128 ± 30 |
| RPE1-E7 | 17 | 37 ± 6 | 102 ± 39 | 138 ± 40 |
| MEFs | ||||
| Wild-type MEFs | 40 | 41 ± 11 | 33 ± 16 | 74 ± 17 |
| Rb−/− MEFs | 40 | 44 ± 15 | 45 ± 27 | 89 ± 38 |
For RPE1 cells, P values of 0.19, 0.66, and 0.54 for time spent in mitosis, cytokinesis, and mitosis plus cytokinesis, respectively, are considered significant. For MEFs, P values of 0.40, 0.01, and 0.03 for time spent in mitosis, cytokinesis, and mitosis plus cytokinesis, respectively, are considered significant.
Rereplication is not a significant source of polyploidy in E7-expressing cells.
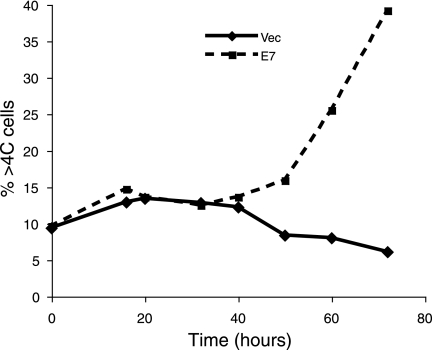
It has been proposed that E7 induces DNA rereplication that occurs prior to mitosis initiation and polyploidy formation (20). Since this possibility has never been directly examined in human epithelial cells, we investigated this mechanism. In PHKs and RPE1 cells expressing E7, cells exit mitosis approximately 16 to 20 h after nocodazole treatment (Fig. 3), but a significant incidence of polyploidy is not seen until approximately 40 h after nocodazole treatment (Fig. 4 and data not shown). These cells have 4C DNA content at mitosis yet do not show 8C DNA content until 24 h later. The kinetics of polyploidization in these E7-expressing cells suggest that rereplication of DNA does not contribute to polyploidy formation.
FIG. 4.
Extensive polyploidy formation in E7-expressing cells occurs after 40 h. RPE1 cells expressing vector (vec) or E7 under nocodazole treatment (50 ng/ml) were harvested at various time points, fixed, stained with PI, and analyzed by flow cytometry. The percentage of cells with DNA content greater than 4C (%>4C cells) was plotted as a function of time.
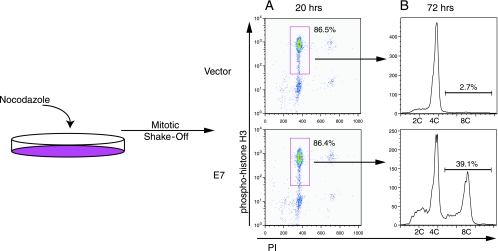
In addition to this observation, we performed a mitotic shake-off experiment to examine the potential contribution of rereplication to polyploidy formation in E7-expressing cells. Cells that have entered mitosis become round and are tenuously attached to the plate. Although some cells in prophase may decondense their chromosomes and return to interphase, the cells that are able to be shaken off the dish are considered to be at “a point of no return” and are committed to entering mitosis (29). Since PHKs do not efficiently detach from the culture dish during mitosis, we used RPE1 cells, which can be readily synchronized and shaken off at mitosis. The mitotic shake-off experiment shown in Fig. 5 was performed by first adding nocodazole to RPE1-vector or E7-expressing cells. After 20 h of nocodazole treatment, a majority of the RPE1 cells (86%) were in mitosis. These mitotic cells were shaken off and replated in nocodazole-containing media, and 39% of these E7-expressing cells became polyploid. Only a small percentage of cells (14%) did not shake off (i.e., were not likely in mitosis); therefore, their potential contribution to polyploidy would be minimal. Since the polyploid population of cells was created from all mitotic, 4C DNA content-containing cells, we can be sure that all of the polyploid cells arose from these cells and not from a subpopulation of rereplicating cells. These results indicate that rereplication is not a significant source of polyploidy in E7-expressing cells. These data are contrary to the previous hypothesis, where it was believed that cells challenged with antimicrotubule drugs do not enter mitosis or pass through a “point of no return” (20).
FIG. 5.
E7-expressing mitotic cells are capable of polyploidy formation. Asynchronous RPE1 cells expressing vector or E7 were treated with nocodazole (50 ng/ml). (A) After 20 h of nocodazole treatment, cells were measured for P-His H3 expression and DNA content. Cells positive for P-His H3 with 4C DNA content are indicated. (B) Mitotic cells were shaken off and replated in nocodazole-containing medium for an additional 52 h. Cells were analyzed for PI content. Data from a representative experiment (of three) are shown.
Expression of HPV E7 abrogates the postmitotic checkpoint.
Next, we tested whether abrogation of the postmitotic checkpoint after adaptation to the spindle assembly checkpoint contributes to polyploidy formation in E7-expressing cells. A previous study using human fibroblast cells suggested that expression of E7 facilitates DNA replication of cells with 4C DNA content in a G1-like stage (18). However, in this study, the cell cycle state after adaptation to mitotic disruption was not fully characterized. Specifically, the molecular markers used (pRb phosphorylation and cyclin B expression) could not differentiate G1 cells from those in G2. Direct evidence of postmitotic checkpoint abrogation leading to polyploidy formation in E7-expressing cells, specifically in human epithelial cells, has not been demonstrated. In order to study this proposed mechanism in more detail, we created a cell population, confirmed it to be in a G1-like state, and examined the postmitotic checkpoint with a BrdU incorporation assay.
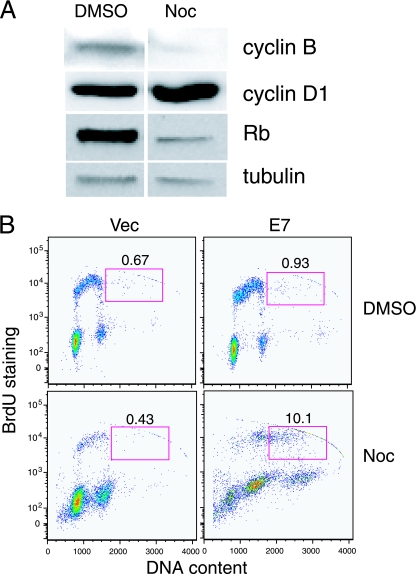
We first treated RPE1-vector cells with nocodazole for 48 h in order to activate the spindle assembly checkpoint and obtain cells with 4C DNA content. We then analyzed in which cell cycle stage these RPE1-vector cells were halted. We measured the steady-state levels of the G1 cyclin D1, the G2 cyclin B, and Rb phosphorylation. Figure 6A shows that after nocodazole treatment in RPE1-vector cells, cyclin D1 was increased approximately twofold, cyclin B was decreased approximately fourfold, and pRb became hypophosphorylated. These results indicate that RPE1 cells are halted in a G1 cell cycle state 2 days after nocodazole treatment.
FIG. 6.
E7 expression in cells with 4C DNA content in a G1-like state results in replication. (A) Asynchronous cultures of RPE1-vector cells were incubated in nocodazole (Noc)-containing medium for 48 h. Total protein extracts were resolved by SDS-PAGE and blotted with antibodies against cyclin B, cyclin D1, pRb, and tubulin. (B) RPE1-vector (vec) and E7-expressing cells were treated with nocodazole or DMSO for a total of 72 h. At 70 h posttreatment, the cells were incubated for the remaining 2 h with BrdU. BrdU-positive cells with DNA content of >4C are indicated. Data from a representative experiment (of three) are shown.
Next, we tested to what extent E7-expressing RPE1 cells are capable of replicating DNA following adaptation to the spindle assembly checkpoint. We incubated nocodazole-treated RPE1-vector and RPE1-E7 cells with BrdU and examined the ability of the cells with 4C DNA content in a G1 state to incorporate BrdU after they adapted to the spindle assembly checkpoint and exited mitosis. As shown in Fig. 6B, 10% of RPE1-E7 cells with DNA content of >4C incorporated BrdU, whereas only 0.4% of vector control cells with DNA content of >4C incorporated BrdU. These results indicate that expression of E7 causes polyploidy in cells with 4C DNA content in a G1 state via abrogation of the postmitotic checkpoint.
Role of Rb in HPV E7-induced polyploidy.
Since the E7 protein from high-risk HPV types binds and targets pRb for the degradation and downregulation of Rb in mouse and human cells and, therefore, leads to polyploidy (6, 14, 16), we are interested in the role of Rb in E7-induced polyploidy. Notably, Patel et al. conducted a mutational analysis for E7-induced polyploidy in response to nocodazole treatment (27). Consistent with a role for pRb family members, the E7 mutant C24G (with a mutation in the pocket protein-binding motif LXCXE) that is defective for association with pRb family members was unable to induce polyploidy. However, interpretation for this result is complicated by the fact that the E7 zinc finger domain mutants (L67R, for example) efficiently induced polyploidy, although they do not derepress Rb function. These results suggest that other Rb family members, namely p107 or p130 instead of pRb, are responsible for the postmitotic checkpoint control. Alternatively, it remains possible that a still unidentified cellular protein(s) that binds to the LXCXE motif is involved in causing the polyploid phenotype. On the other hand, although a previous study has implicated a role for Rb in the postmitotic checkpoint (18), a more recent study suggested that Rb downregulation affects the spindle checkpoint (14). Such a result argues against a role for Rb in the postmitotic checkpoint (40).
To clarify the role of Rb in mitotic checkpoints and ploidy control, we used Rb-null MEFs. Figure 3D shows that the mitotic index curves for wild-type MEFs and Rb−/− MEFs are not significantly different. We also used time-lapse videomicroscopy to compare the lengths of normal mitosis and cytokinesis for wild-type MEFs versus those of Rb−/− MEFs. Table 1 shows that the time spent in mitosis was not significantly different between wild-type MEFs and Rb−/− MEFs (41 min plus or minus 11 min versus 44 min plus or minus 15 min, respectively). These data are consistent with the mitotic index curves in Fig. 3D, supporting the hypothesis that a lack of Rb does not have a significant effect on the duration of mitosis, an observation that is different from the previous study by Hernando et al. (14). In agreement with what was described by Hernando et al. (2004), our time-lapse data show that Rb−/− MEFs did spend significantly more time completing cytokinesis than did wild-type MEFs (14). However, the cells did eventually cleave into two daughter cells. Therefore, cleavage failure in response to spindle disruption does not appear to be a mechanism for polyploidization.
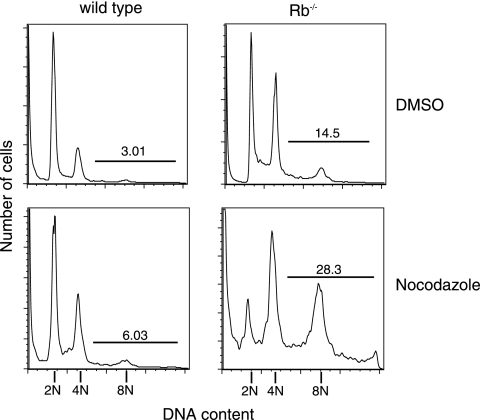
As shown in Fig. 7, Rb−/− MEFs have fivefold more cells with DNA content greater than 4C compared to that of wild-type controls (14.5% versus 3.0%, respectively). We also found that arresting Rb−/− MEFs in mitosis with nocodazole, where they eventually adapt to the spindle assembly checkpoint, results in double the amount of polyploidy compared to the amount in untreated cells (28.3% versus 14.5%, respectively; Fig. 7). This confirms that a lack of Rb in MEFs can lead to polyploidy in our cells, both spontaneously and in response to microtubule-depolymerizing drugs.
FIG. 7.
Absence of Rb in MEFs can lead to polyploidy formation. Asynchronous cultures of wild-type and Rb−/− MEFs were treated with DMSO or nocodazole (50 ng/ml) for 48 h. Cells were collected, fixed, stained with PI, and analyzed by flow cytometry. Data from a representative experiment (of three) are shown. The percentages of cells with more than 4C DNA content are indicated.
Taken together, our results indicate that Rb-defective cells undergo normal mitoses with an intact spindle assembly checkpoint. Our results suggest that downregulation of Rb is important for E7 to induce polyploidy and subsequent abrogation of the postmitotic checkpoint that leads to polyploidization.
DISCUSSION
This study explores the mechanism underlying the generation of polyploidy in HPV E7-expressing human epithelial cells. We tested the conflicting possibilities, and our data indicate that E7 does not have an effect on the spindle assembly checkpoint or on the length of time cells spent in mitosis. Our data also suggest that DNA rereplication is not a significant cause of polyploidy in E7-expressing human epithelial cells in response to microtubule disruption. Rather, we have shown that wild-type epithelial cells treated with low doses of nocodazole adapt to the spindle assembly checkpoint and exit mitosis. They then proceed into G1 with 4C DNA content, where they halt at the postmitotic checkpoint. Our results then demonstrate that E7-expressing cells are able to proceed through this checkpoint, replicate their DNA, and become polyploid.
Our results also demonstrate that Rb downregulation does not have a major effect on the spindle checkpoint. This is in contrast to the work of Hernando et al., which suggests that Rb downregulation and the subsequent effects on the Rb pathway, including upregulation of Mad2, result in a hyperactive spindle checkpoint (14) and may predispose cells to additional genomic instability. We believe that the discrepancy may be due to our interpretation of the time-lapse videomicroscopy results, which differs from theirs. They measured the length of mitosis from prometaphase to late anaphase by examining chromosome condensation and cell division (14). On the other hand, we measured mitosis from nuclear envelope breakdown to nuclear envelope reformation, which are more distinctly defined and easily seen parameters. Another difference between our experiments and those of Hernando et al. is the types of cells used. We have used primary MEFs and the immortalized RPE1 cells, while Hernando et al. used the established mouse NIH 3T3 cells as well as the human colon carcinoma line HCT116. A more recent study with Mad2-inducible upregulation demonstrates an increase in polyploid cells and a link to chromosomal instability upon Mad2 overexpression (33). However, cells overexpressing Mad2 do not appear to be equivalent to Rb-downregulated cells, as Mad2 overexpression led to a partial mitotic block of proliferating cells, a phenomenon never seen in Rb−/− cells.
As previously suggested in studies with fibroblasts and as our data indicate, Rb plays an important role in postmitotic checkpoint control (19). However, our results do not rule out the role of inactivation of p107 or p130 in E7-induced polyploidy, nor can we explain how the E7 zinc finger domain mutants that are defective for Rb derepression induce polyploidy (27). Notably, Patel et al. tested the E7 mutants in the presence of E6, which by themselves could induce polyploidy under our experimental conditions. In addition, we examined the role of Rb by using MEFs, which might be different from or less stringent than using human keratinocytes in regulation of the postmitotic checkpoint. The conclusions from our study and those by Patel et al. suggest that E7 possesses both Rb-dependent and Rb-independent activities in the abrogation of the postmitotic checkpoint.
p53 appears to play a key role in mediating the postmitotic checkpoint (1, 2, 19), and p21 is responsible for at least part of this p53-mediated postmitotic arrest (18, 19, 34). The cdk inhibitor p21 binds to and inactivates cyclin/cdk complexes, including cyclinE1/cdk2 and cyclinA2/cdk2, resulting in pRb hypophosphorylation that inhibits E2F activity and arrests the cell cycle at the G1/S transition (35, 41). Consistent with observations made by others, we have previously shown that p53 levels were upregulated in E7-expressing PHKs (21). E7 may abrogate the transcriptional activity of p53 (23). E7 has also been shown to inactivate the Cdk inhibitory activity of p21 (10, 17). These functions may contribute to E7-induced polyploidy.
This study has important implications for cancer in general, and for cervical cancer in particular, because of our results with the HPV oncogene E7. HPV E6 and E7 are the oncogenes necessary for transformation in high-risk HPVs, which are associated with cervical lesions that eventually may lead to cervical carcinomas. Although E6 and E7 are able to immortalize primary human epithelial cells, they are not sufficient to induce the transformation of human cells (7). Instead, it is believed that genomic instability caused by E6 and E7 predisposes the cells to accumulate additional genomic aberrations necessary for malignant transformation (4, 5). As shown here and in other studies, HPV E7-expressing cells are able to become polyploid (27, 37, 38), and Olaharski et al. concluded that polyploidy and chromosomal instability are related events that predispose cells to aneuploidy and subsequently to malignancy (26). Our finding that E7 expression results in polyploidy formation through abrogation of the postmitotic checkpoint has important implications for cancer prevention and/or treatment. Understanding this mechanism of polyploidy formation will be useful in developing therapies against its accumulation in E7-expressing, untransformed cells in order to prevent the accumulation of additional genomic aberrations that could lead to carcinogenesis.
Acknowledgments
We thank Elliot Androphy and the members of his lab for helpful discussions. We thank Stephen Jones for providing the wild-type and Rb−/− MEFs.
The project described was supported by a Developmental Award from the University of Massachusetts Center for AIDS Research (award P30 AI042845), grant R21AI070772 from the National Institute of Allergy and Infectious Diseases (NIAID), and award R01CA119134 from the National Cancer Institute (NCI).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID, NCI, or NIH.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Andreassen, P. R., O. D. Lohez, F. B. Lacroix, and R. L. Margolis. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell 121315-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Leonardo, A., S. H. Khan, S. P. Linke, V. Greco, G. Seidita, and G. M. Wahl. 1997. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 571013-1019. [PubMed] [Google Scholar]
- 3.Duelli, D. M., H. M. Padilla-Nash, D. Berman, K. M. Murphy, T. Ried, and Y. Lazebnik. 2007. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr. Biol. 17431-437. [DOI] [PubMed] [Google Scholar]
- 4.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Piboonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 9710002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duensing, S., and K. Munger. 2002. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 627075-7082. [PubMed] [Google Scholar]
- 6.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243934-937. [DOI] [PubMed] [Google Scholar]
- 7.Fan, X., and J. J. Chen. 2004. Regulation of cell cycle progression and apoptosis by the papillomavirus E6 oncogene. Crit. Rev. Eukaryot. Gene Expr. 14183-202. [DOI] [PubMed] [Google Scholar]
- 8.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 746622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara, T., M. Bandi, M. Nitta, E. V. Ivanova, R. T. Bronson, and D. Pellman. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 4371043-1047. [DOI] [PubMed] [Google Scholar]
- 10.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 112090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galipeau, P. C., D. S. Cowan, C. A. Sanchez, M. T. Barrett, M. J. Emond, D. S. Levine, P. S. Rabinovitch, and B. J. Reid. 1996. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc. Natl. Acad. Sci. USA 937081-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganem, N. J., Z. Storchova, and D. Pellman. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17157-162. [DOI] [PubMed] [Google Scholar]
- 13.Goto, H., Y. Tomono, K. Ajiro, H. Kosako, M. Fujita, M. Sakurai, K. Okawa, A. Iwamatsu, T. Okigaki, T. Takahashi, and M. Inagaki. 1999. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 27425543-25549. [DOI] [PubMed] [Google Scholar]
- 14.Hernando, E., Z. Nahle, G. Juan, E. Diaz-Rodriguez, M. Alaminos, M. Hemann, L. Michel, V. Mittal, W. Gerald, R. Benezra, S. W. Lowe, and C. Cordon-Cardo. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430797-802. [DOI] [PubMed] [Google Scholar]
- 15.Incassati, A., D. Patel, and D. J. McCance. 2006. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene. 252444-2451. [DOI] [PubMed] [Google Scholar]
- 16.Iovino, F., L. Lentini, A. Amato, and A. Di Leonardo. 2006. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol. Cancer 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 112101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, S. H., and G. M. Wahl. 1998. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 58396-401. [PubMed] [Google Scholar]
- 19.Lanni, J. S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 181055-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lentini, L., L. Pipitone, and A. Di Leonardo. 2002. Functional inactivation of pRB results in aneuploid mammalian cells after release from a mitotic block. Neoplasia 4380-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Y., S. A. Heilman, D. Illanes, G. Sluder, and J. J. Chen. 2007. p53-independent abrogation of a postmitotic checkpoint contributes to HPV E6-induced polyploidy. Cancer Res. 672603-2610. [DOI] [PubMed] [Google Scholar]
- 22.Margolis, R. L., O. D. Lohez, and P. R. Andreassen. 2003. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J. Cell. Biochem. 88673-683. [DOI] [PubMed] [Google Scholar]
- 23.Massimi, P., D. Pim, and L. Banks. 1997. Human papillomavirus type 16 E7 binds to the conserved carboxy-terminal region of the TATA box binding protein and this contributes to E7 transforming activity. J. Gen. Virol. 782607-2613. [DOI] [PubMed] [Google Scholar]
- 24.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasmyth, K. 2002. Segregating sister genomes: the molecular biology of chromosome separation. Science 297559-565. [DOI] [PubMed] [Google Scholar]
- 26.Olaharski, A. J., R. Sotelo, G. Solorza-Luna, M. E. Gonsebatt, P. Guzman, A. Mohar, and D. A. Eastmond. 2006. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 27337-343. [DOI] [PubMed] [Google Scholar]
- 27.Patel, D., A. Incassati, N. Wang, and D. J. McCance. 2004. Human papillomavirus type 16 E6 and E7 cause polyploidy in human keratinocytes and up-regulation of G2-M-phase proteins. Cancer Res. 641299-1306. [DOI] [PubMed] [Google Scholar]
- 28.Plug-Demaggio, A. W., and J. K. McDougall. 2002. The human papillomavirus type 16 E6 oncogene induces premature mitotic chromosome segregation. Oncogene 217507-7513. [DOI] [PubMed] [Google Scholar]
- 29.Rieder, C. L., and R. W. Cole. 1998. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J. Cell Biol. 1421013-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 31.Shi, Q., and R. W. King. 2005. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 4371038-1042. [DOI] [PubMed] [Google Scholar]
- 32.Sluder, G., and J. J. Nordberg. 2004. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 1649-54. [DOI] [PubMed] [Google Scholar]
- 33.Sotillo, R., E. Hernando, E. Diaz-Rodriguez, J. Teruya-Feldstein, C. Cordon-Cardo, S. W. Lowe, and R. Benezra. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 119-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart, Z. A., S. D. Leach, and J. A. Pietenpol. 1999. p21(Waf1/Cip1) inhibition of cyclin E/Cdk2 activity prevents endoreduplication after mitotic spindle disruption. Mol. Cell. Biol. 19205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart, Z. A., and J. A. Pietenpol. 2001. p53 signaling and cell cycle checkpoints. Chem. Res. Toxicol. 14243-263. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, S. S., and F. McKeon. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89727-735. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, J. T., and L. A. Laimins. 1998. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J. Virol. 721131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, D. A., G. Belinsky, T. H. Chang, D. L. Jones, R. Schlegel, and K. Munger. 1997. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene 153025-3035. [DOI] [PubMed] [Google Scholar]
- 39.Uetake, Y., and G. Sluder. 2004. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J. Cell Biol. 165609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel, C., A. Kienitz, I. Hofmann, R. Muller, and H. Bastians. 2004. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene 236845-6853. [DOI] [PubMed] [Google Scholar]
- 41.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408307-310. [DOI] [PubMed] [Google Scholar]
- 42.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2342-350. [DOI] [PubMed] [Google Scholar]