Abstract
The regulatory regions surrounding many genes may be large and difficult to study using standard transgenic approaches. Here we describe the use of bacterial artificial chromosome clones to rapidly survey hundreds of kilobases of DNA for potential regulatory sequences surrounding the mouse bone morphogenetic protein-5 (Bmp5) gene. Simple coinjection of large insert clones with lacZ reporter constructs recapitulates all of the sites of expression observed previously with numerous small constructs covering a large, complex regulatory region. The coinjection approach has made it possible to rapidly survey other regions of the Bmp5 gene for potential control elements, to confirm the location of several elements predicted from previous expression studies using regulatory mutations at the Bmp5 locus, to test whether Bmp5 control regions act similarly on endogenous and foreign promoters, and to show that Bmp5 control elements are capable of rescuing phenotypic effects of a Bmp5 deficiency. This rapid approach has identified new Bmp5 control regions responsible for controlling the development of specific anatomical structures in the vertebrate skeleton. A similar approach may be useful for studying complex control regions surrounding many other genes important in embryonic development and human disease.
Many developmental genes that control the formation of particular anatomical structures or body regions are expressed in highly specific patterns. These expression patterns often are created by the combined action of a complex array of cis-acting regulatory sequences surrounding the coding region of the gene. For example, detailed molecular and genetic studies have shown that different aspects of Drosophila melanogaster decapentaplegic (dpp) gene expression are regulated by distinct sequences. Expression of the gene in the midgut is controlled by sequences located 5′ of the gene (1, 2), early embryonic expression by sequences in the intron (3), and imaginal disk expression by sequences located 3′ of the coding sequences (4–6). Even in the relatively compact fly genome, some of the dpp enhancers are located more than 30 kb from the promoter. Other genes in flies and mammals, such as Ubx (7) and β-globin (8, 9), also are regulated by sequences located far from transcription start sites. The existence of distant regulatory regions often is suggested first by mutations that disrupt the function of a gene, but map at some distance from the coding regions. In vertebrates, several examples of disease-causing mutations that map great distances from the gene are known (10, 11) or suggested (12–16).
We recently have described an interesting example of long-distance gene regulation in our studies of the mouse bone morphogenetic protein-5 (Bmp5) gene (17). Bone morphogenetic proteins (BMPs) are secreted signaling molecules (18, 19) that have the ability to induce ecotopic bone when placed under the skin of an animal (20, 21). We have shown previously that the classical mouse short ear mutation disrupts the Bmp5 gene, which encodes one member of this large family of related proteins (21). Null mutations at the short ear/Bmp5 locus reduce the size and shape of external ear, the number of ribs along the vertebral column, and the size of bones at the anterior and posterior ends of the sternum and lead to loss of specific bony processes on individual vertebrae (21–24). The Bmp5 gene is expressed at the earliest stages of skeletal development in small, local patterns that prefigure the shapes of future skeletal elements (25). A combination of biochemical, genetic, and expression data suggest that local expression of the Bmp5 gene is one of the signals normally used to induce the formation of particular anatomical features in the skeleton (26).
The short ear/Bmp5 locus was one of a handful of mouse loci previously subjected to extensive mutagenesis experiments (27). These experiments have resulted in the isolation of hundreds of mutations that disrupt Bmp5 function. Molecular studies have shown that some of these mutations are chromosome rearrangements that disrupt DNA sequences located far outside of the coding region of the gene (17). We previously have used these regulatory mutations to define an extensive control region located 3′ of all Bmp5 coding regions. Reporter gene studies in the region were used to detect important enhancers located up to 270 kb downstream of the transcription start site (17).
These regulatory mutations did not affect all endogenous Bmp5 expression patterns, suggesting that other regulatory sequences controlling Bmp5 expression may be located outside the 3′ region. To efficiently screen large amounts of DNA for potential regulatory regions, we have developed a coinjection reporter gene approach by using bacterial artificial chromosomes (BACs). This technique has made it possible to efficiently screen hundreds of kilobases of DNA for new Bmp5 control elements, rapidly test the effect of control regions on different promoters, and directly rescue short ear mutant phenotypes. Similar techniques may greatly speed the search for control elements in many other vertebrate genes.
Materials and Methods
BAC Isolation and Characterization.
A Research Genetics BAC Library was screened by PCR by using primers located at the 3′ end of the Bmp5 gene (position 310 of Fig. 1; forward primer, 5′-CTAGTGTTTC CAGACCATG -3′; reverse primer, 5′-ATGAAATGACCCAGGTTGAT-3′). This resulted in the isolation of both BAC 199 (100 kb) and BAC 551 (130 kb). A second set of screening primers was derived from sequencing the 3′ end (away from the Bmp5 exons) of BAC 551. BAC library screening with these primers (forward primer, 5′-CCAGTGCGTCGTGTGTCATG-3′; reverse primer, 5′-ATGGGAGTTGGATGCTTTTC-3′) led to the isolation of BAC 178 (135 kb). The overlaps and map positions of the BAC clones were determined by hybridization with probes from positions 222, 340, 372, 420, and 430 kb of the previously described chromosome walk (21).
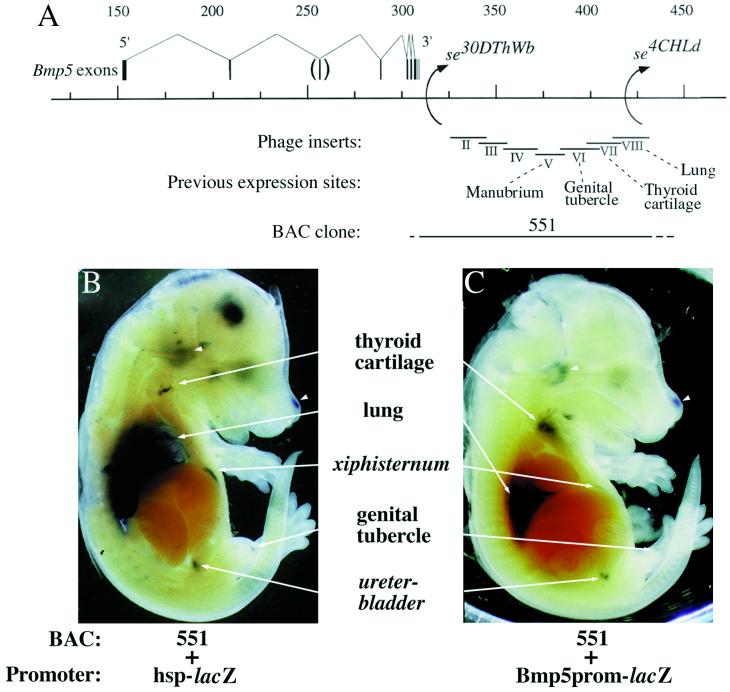
Figure 1.
BAC coinjections drive expression of a lacZ reporter gene in previously detected Bmp5 expression domains. (A) The position of BAC 551 relative to the Bmp5 gene. Dotted lines at the end of the insert bracket minimal and maximal extent of the insert based on restriction mapping and probe hybridization. The arrows on the 3′ side of the Bmp5 exons indicate positions of chromosome breakpoints in two short ear regulatory mutations, se30DThWb and se4CHLd (17). Enhancers previously detected by reporter transgenic work with phage clones (17) are labeled and indicated by dotted lines. (B) Embryos (E14.5–15.0) transgenic for BAC 551 and hsp-lacZ transgenes show reporter gene expression (arrowheads and small arrows) in areas previously detected by phage clone reporter constructs and in the xiphisternum and ureter–bladder (italics), locations also known to express Bmp5 (25). (C) Embryos transgenic for BAC 551 and the Bmp5prom-lacZ transgenes show a very similar overall expression pattern.
Preparation of BAC DNA for Microinjection.
BAC DNA was prepared by using a standard cesium chloride protocol, with all steps done as gently as possible to avoid shearing. Approximately 5 μg of purified DNA was digested with restriction enzyme (SalI or NotI) and prepared for injection by using buffer exchange with Centriprep-30 columns as described previously (17). The entire buffer exchange sequence was done at 4°C to minimize degradation. The integrity of all DNA preparations was evaluated by using pulsed-field gel electrophoresis. For coinjections, 200 ng of BAC DNA was combined with 200 ng of reporter gene DNA for a final concentration of 1.0–1.25 ng/μl. Before injection, the solution was filtered by using Millipore spin columns (Filter HV; 0.45 μM).
Transgenic Mouse Production.
The oocytes used for most injections were derived from female FVB/N mice crossed to B6DBAF1/J males. These oocytes retain the clarity and large pronucleus of the FVB/N strain but may have better survival after injections when compared with oocytes derived from FVB/N × FVB/N matings (unpublished observations). The injected oocytes were transferred into the oviducts of pseudopregnant CD-1 mice. All techniques followed standard protocols (28). Embryos were collected at approximately embryonic day 15 (E15) and genotyped and processed for lacZ staining as described previously (17).
Mapping of the Mouse Bmp5 Transcription Start Site.
A 246-bp Bmp5 probe (figure 7 of ref. 21) was used to screen a mouse E8.5 cDNA library (gift from B. Hogan, Vanderbilt University, Nashville, TN). The resulting cDNA clones covered 3.8 kb of the 4.0-kb Bmp5 transcript detected previously by Northern blotting (ref. 25 and unpublished data). The 5′ rapid amplification of cDNA ends system (GIBCO/BRL) was used to locate the 5′ end of Bmp5 in poly(A)+ mRNA samples from adult lung (first reverse primer, 5′-ATCTACGTCTGATCTGCACC-3′; second reverse primer, 5′-CCCAAGGTTATAGTGTTCAAGT-3′). RNase protection experiments (29) were used to confirm this approximate position, and primer extension experiments (30) were used to determine the exact starting nucleotide (primer, 5′-CAATCCTCCCTGTAAATCCATTCAATCTC-3′).
Cross-Species Bmp5 Clone Isolation and Sequencing.
Genomic lambda phage libraries from both chicken (Stratagene) and human (gift from U. Franke, Stanford University, Stanford, CA) were screened with a 246-bp probe (21) from the 5′ untranslated region of the mouse Bmp5 gene. Genomic fragments containing the cross-hybridizing sequences were subcloned from the phage and sequenced by using the Sequenase II protocol (United States Biochemical).
Bmp5 cDNA Construct and Functional Rescue Studies.
A construct replacing the lacZ gene of the hsp68lacZ plasmid with Bmp5 coding regions was constructed by ligation of a full-length Bmp5 cDNA into a modified hsp vector (phspGM). The phspGM construct was created by modification of the original (31) hsp68lacZpA vector construct. First, hsp68lacZpA was digested with Asp718 and SacI to remove the translation initiation sequences and most of the lacZ gene. A linker containing Asp718- and SacI-compatible ends and an internal BglII site was inserted to create phspMOD. A separate hsp68lacZpA construct was digested with NcoI followed by exonuclease digestion and addition of BglII linkers. This fragment then was digested with Asp718 and BglII to liberate the heat shock promoter from the rest of the vector. This heat shock promoter was inserted into the phspMOD vector that had been digested with Asp718 and BglII to create the hspGM vector. Full-length Bmp5 was amplified from RNA and cloned into pCDNA3 vector (Andrew Ho, unpublished data). This construct was digested with BamHI, and the Bmp5 sequence was inserted into the BglII site of the hspGM vector to create the final hspGMB5 construct. hspGMB5 was microinjected into oocytes derived from a cross between se2OZb/se2OZb males and FVB/N superovulated females. The se2OZb allele is a complete deletion of the Bmp5 gene (21). Founder mice carrying the transgene and heterozygous for the se2OZb mutation were detected by typing with primers specific to the BAC vector (forward primer, 5′-GTGTCGGGGCTGGCTTAA-3′; reverse primer, 5′-TGCCTGCAGGTCGACTCT-3′). Positives were confirmed by genomic Southern blots and hybridizations with the BAC vector probe or BAC 551 end probe. To test for rescue of the short ear phenotype, the founders were crossed to se2OZb/se2OZb animals. The progeny then were typed by using PCR with primers specific for the BAC vector. Mice homozygous for the se2OZb allele were identified by PCR and Southern blots by using probes from positions 152 and 445 of the chromosome walk (21).
Skeletal Preparations.
Eight-week-old mice were sacrificed, eviscerated, and rinsed in 1× PBS. The skeletons then were fixed in 95% ethanol for 2–3 days. The cartilage was stained in 76% ethanol/20% acetic acid/4% water/0.015% Alcian blue solution for 5–7 days followed by a 95% ethanol rinse for 2–3 days. The skeletons then were incubated in 1.2% KOH for 7–10 days until the bones were completely white. Alizarin Red then was added at approximately 2 μg/ml, and the skeletons were stained until the desired amount of red was seen (about 2–3 days). The final skeletons were cleared and photographed in 100% glycerol.
In Situ Hybridizations.
All in situ hybridizations were done as described previously (17).
Results
A Novel BAC Coinjection Approach to Detect Control Elements.
We previously have used regulatory alleles at the mouse short ear locus (se30DThWb and se4CHLd) to identify a large region 3′ of all Bmp5 exons that is required for normal expression of the endogenous Bmp5 gene at particular anatomical locations during development. Reporter studies in transgenic mice confirmed the existence of separate regulatory elements between the se30DThWb and se4CHLd mutant breakpoints (17). These studies required the production of large numbers of constructs carrying overlapping 15- to 20-kb inserts fused to a minimal heat-shock promoter–lacZ reporter. To test whether a simpler and more efficient method could be used to study this and other large regulatory regions, we isolated a BAC clone containing a DNA insert that spans the entire interval between the se30DThWb and se4CHLd breakpoints (Fig. 1A). This BAC clone (551) then was coinjected into fertilized mouse eggs with a separate DNA construct containing a minimal heat-shock promoter driving a lacZ reporter gene (31). The coinjection approach avoids complex cloning steps with large DNA fragments and takes advantage of the fact that coinjected DNA fragments usually cointegrate at a common site (32–34). The coinjection of BAC 551 and the hsplacZ cassette yielded embryos that show reporter expression simultaneously in all of the sites detected previously by individual phage subclones (Fig. 1B). The BAC drove expression consistently in the lung, thyroid cartilage, genital tubercle, cochlea, and the tip of the nasal region, all sites that were also seen in previous reporter mice created by standard techniques (ref. 17 and unpublished data) and all sites at which the endogenous Bmp5 gene is also known to be expressed (17, 25). In addition, the BAC clone also drove expression in the xiphisternum and ureter–bladder junction, two new sites at which the Bmp5 gene is also known to be expressed (25). Similar patterns were seen in three of three independent embryos transgenic for both BAC 551 and hsplacZ. Ectopic sites of expression also were observed (such as the brain in Fig. 1B), but were easy to distinguish from the major patterns because they were not reproducible in multiple embryos.
Similar Effects on Different Promoters.
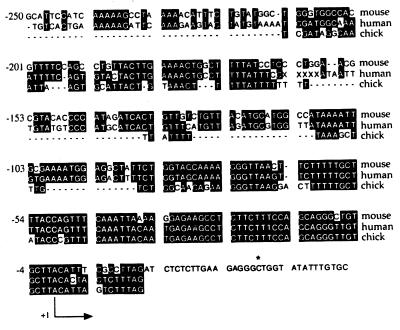
This simple coinjection assay made it possible to rapidly test the effect of Bmp5 regulatory regions on other reporter constructs. All of our previous transgenic work with Bmp5 control regions has utilized the hsplacZ reporter construct to detect enhancer activity. The minimal promoter sequences in this construct are derived from the heat shock 68 gene (31). Because some control elements are known to show specificity for different promoters (35), we also wanted to test the effects of potential Bmp5 control regions on constructs carrying the endogenous Bmp5 promoter. The Bmp5 transcription initiation site therefore was defined by a combination of 5′ rapid amplification of cDNA ends, RNase protection, and primer extension experiments (data not shown). The transcription initiation site occurs 31 bp upstream of the end of previously described sequence from the mouse and human Bmp5 cDNA clones (21, 36). The sequence surrounding the Bmp5 transcription start site matches a consensus transcription start site sequence seen in mammals (37). A region upstream of the transcription initiation site shows very high sequence conservation in mice, humans, and chickens (Fig. 2). This conservation is higher than that seen in other promoters sequenced in multiple species (e.g., refs. 38 and 39), suggesting that sequences in this region may serve an important conserved function. A construct containing the transcription initiation site and 5 kb of upstream sequences (Bmp5prom-lacZ) was fused to lacZ and injected into mice. The Bmp5prom-lacZ construct does not show any reporter gene activity when injected alone (data not shown). However, coinjection with the BAC 551 results in lacZ expression at all of the sites detected previously in the coinjection experiments with the hsp-lacZ constructs (Fig. 1B; three of three embryos). These data suggest that the Bmp5 control region located 3′ of the gene is able to function similarly with either the heat shock 68 promoter or the endogenous Bmp5 promoter.
Figure 2.
Promoter conservation among the mouse, human, and chicken Bmp5 genes. The transcription start site in mice is shown as the +1 arrow at the bottom. The blackened boxes indicate base pairs that are identical in all three species. Previously published Bmp5 sequence (21) begins at base +32 marked with an asterisk.
Surveys for Additional Control Sequences in the Bmp5 Gene.
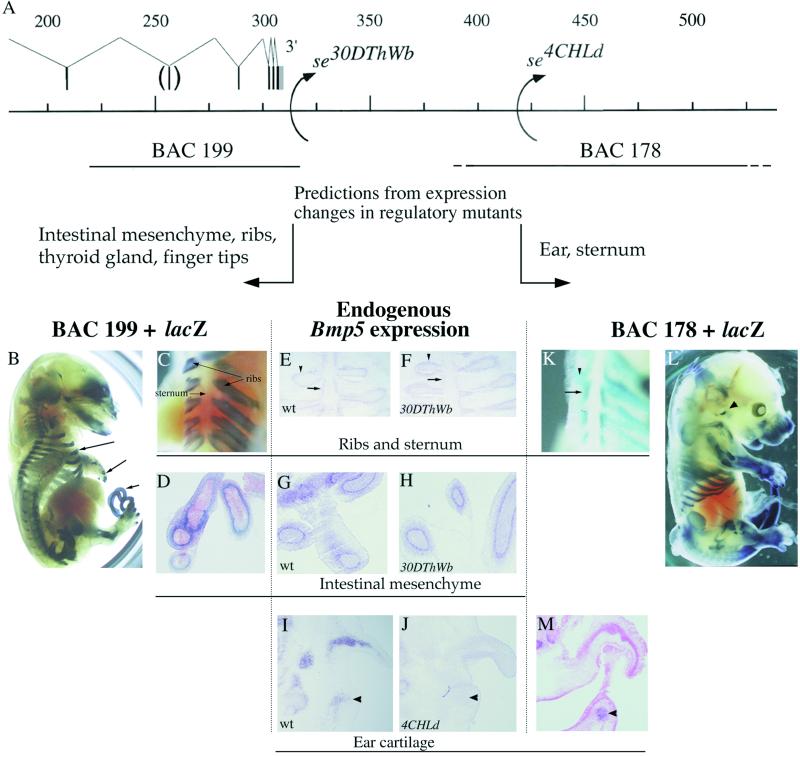
The localized effects of the se30DThWb and se4CHLd mutations on endogenous Bmp5 expression patterns suggest that additional Bmp5 control elements map outside the region that has been surveyed in previous transgenic studies (17). For example, the expression of the Bmp5 gene in intestinal mesenchyme, finger tips, ribs, and thyroid gland is not disrupted by either the se30DThWb or the se4CHLd chromosome rearrangements, suggesting that elements driving expression at these locations are located 5′ of both breakpoints (ref. 17 and Fig. 3). Conversely, Bmp5 expression in the ear and sternum is disrupted by both the se30DThWb and se4CHLd mutations, suggesting that some elements driving expression would be located distal to the se4CHLd mutation (ref. 17 and Fig. 3 I and J). To search for the predicted elements, we isolated two additional BAC clones (199 and 178) that extend either 5′ of the se30DThWb breakpoint or 3′ of the se4CHLd breakpoint (Fig. 3). Coinjection of BAC 199 with either the hsplacZ reporter or the Bmp5prom-lacZ construct produced mice with high levels of expression in sites predicted from previous studies, including ribs, digit tips, intestines, and a number of other skeletal elements (Fig. 3 B– D; four of five embryos). Expression in the ribs is particularly striking because it is restricted to the ribs and excluded from the adjacent sternum (Fig. 3C). This is consistent with the expression data of se30DThWb embryos, which show preferential disruption of Bmp5 expression in the sternum whereas the rib expression remains at wild-type levels (Fig. 3 E and F). Also consistent with expression data in se30DThWb is the strong reporter gene expression in mesenchyme surrounding the epithelial cells of the intestine (Fig. 3D). This matches the endogenous expression of Bmp5 mRNA in intestines (Fig. 3G), a site not disrupted by the se30DThWb mutation (Fig. 3H).
Figure 3.
BACs 199 and 178 drive expression of lacZ at sites predicted from effects of regulatory mutations on endogenous Bmp5 expression. (A) BACs 199 and 178 extend proximal of the se30DThWb and distal of the se4CHLd breakpoint, respectively. Previous expression studies suggest that control elements for the indicated sites also should be located proximal or distal of these breakpoints. (B) BAC 199 drives expression in three sites predicted by previous studies: the ribs (top arrow), the finger tips (middle arrow), and the intestines (bottom arrow). A frontal view (C) shows that the ribs, but not the sternum, display robust reporter expression, consistent with normal rib expression (arrowhead) and reduced sternum expression (arrow) of Bmp5 in se30DThWb embryos (E and F). In contrast, BAC 178 (L) drives strong expression in the sternal bands as well as some expression in the ribs (K). This is consistent with regulatory mutant data that place the sternal enhancer 3′ of the se30DThWb breakpoint and BAC 551 data that place it further 3′ as shown in A. The intestinal mesenchyme enhancer activity in BAC 199 (B and D) matches Bmp5 expression at the same site (G) and is consistent with normal Bmp5 expression in se30DThWb embryos (H). The BAC 178-driven reporter expression in the base of the ear (arrowheads in L and M) coincides with one part of the endogenous Bmp5 ear expression domain (arrowhead in I). The position of this enhancer in the 3′ BAC 178 is consistent with the effects of se4CHLd on Bmp5 expression at that site (arrowhead in J).
Coinjection of the most distally located BAC clone 178 (either with the hsplacZ reporter or the Bmp5 promoter-lacZ construct) produced consistent expression in (i) the base of the ear (Fig. 3 L and M; three of five embryos) and (ii) the sternum and ribs (Fig. 3 K and L; four of five embryos). The expression at the base of the ear recapitulates one portion of the Bmp5 mRNA expression in ear cartilage that is disrupted by the distal mutation (Fig. 3 I, J, and M, arrowheads). The expression in the sternum is consistent with the effects of the se30DThWb mutation on Bmp5 expression (Fig. 3 E and F) and with reporter gene studies that did not detect sternal enhancers in the region between the se30DThWb and se4CHLd breakpoints (ref. 17 and Fig. 2).
Rescue of Bmp5 Phenotypes with Bmp5 Control Elements.
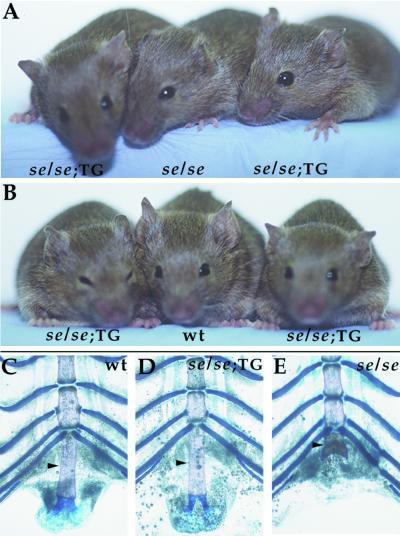
The coinjection technique also makes it possible to test whether the regulatory regions from the Bmp5 gene can rescue the phenotypic defects normally seen in short ear mice. For these experiments, BAC 551 was coinjected with a construct containing the complete ORF of mouse BMP5 fused downstream of the heat-shock promoter (hspGMB5). Transgenic founders that were heterozygous for a short ear null mutation were crossed to homozygous short ear mutant animals. At 8 weeks of age, the progeny were scored for phenotypes and typed for the presence of the Bmp5 locus and the presence of the BAC 551 and hspGMB5 transgenes. Fifteen of 15 progeny were either negative for both the BAC and hspGMB5 constructs or positive for both, suggesting that the two injected constructs have cointegrated near each other on a single chromosome. Progeny that were homozygous for the Bmp5 mutation and did not inherit the transgenes showed typical short ear phenotypes, including short external ear, loss or truncation of the xiphoid process at the end of the sternum, and reduction of the peroneal process of the fibula. In contrast, transgenic progeny that were homozygous for the Bmp5 mutation showed markedly longer ears (Fig. 4A) and rescue of the xiphisternum phenotype (Fig. 4 C–E), but still had reduced peroneal processes (data not shown). Rescue of these short ear phenotypes is consistent with previous studies showing that enhancers controlling expression in the xiphisternum are present in BAC 551 (Fig. 1). The incomplete rescue of ear length (see Fig. 4B) also is consistent with previous studies showing that the ears of se30DThWb mice are shorter than those of se4CHLd mice (ref. 17 and Fig. 1), suggesting that some but not all of the control elements required for the development of full-length ears are located between the two mutant breakpoints.
Figure 4.
Mice transgenic for BAC 551 and a hsp-BMP5 construct show rescue of short ear phenotypes. Two transgenic short ear mice (se/se;TG) are shown on either side of a nontransgenic short ear littermate (se/se) in A or a se/+ long ear control (wt) in B. The ear length is rescued substantially in transgenic mice, but only to 80–90% of total ear length seen in wild-type mice. (C–E) Xiphisternums (arrowheads) from wild-type, se/se;TG, and se/se mice show that the presence of the transgene restores the xiphisternum to wild-type length. Alcian blue- and Alizarin red-stained regions correspond to regions of cartilage and bone, respectively.
Discussion
Previous work at the Bmp5 locus has demonstrated a complex and large array of regulatory sequences located 3′ of the gene exons (17). However, analysis of short ear regulatory mutants suggested that additional control elements should be located outside this region. A large-fragment transgenic approach has allowed us to characterize further this complex locus and to confirm many of the predictions made from studies of regulatory mutations at the Bmp5 locus.
The BAC coinjections have provided direct experimental evidence for new regulatory regions that control expression in intestine, ribs, and finger tips (BAC 199) and in ear, ribs, and sternum (BAC 178). The results of these transgenic studies are consistent with the effects of regulatory mutations on specific subsets of the endogenous Bmp5 expression pattern (ref. 17 and Fig. 3). These studies provide further evidence that the normal expression pattern of the Bmp5 gene is controlled by a large number of different regulatory elements, some of which show surprising specificity for particular subsets of the skeleton. Further study of such elements should provide new insights into the molecular mechanisms used by vertebrate embryos to control the morphology of different parts of the skeleton.
The coinjection procedure has greatly simplified tests of the effects of Bmp5 control regions on different reporter gene constructs with distinct promoter sequences. Despite the high conservation of sequence seen in the Bmp5 promoter of different species (Fig. 3), we can detect no major differences when using the Bmp5 promoter or the heat-shock promoter in our reporter gene experiments (Fig. 2). However, we cannot rule out the possibility that significant differences may be found for other Bmp5 control regions not yet analyzed or that different promoters may direct different quantitative levels of gene expression.
Coinjection of BAC 551 and a full-length Bmp5 cDNA rescues some of the phenotypic defects seen in short ear (Bmp5 mutant) mice. The xiphisternum of short ear mice was rescued significantly by the coinjection (Fig. 4 C–E), consistent with the detection of the xiphisternum enhancer in BAC 551 (Fig. 1 B and C). The external ear length of short ear mutant mice was also partially rescued (Fig. 4A). Although BAC 551 did not drive reporter gene expression in embryonic ears (Fig. 1 B and C), previous studies have shown that the pinna abnormalities in short ear mice develop only after birth and therefore are likely to depend on Bmp5 expression at later stages (22).
Previous studies also have shown that mutations that break farther away from the Bmp5 gene have milder effects on ear length (17). These results suggest that the 3′ region may contain multiple elements that define ear length, some located between the se30DThWb and se4CHLd breakpoints and some located distal to the se4CHLd mutation. A similar situation has been described for the Drosophila BMP-related dpp gene. Expression of dpp in imaginal discs is also controlled by a large 3′ regulatory region that contains a number of different, smaller disc control elements (6, 40). Mutations that break progressively further away from the dpp coding region disrupt fewer elements and have milder effects on the length of the appendages (4, 5).
The studies reported here demonstrate the utility of using large-insert clones for transgenic reporter analysis in mice. Although the use of BACs provides less coverage per clone when compared with yeast artificial chromosomes (YACs), it simplifies the injection procedures and DNA handling. The simplicity of the coinjection strategy allows for efficient testing of multiple BACs for enhancer and rescue activity. More detailed studies then can be conducted by using homologous recombination in BACs (41, 42) to delete or alter specific sequences to observe the effect on expression patterns. The BAC coinjection technique should facilitate the studies of control elements in many different genes. It will be particularly useful for further studies of a growing number of important mouse and human disease mutations that map large distances from the coding regions of the gene that they disrupt (e.g., refs. 14–16). As in the case of the Bmp5 gene, such mutations are likely to define long-distance regulatory elements crucial for normal expression of the corresponding gene product.
Acknowledgments
We thank Andrew Ho for the full-length mouse Bmp5 cDNA and members of the Kingsley lab for useful discussions and comments on the manuscript. The BAC rescue injections were done by the Stanford Transgenic Facility. This work was supported by a grant from the National Institutes of Health (D.M.K.) as well as by fellowships from the National Science Foundation (R.J.D.) and the American Cancer Society (G.A.M.).
Abbreviations
- BMP
bone morphogenetic protein
- BAC
bacterial artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Manak J R, Mathies L D, Scott M P. Development. 1994;120:3605–3619. doi: 10.1242/dev.120.12.3605. [DOI] [PubMed] [Google Scholar]
- 2.Capovilla M, Brandt M, Botas J. Cell. 1994;76:461–475. doi: 10.1016/0092-8674(94)90111-2. [DOI] [PubMed] [Google Scholar]
- 3.Huang J D, Schwyter D H, Shirokawa J M, Courey A J. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 4.St. Johnston R D, Hoffmann F M, Blackman R K, Segal D, Grimaila R, Padgett R W, Irick H A, Gelbart W M. Genes Dev. 1990;4:1114–1127. doi: 10.1101/gad.4.7.1114. [DOI] [PubMed] [Google Scholar]
- 5.Masucci J D, Miltenberger R J, Hoffmann F M. Genes Dev. 1990;4:2011–2023. doi: 10.1101/gad.4.11.2011. [DOI] [PubMed] [Google Scholar]
- 6.Blackman R K, Sanicola M, Raftery L A, Gillevet T, Gelbart W M. Development. 1991;111:657–666. doi: 10.1242/dev.111.3.657. [DOI] [PubMed] [Google Scholar]
- 7.Irvine K D, Helfand S L, Hogness D S. Development. 1991;111:407–424. doi: 10.1242/dev.111.2.407. [DOI] [PubMed] [Google Scholar]
- 8.Goldhamer D J, Faerman A, Shani M, Emerson C P. Science. 1992;256:538–542. doi: 10.1126/science.1315077. [DOI] [PubMed] [Google Scholar]
- 9.Grosveld F, Antoniou M, Berry M, de Boer E, Dillon N, Ellis J, Fraser P, Hurst J, Imam A, Meijer D, et al. Cold Spring Harbor Symp Quant Biol. 1993;58:7–13. doi: 10.1101/sqb.1993.058.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Van der Ploeg L H, Konings A, Oort M, Roos D, Bernini L, Flavell R A. Nature (London) 1980;283:637–642. doi: 10.1038/283637a0. [DOI] [PubMed] [Google Scholar]
- 11.Driscoll M C, Dobkin C S, Alter B P. Proc Natl Acad Sci USA. 1989;86:7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 13.Vortkamp A, Gessler M, Grzeschik K H. Nature (London) 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 14.Foster J W, Dominguez-Steglich M A, Guioli S, Kowk C, Weller P A, Stevanovic M, Weissenbach J, Mansour S, Young I D, Goodfellow P N, et al. Nature (London) 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 15.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell H F, Donis-Keller H, Helms C, Hing A V, et al. Nat Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 16.Kluppel M, Nagle D L, Bucan M, Bernstein A. Development. 1997;124:65–77. doi: 10.1242/dev.124.1.65. [DOI] [PubMed] [Google Scholar]
- 17.DiLeone R J, Russell L B, Kingsley D M. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 19.Hogan B L. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 20.Sampath T K, Maliakal J C, Hauschka P V, Jones W K, Sasak H, Tucker R F, White K H, Coughlin J E, Tucker M M, Pang R H, et al. J Biol Chem. 1992;267:20352–20362. [PubMed] [Google Scholar]
- 21.Kingsley D M, Bland A E, Grubber J M, Marker P C, Russell L B, Copeland N G, Jenkins N A. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 22.Green E L, Green M C. J Morphol. 1942;70:1–19. [Google Scholar]
- 23.Green M C. J Morphol. 1951;88:1–22. doi: 10.1002/jmor.1050880102. [DOI] [PubMed] [Google Scholar]
- 24.Green M C. J Exp Zool. 1968;167:129–150. doi: 10.1002/jez.1401670202. [DOI] [PubMed] [Google Scholar]
- 25.King J A, Marker P C, Seung K J, Kingsley D M. Dev Biol. 1994;166:112–22. doi: 10.1006/dbio.1994.1300. [DOI] [PubMed] [Google Scholar]
- 26.Kingsley D M. Trends Genet. 1994;10:16–21. doi: 10.1016/0168-9525(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 27.Russell L B. Mutat Res. 1971;11:107–123. doi: 10.1016/0027-5107(71)90036-4. [DOI] [PubMed] [Google Scholar]
- 28.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 29.Bordonaro M, Saccomanno C F, Nordstrom J L. BioTechniques. 1994;16:428–430. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Kothary R, Clapoff S, Darling S, Perry M D, Moran L A, Rossant J. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 32.Methot D, Reudelhuber T L, Silversides D W. Nucleic Acids Res. 1995;23:4551–4556. doi: 10.1093/nar/23.22.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migchielsen A A, Breuer M L, Hershfield M S, Valerio D. Hum Mol Genet. 1996;5:1523–1532. doi: 10.1093/hmg/5.10.1523. [DOI] [PubMed] [Google Scholar]
- 34.Kucera G T, Bortner D M, Rosenberg M P. Dev Biol. 1996;173:162–173. doi: 10.1006/dbio.1996.0014. [DOI] [PubMed] [Google Scholar]
- 35.Merli C, Bergstrom D E, Cygan J A, Blackman R K. Genes Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- 36.Celeste A J, Iannazzi J A, Taylor R C, Hewick R M, Rosen V, Wang E A, Wozney J M. Proc Natl Acad Sci USA. 1990;87:9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortini M E, Rubin G M. Genes Dev. 1990;4:444–463. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- 39.Jaworski C J, Chepelinsky A B, Piatigorsky J. J Mol Evol. 1991;33:495–505. doi: 10.1007/BF02102802. [DOI] [PubMed] [Google Scholar]
- 40.Sanicola M, Sekelsky J, Elson S, Gelbart W M. Genetics. 1995;139:745–756. doi: 10.1093/genetics/139.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X W, Model P, Heintz N. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 42.Muyrers J P, Zhang Y, Testa G, Stewart A F. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]