Abstract
Objectives
To evaluate the auditory, vestibular, and retinal characteristics of a large American DFNA11 pedigree with autosomal dominant progressive sensorineural hearing loss that first impacts the low and mid-frequency auditory range. The pedigree (referred to as the HL2 family) segregates a myosinVIIA (MYO7A) mutation in exon 17 at DNA residue G2164C (MYO7AG2164C) that appears to be influenced by a genetic modifier that either rescues or exacerbates the MYO7AG2164C alteration. DNA analysis to examine single nucleotide polymorphisms (SNPs) in two candidate modifier genes (ATP2B2 and WFS1) is summarized in this report.
Study Design
Family study.
Results
The degree of low and mid-frequency hearing loss in HL2 family members segregating the MYO7AG2164C mutation varies from mild to more severe with approximately the same number of HL2 family members falling at each end of the severity spectrum. The extent of hearing loss in HL2 individuals can vary between family generations. Differences in the degree of hearing loss in MYO7AG2164C HL2 family members may be mirrored by vestibular function in at least two of these same individuals. The SNPs examined within ATP2B2 and WFS1 did not segregate with the mild versus more severe auditory phenotype.
Conclusions
The severity of the auditory and vestibular phenotypes in MYO7AG2164C HL2 family members may run in parallel suggesting a common modifier gene within the inner ear. The putative MYO7AG2164C genetic modifier is likely to represent a common polymorphism that is not linked tightly to the MYO7A mutation on the MYO7A2164C allele.
Keywords: Single nucleotide polymorphisms, DFNA11, MYO7A, low frequency hearing loss, electroretinography
INTRODUCTION
Hearing impairment is a common clinical finding with both genetic and environmental origins (1). Hearing loss with a genetic etiology can be syndromic associated with diagnoses compromising other body systems or non-syndromic restricted to deficits of the inner ear. Large human pedigrees segregating monogenic syndromic and non-syndromic hearing loss have led to the discovery of at least 100 chromosomal locations harboring auditory-related genes (2). Mutations within these genes cause the inherited sensory challenge within the pedigrees.
Mutations within seven different myosin genes are known to underlie auditory dysfunction: two conventional myosins, non-muscle myosin, heavy polypeptide 9 (MYH9) (3) and 14 (MYH14) (4); and five unconventional myosins, myosin IA (MYO1A) (5), myosin IIIA (MYO3A) (6), myosin VI (MYO6) (7), myosin VIIA (MYO7A) (8), and myosin XV (MYO15A) (9). Myosins constitute a family of motor proteins that share a conserved globular head domain with actin- and ATP-binding sites coupled to varied amino and carboxy-terminus regions. These variable regions determine the unique cellular function of each motor protein giving rise to diverse roles in biological processes such as muscle contraction, cell adhesion, organelle translocation, cytokinesis, and cell movement (10,11).
The MYO7A gene is expressed in the testis, lung, kidney, inner and outer hair cells of the cochlea, and retina (12). Within the retina, MYO7A is expressed in the retinal pigment epithelium (RPE) and rod and cone photoreceptor cells (12-14). In hair cells, MYO7A is found in the actin-rich stereocila bundles, cuticular plate, pericuticular necklace, and cell body (15). MYO7A is also expressed in both type I and type II hair cells of the semicircular canals and utricle (15).
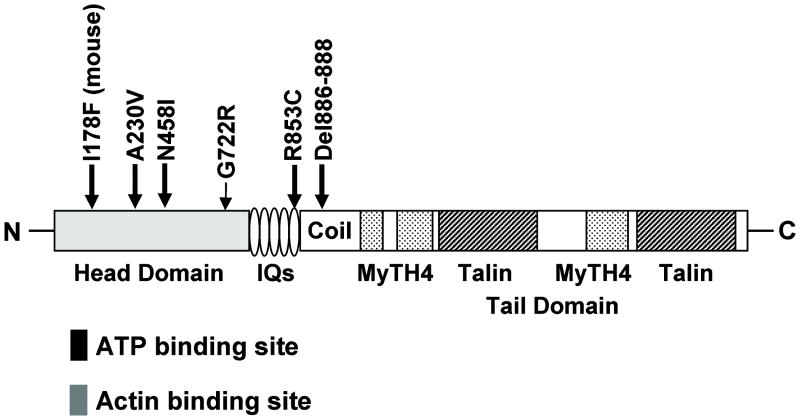
Mutations within MYO7A can lead to both syndromic and non-syndromic hearing impairment in humans. Syndromic MYO7A mutations are inherited in a recessive fashion with over 80 identified MYO7A mutations (16) leading to a diagnosis of Usher type 1B (USH1B), a disease characterized by profound, congenital, sensorineural deafness with progressive retinitis pigmentosa leading to visual loss and vestibular areflexia. Non-syndromic MYO7A mutations can be inherited in either a recessive (DFNB2, the 2nd autosomal recessive deafness locus identified) or dominant (DFNA11, the 11th autosomal dominant deafness locus identified) manner. Five DFNA11 mutations have been characterized: p.delA886-K887-K888 in a Japanese pedigree (17); p.G772R in an American pedigree (18); N458I in a Dutch pedigree, (19); p.R853C in a German pedigree (20); and p.A230V in an Italian pedigree (21) (Figure 1).
FIG. 1.
Dominant mutations within Myosin VIIA. Schematic representation of MYO7A functional regions indicates the location of five DFNA11 mutations and the dominant Hdb mouse mutant. The head region (light gray shading) contains ATP (black box) and actin (dark gray box) binding sites. The tail domain contains four notable regions: 1) IQs represent five light-chain-binding repeats; 2) coil indicates coiled-coil domain that may be involved in dimerization; 3) MyTH4 indicates myosin tail homology-4 domains that are regions conserved between myosins; and 4) talin represents talin-like homology domains which are predicted to bind actin.
We previously mapped the large hearing impaired DFNA11 American pedigree [referred to as HL2, for hearing loss family 2] to the long arm of chromosome 11 in band 13.5. A MYO7A mutation in exon 17 at DNA residue G2164C was discovered in the HL2 family. The MYO7AG2164C alteration leads to a predicted non-conservative glycine-to-arginine (G772R) amino acid substitution at a highly conserved glycine residue (18). The MYO7AG2164C mutation is unique as it was the first alteration in MYO7A associated with the uncommon clinical finding of progressive low-frequency hearing loss (18). We previously showed that the degree of low and mid-frequency hearing loss within the HL2 family varies markedly suggesting the presence of a genetic modifier that either rescues or exacerbates the primary MYO7AG2164C mutation (18).
The appreciation of genetic modifiers in the human auditory system has been increasing over the past several years. For example, the severity of a homozygous mutation in exon 42 of CDH23 (encoding cadherin 23) predicting a F1888S amino acid substitution appears to be modified by a heterozygous single nucleotide polymorphism (SNP) in exon 12 of ATP2B2 (encoding PMCA2) predicting a V586M amino acid substitution. Specifically, the PMCA2V586M allele significantly exacerbates the degree of hearing impairment in the homozygous CDH231888S individuals (22). The PMCA2V586M allele also worsens the extent of low-frequency hearing loss in a family segregating a dominant H246R mutation in the head domain of MYO6 (22,23). The PMCA2V586M allele by itself does not appear to cause auditory impairment but rather modulates the severity of hearing loss in families with mutations in either CDH23 or MYO6.
In this report, we characterize in greater detail the extent of hearing loss variation in the HL2 pedigree within and between family generations which supports the prediction that the putative genetic modifier is a SNP commonly found in the general Caucasian population. The candidacy of SNPs within the ATP2B2 and WFS1 (encoding wolframin) genes as potential modifiers of the MYO7AG2164C mutation was analyzed in the HL2 family. Members of the HL2 pedigree carrying the MYO7AG2164C mutation also completed formal vestibular testing and ERG evaluations.
MATERIALS AND METHODS
Research subjects and controls
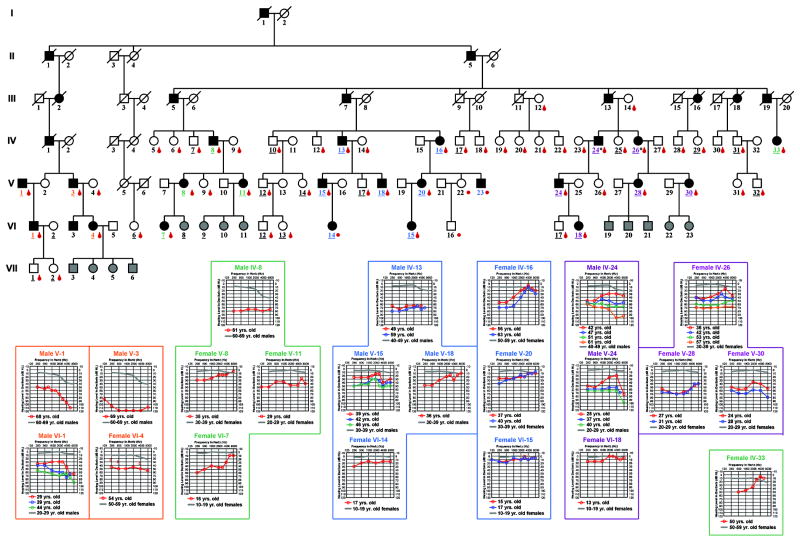
Under a protocol of informed consent approved by the Institutional Review Board of the University of Washington, Seattle, 5 mls of blood were obtained by venipuncture for high molecular weight DNA isolation using standard techniques. Control DNA samples were taken from a predominantly Caucasian population as described previously (18). The HL2 pedigree is of English decent. Male-to-male transmission is observed confirming autosomal dominant inheritance (Figure 2).
FIG. 2.
Audiologic characterization of the HL2 pedigree. Each individual in the pedigree is assigned a number by generation. Underlined numbers indicate the person completed an auditory evaluation. Affected individuals are denoted by blackened symbols, males by squares, females by circles, and deceased persons are indicated by a diagonal line through the symbol. If the auditory phenotype of a child is unknown, the symbol is filled in gray. Audiograms for affected individuals (shown for right ear only) are grouped as color-coded family clusters and positioned near the appropriate family branch. Frequency in hertz (Hz) is plotted on the x-axis and hearing level in decibels (dB HL) on the y-axis. Plotted on each audiogram (gray line) are the average pure-tone air conduction thresholds for a person with normal hearing (24) matched in age to the earliest audiogram collected for the HL2 family member.
Auditory, vestibular, and ERG assessment
Audiologic evaluations were conducted as described previously (18). Symmetrical hearing loss was detected in all affected HL2 family members. For clarity, only right ear responses are plotted on the audiograms in Figure 2. Audiometric data for a normal hearing individual of similar age to the research subject are included on each audiogram plot (24). The vestibular evaluations were conducted in the Otolaryngology-HNS Clinic testing suites at the University of Washington. Oculomotor testing and computerized dynamic posturography were conducted as described previously (25). Caloric testing was performed with an ICS air caloric irrigator (60 second irrigations at 24 and 50°C) and an Interacoustics 2D VOG eye tracker. The electroretinography (ERG) assessments were conducted at Children’s Hospital and Regional Medical Center in Seattle, Washington. Recording procedures followed the International Society for Clinical Electrophysiology of Vision (ISCEV) recommendations as closely as possible for full-field electroretinograms (26) and were performed as described previously (27).
Single nucleotide polymorphism analysis
For SNP analysis, PCR incubation mixture, thermocycling, and purification parameters were performed as noted previously (28) with ATP2B2 and WFS1 primers designed with the Primer 3 web-based program (29). Electropherograms were analyzed using the DNASTAR software package (30) (DNASTAR, Inc) or the CodonCode Aligner software package (CodonCode Corporation, Dedham, MA).
RESULTS
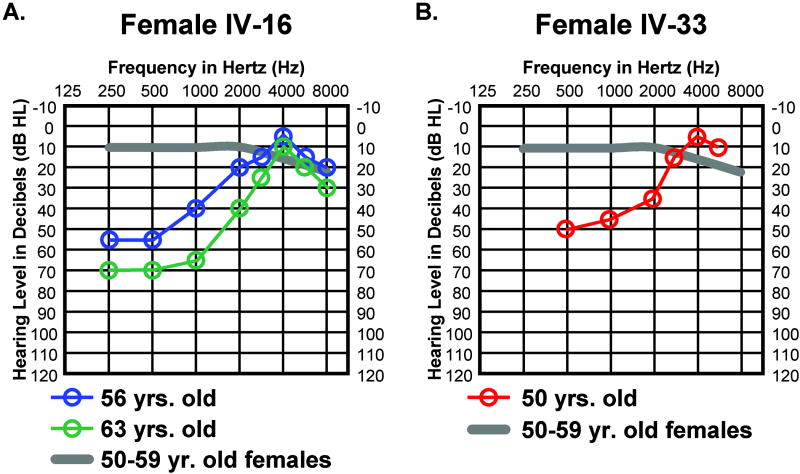
Two MYO7AG2164C females maintain normal high-frequency hearing in 5th and 6th decade of life
Females IV-16 and IV-33 both carry the MYO7AG2164C mutation which is consistent with their notable hearing loss between 250 and 2000 Hz (Figs. 3A, 3B). Remarkably, between 3000 and 8000 Hz they both have normal or above average hearing for their age (Figs. 3A, 3B) resulting in rising audiogram contours. Maintenance of normal high frequency hearing in females IV-16 and IV-33 may be attributed to a self-reported lack of significant noise or ototoxic drug exposure or perhaps to a genetic component that acts to protect hearing in the higher frequencies.
FIG. 3.
High-frequency hearing is well preserved in (A) female IV-16 and (B) female IV-33. Auditory thresholds are shown for the right ears only. Responses between the right and left ears were symmetrical. Frequency in hertz (Hz) is plotted on the x-axis and hearing level in decibels (dB HL) on the y-axis. Plotted on each audiogram (gray line) are the average pure-tone air conduction thresholds for a person with normal hearing matched in age to the earliest audiogram collected for the HL2 family member.
Genetic modifier may control severity differences in the low and mid-frequency ranges
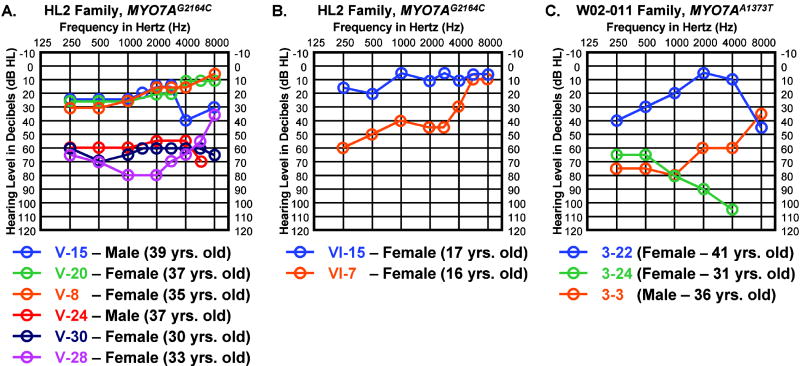
While most HL2 family members exhibit similar patterns in their audiologic profiles, some members of the HL2 pedigree show marked variation in their level of hearing impairment particularly in the low and mid-frequency ranges. Individuals V-15 (male), V-20 (female), and V-8 (female) are all mildly affected MYO7AG2164C family members. Individuals V-24 (male), V-30 (female), and V-28 (female) are all more severely affected MYO7AG2164C family members. Figure 4A compares the hearing sensitivity of these six individuals between the ages of 30-39 years old. Between 250 and 2000 Hz the pure tone auditory thresholds vary from 30 to 65 dB between these two distinct groups of HL2 family members. None of these six individuals reports a significant medical or noise exposure history that could account for these marked threshold differences in the low and mid-frequency ranges. These threshold differences can also be noted when comparing the auditory thresholds in younger MYO7AG2164C family members such as females VI-15 and VI-7 at 17 and 16 years of age, respectively (Fig. 4B). Salient variations in the degree of hearing loss between similarly-aged MYO7AG2164C individuals with comparable histories suggest the presence of a modifier gene that either rescues or exacerbates the primary MYO7AG2164C mutation. Given that approximately equal numbers of mild versus more severely affected MYO7AG2164C individuals are noted in the HL2 pedigree, the modifier alleles underlying these differences are likely to represent common polymorphisms. Individuals with auditory thresholds falling between the two distinct groups highlighted in Figure 4A are also found in the HL2 pedigree (e.g. male V-18 and female V-11). Interestingly, variations in the degree of hearing loss between similarly-aged family members is also seen in the Dutch DFNA11 pedigree carrying the MYO7AA1373G mutation predicting the N458I amino acid substitution (Fig. 4C) (31).
FIG. 4.
Variation in clinical severity between similarly aged HL2 family members. (A) Three MYO7AG2164C individuals with mild hearing loss versus three MYO7AG2164C individuals with more severe hearing loss in the low and mid-frequency ranges. All six individuals are between the ages of 30-39 years old. (B) Two MYO7AG2164C teenage females showed marked differences in low and mid-frequency auditory thresholds. Auditory thresholds are shown for the right ears only. Responses between the right and left ears were symmetrical. Frequency in hertz (Hz) is plotted on the x-axis and hearing level in decibels (dB HL) on the y-axis. (C) Three MYO7AA1373T individuals between the ages of 31 to 41 years old from the Dutch N458I family demonstrate variation in their auditory thresholds.
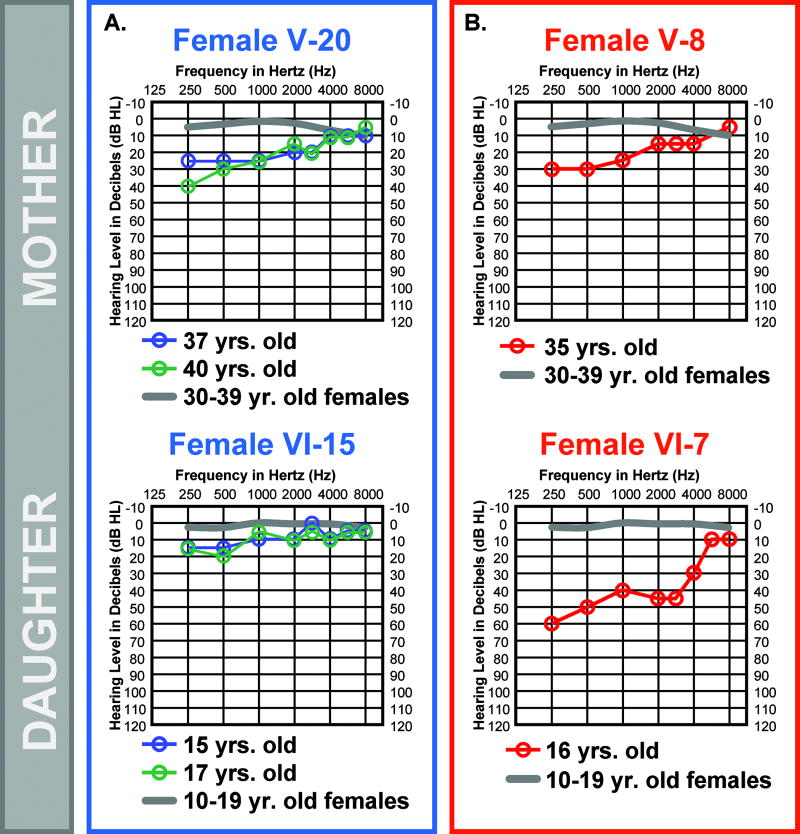
Degree of Hearing Loss can Vary between Generations
The audiologic profiles of females V-20 (mother) and VI-15 (daughter) show low-frequency hearing loss greater than expected for their ages (Fig. 5A). However, by definition at 37 and 17 years-of-age females V-20 and VI-15, respectively, do not have abnormal hearing (threshold elevations >25 dB HL) highlighting how subtly the MYO7AG2164C mutation can impact the auditory system in some individuals. The mother-daughter pair (females V-20 and VI-15) both demonstrate relatively mild hearing loss (Fig. 5A), suggesting that they both carry the same modifier SNP. In the case of the affected mother-daughter pair (female V-8 and female VI-7), the mother at 35 years of age is mildly affected by the MYO7AG2164C mutation while the daughter at 16 years of age demonstrates remarkably elevated auditory thresholds in the low and mid-frequencies compared to her mother (Fig. 5B) suggesting that they do not carry the same modifier SNP. The ability to frequently switch the severity of hearing loss between generations suggests that the modifier is not linked tightly to the MYO7A mutation on the MYO7A2164C allele and that the modifier is likely represented by a SNP commonly found in the general Caucasian population.
FIG. 5.
Comparison of hearing loss severity between HL2 family generations. Auditory thresholds are shown for the right ears only. Responses between the right and left ears were symmetrical. Frequency in hertz (Hz) is plotted on the x-axis and hearing level in decibels (dB HL) on the y-axis. Plotted on each audiogram (gray line) are the average pure-tone air conduction thresholds for a person with normal hearing matched in age to the earliest audiogram collected for the HL2 family member. (A) Mild HL2 auditory phenotype is maintained between mother (V-20) and daughter (VI-15). (B) Mild HL2 auditory phenotype is not maintained between mother (V-8) and daughter (VI-7).
Normal ERG Test Results in the HL2 Family
Next, we wanted to determine if a retinal phenotype segregates in the HL2 family and if the extent of retinal involvement follows the degree of hearing loss between MYO7AG2164C individuals. Therefore, male V-15 (at 42 years-of-age) with mild hearing loss and female V-28 (at 37 years-of-age) with more severe hearing loss underwent electroretinography (ERG) to assay for changes in retinal photoreceptor function. In both individuals, the scotopic and photopic responses had overall normal appearing waveforms with normal amplitude and latencies to the a- and b-waves. Under dark adaptation, an isolated b-wave of large amplitude was elicited to dim flashes of 0.06 and 0.14 cd · sec/m2 of white light and a 1.8 cd · sec/m2 blue flash. Red flashes (0.22 cd · sec/m2) elicited a small early cone peak prior to the scotopic b-wave, demonstrating a cone contribution under scotopic adaptation. With increasing flash intensity (3.1 to 60.9 cd · sec/m2), normal amplitude a- and b-waves are present. Photopic ERGs to the single flash and 30 Hz flicker also showed normal appearing a- and b-wave components.
Vestibular Phenotype May Mirror Degree of Hearing Loss
Finally, we wanted to determine if a clinically detectable vestibular phenotype segregates in the HL2 family and if the extent of vestibular involvement parallels the degree of hearing loss between MYO7AG2164C individuals. Therefore, male V-15 with mild hearing loss and female V-28 with more severe hearing loss underwent vestibular assessment (Table). Computerized dynamic posturography. Sensory organization testing: Male V-15 displayed a normal SOT composite score of 82. Female V-28 had an abnormal SOT composite score of 69 with reliance on visual cues to maintain her balance. The COG (center of gravity) alignment indicated that female V-28 had a weight shift to the left during testing. Motor control test. Male V-15 displayed normal motor control test scores. Female V-28 demonstrated abnormal responses on the toes down adaptation test. Oculomotor testing was normal for both male V-15 and female V-28. Bithermal caloric testing was normal for male V-15 (>10°/sec). Female V-28 displayed a 17% unilateral weakness in the left ear indicating that she is receiving more vestibular information from her right inner ear than her left. As summarized in the Table, male V-15 with more mild hearing loss generated normal vestibular test results while female V-28 with more severe hearing loss demonstrated abnormal vestibular findings. Interestingly, vestibular impairment may also parallel the degree of hearing loss in two individuals from the Dutch DFNA11 pedigree carrying the MYO7AA1373G mutation predicting the N458I amino acid substitution (31). Individual 3-22 with more mild hearing loss (Fig. 4C) is reported to have normal clinical vestibular responses while individual 3-3 with more severe hearing loss (Fig. 4C) is reported to have bilateral caloric weakness (31).
TABLE.
Phenotype Comparison
| Male V-15 | Female V-28 | |
|---|---|---|
| Auditory | Mild HL | More Severe HL |
| ERG | Normal | Normal |
| Vestibular | ||
| SOT | 82 | 69*, abnormal reliance on visual cues |
| COG | Normal | Weight shift to left |
| MCT | Normal | Abnormal on toes down adaptation test |
| Calorics | Normal | 17% unilateral weakness to the left ear |
| Oculomotor | Normal | Normal |
SOT = Sensory organization test composite score, * abnormal score
COG = Center of gravity measurement during SOT exam
MCT = Motor control test
Investigating Candidate Modifier Gene SNPs
Given that the PMCA2V586M allele can modulate the severity of hearing loss in families with mutations in either CDH23 or MYO6, we analyzed the PMCA2V586M allele in all MYO7AG2164C HL2 family members affected by hearing loss for which a DNA sample was available (21 individuals). The MYO7AG2164C HL2 individuals were homozygous for the PMCA2V586 allele indicating that the PMCA2V586M polymorphism was not responsible for the difference in hearing loss severity in the HL2 pedigree. We also compared the open-reading-frame (ORF) sequence of WFS1, another gene known to cause low-frequency hearing loss, between DNAs from HL2 individuals V-20 and V-24. All WFS1 SNPs detected in exons 1-8 were shared in common between these two family members.
DISCUSSION
In this report, we have characterized the auditory, vestibular, and retinal characteristics of a large American DFNA11 MYO7AG2164C pedigree with autosomal dominant progressive sensorineural hearing loss that generally first impacts the low and mid-frequency auditory range. Hearing loss within members of the HL2 pedigree segregating the MYO7AG2164C alteration can range from mild to severe with similar numbers of HL2 family members falling at the end of each severity scale. The degree of hearing loss severity can switch between family generations. These findings suggest the presence of a genetic modifier acting on the MYO7AG2164C mutation that is likely to represent a common polymorphism in the Caucasian population that is not linked tightly to the MYO7A2164C allele. Vestibular test results from two MYO7AG2164C HL2 family members and two individuals with the MYO7AA1373G mutation suggest that the severity of the auditory and vestibular phenotype may run in parallel implicating a common modifier gene within the inner ear.
The HL2 audiometric configuration with hearing loss first impacting the low and mid-frequencies is noteworthy as the vast majority of non-syndromic deafness initiate with high-frequency hearing loss (32). The other four DFNA11 mutations result in either a flat or downward sloping audiogram contour with the exception of a few affected individuals with the MYO7AN458I alteration (31). Interestingly, the dominant Headbanger (Hdb) mouse mutant has been shown to demonstrate low frequency hearing loss associated with a Myo7aI178F mutation (33). These three different predicted amino acid substitutions within the myosin VIIA head domain (Myo7aI178F, MYO7AN458I, MYO7AG722R) are all capable of manifesting the phenotype of low-frequency hearing loss. However, this phenotype-to-genotype correlation with the head domain is not absolute as the audiograms reported for the MYO7AA230V mutation demonstrate primarily flat or downward sloping configurations (21). The HL2 audioprofile is most similar to the progressive non-syndromic low frequency hearing loss characteristic of DFNA1 and DFNA6/DFNA14/DFNA38 caused by heterozygous mutations in the diaphanous 1 (DIAPH1) (34) and Wolfram syndrome 1 (WFS1) (35) genes, respectively.
The HL2 pedigree is also notable due to the striking difference in hearing loss severity seen between affected family members; a phenotypic difference that may extend to the vestibular system. Clinical vestibular assessment was normal in a MYO7AG2164C HL2 family member with mild hearing loss (male V-15) and abnormal in a MYO7AG2164C HL2 family member (female V-28) with more pronounced auditory dysfunction. Vestibular testing of additional HL2 family members falling at different ends of the hearing loss spectrum in the family will be useful in expanding these findings. The MYO7AG2164C mutation does not appear to impact the retina as individuals with both mild and more severe low-frequency hearing loss generated normal ERG responses.
The variation in the degree of hearing loss in the HL2 family suggests the presence of a genetic modifier that may be represented by a SNP commonly found in the general population. While the ATP2B2 and WFS1 SNPs examined in this report and the MYO7A and GJB2 SNPs studied previously (18) do not segregate with the phenotypic differences in the HL2 pedigree, SNPs within the MYO7A promoter and other gene products known to interact with MYO7A in the ear should also be considered as candidates. Identification of SNPs regulating the clinical severity of hearing loss and vestibular deficits will enhance understanding of gene product interactions within the inner ear and may provide predictive value in counseling patients carrying these mutations and genetic variations.
Acknowledgments
Sources of Support that Require Acknowledgment: This work was funded by NIH grants DC04945 (V.A.S.) and P30 DC04661 (V. M. Bloedel Core), and the following three private funds, William O. Rogers Trust Fund, Anderson Charitable Contribution, and the Peter G. La Haye Fund (J.P.K). We are grateful to the HL2 family for their cooperation throughout this study. We thank Drs. Bruce Tempel, Helen Brew, and Donald Farrell for comments on this project. We thank Evan Thilo for his comments on this manuscript. We appreciate the cooperation of audiology clinics across the country that participated in the hearing evaluations.
References
- 1.Morton NE. Genetic epidemiology of hearing impairment. Ann NY Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 2.Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. 2008 World Wide Web URL: http://dnalab-www.uia.ac.be/dnalab/hhh/
- 3.Lalwani AK, Goldstein JA, Kelley MJ, et al. Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet. 2000;67:1121–8. doi: 10.1016/s0002-9297(07)62942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaudy F, Snoeckx R, Pfister M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74:770–6. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaudy F, Ferrara A, Esposito L, et al. Multiple mutations of MYO1A, a cochlear-expressed gene, in sensorineural hearing loss. Am J Hum Genet. 2003;72:1571–7. doi: 10.1086/375654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, Walsh V, Vreugde S, et al. From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–23. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melchionda S, Ahituv N, Bisceglia L, et al. MYO6, the human homologue of the gene responsible for deafness in Snell’s walter mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet. 2001;69:635–40. doi: 10.1086/323156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weil D, Blanchard S, Kaplan J, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–1. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang A, Liang Y, Fridell RA, et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–51. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- 10.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–65. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 11.Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–33. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 12.Hasson T, Heintzelman MB, Santos-Sacchi J, et al. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci USA. 1995;92:9815–9. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Amraoui A, Sahly I, Picaud S, et al. Human Usher 1B/mouse shaker-1: the retinal phenotype discrepancy explained by the presence/absence of myosin VIIA in the photoreceptor cells. Hum Mol Genet. 1996;5:1171–8. doi: 10.1093/hmg/5.8.1171. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Vansant G, Udovichenko IP, et al. Myosin VIIa, the product of the Usher 1B sydrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil Cytoskeleton. 1997;37:240–52. doi: 10.1002/(SICI)1097-0169(1997)37:3<240::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 15.Hasson T, Gillespie PG, Garcia JA, et al. Unconventional myosins in inner-ear sensory epithelia. J Cell Biol. 1997;137:1287–307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson PD, Ball EV, Mort M, et al. Human gene mutation database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 17.Tamagawa Y, Kitamura K, Ishida T, et al. Sensorineural hearing impairment non-syndromic, dominant DFNA11. Adv Otorhinolaryngol. 2000;56:103–6. doi: 10.1159/000059092. [DOI] [PubMed] [Google Scholar]
- 18.Street VA, Kallman JC, Kiemele KL. Modifier controls severity of a novel dominant low frequency Myosin VIIA (MYO7A) auditory mutation. J Med Genet. 2004;41:e62. doi: 10.1136/jmg.2003.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luijendijk MW, Van Wijk E, Bischoff AM, et al. Identification and molecular modelling of a mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11) Hum Genet. 2004;115:149–56. doi: 10.1007/s00439-004-1137-3. [DOI] [PubMed] [Google Scholar]
- 20.Bolz H, Bolz SS, Schade G, et al. Impaired calmodulin binding of myosin-7A causes autosomal dominant hearing loss (DFNA11) Hum Mutat. 2004;24:274–5. doi: 10.1002/humu.9272. [DOI] [PubMed] [Google Scholar]
- 21.Di Leva F, D’Adamo P, Cubellis MV, et al. Identification of a novel mutation in the myosin VIIA motor domain in a family with autosomal dominant hearing loss (DFNA11) Audiol Neurootol. 2006;11:157–64. doi: 10.1159/000091199. [DOI] [PubMed] [Google Scholar]
- 22.Schultz JM, Yang Y, Caride AJ, et al. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N Engl J Med. 2005;352:1557–64. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- 23.Mohiddin SA, Ahmed ZM, Griffith AJ, et al. Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6) J Med Genet. 2004;41:309–14. doi: 10.1136/jmg.2003.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterhammel D, Osterhammel P. High-frequency audiometry. Scand Audiol. 1979;8:73–81. doi: 10.3109/01050397909076304. [DOI] [PubMed] [Google Scholar]
- 25.Street VA, Kallman JC, Robertson NG, et al. A novel DFNA9 mutation in the vWFA2 domain of COCH alters a conserved cysteine residue and intrachain disulfide bond formation resulting in progressive hearing loss and site-specific vestibular and central oculomotor dysfunction. Am J Hum Genet A. 2005;139:86–95. doi: 10.1002/ajmg.a.30980. [DOI] [PubMed] [Google Scholar]
- 26.Marmor MF, Arden GB, Nilsson SEG, et al. Standard for clinical electroretinography. Arch Ophthalmol. 1989;107:816–9. doi: 10.1001/archopht.1989.01070010838024. [DOI] [PubMed] [Google Scholar]
- 27.Marmor MF, Holder GE, Seeliger MW, et al. International society for clinical electrophysiology of vision. Standard for clinical electroretinography. Doc Opththalmol. 2004;108:107–14. doi: 10.1023/b:doop.0000036793.44912.45. [DOI] [PubMed] [Google Scholar]
- 28.Street VA, Robinson LC, Erford SK, et al. Molecular genetic analysis of distal mouse chromosome 6 defines gene order and positions of the deafwaddler and opisthotonos mutations. Genomics. 1995;29:123–30. doi: 10.1006/geno.1995.1222. [DOI] [PubMed] [Google Scholar]
- 29.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–86. [DOI] [PubMed] [Google Scholar]
- 30.Burland T. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 31.Bischoff AM, Pennings RJ, Huygen PL, et al. Cochleovestibular and ocular features in a Dutch DFNA11 family. Otol Neurotol. 2006;27:323–31. doi: 10.1097/00129492-200604000-00006. [DOI] [PubMed] [Google Scholar]
- 32.Smith RJ, Huygen PL. Making sense of nonsyndromic deafness. Arch Otolaryngol Head Neck Surg. 2003;129:405–6. doi: 10.1001/archotol.129.4.405. [DOI] [PubMed] [Google Scholar]
- 33.Rhodes CR, Hertzano R, Fuchs H, et al. A Myo7a mutation cosegregates with stereocilia defects and low-frequency hearing impairment. Mamm Genome. 2004;15:686–97. doi: 10.1007/s00335-004-2344-x. [DOI] [PubMed] [Google Scholar]
- 34.Lynch ED, Lee MK, Morrow JE, et al. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–8. [PubMed] [Google Scholar]
- 35.Lesperance MM, Hall JWr, San Agustin TB, et al. Mutations in the Wolfram syndrome type 1 gene (WFS1) define a clinical entity of dominant low-frequency sensorineural hearing loss. Arch Otolaryngol Head Neck Surg. 2003;129:411–20. doi: 10.1001/archotol.129.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]