Abstract
Integrating laterally acquired virulence genes into the backbone regulatory network is important for the pathogenesis of Escherichia coli O157:H7, which has captured many virulence genes through horizontal transfer during evolution. GadE is an essential transcriptional activator of the glutamate decarboxylase (GAD) system, the most efficient acid resistance (AR) mechanism in E. coli. The full contribution of GadE to the AR and virulence of E. coli O157:H7 remains largely unknown. We inactivated gadE in E. coli O157:H7 Sakai and compared global transcription profiles of the mutant with that of the wild type in the exponential and stationary phases of growth. Inactivation of gadE significantly altered the expression of 60 genes independently of the growth phase and of 122 genes in a growth phase-dependent manner. Inactivation of gadE markedly downregulated the expression of gadA, gadB, and gadC and of many acid fitness island genes. Nineteen genes encoded on the locus of enterocyte effacement (LEE), including ler, showed a significant increase in expression upon gadE inactivation. Inactivation of ler in the ΔgadE strain reversed the effect of gadE deletion on LEE expression, indicating that Ler is necessary for LEE repression by GadE. GadE is also involved in downregulation of LEE expression under conditions of moderately acidic pH. Characterization of AR of the ΔgadE strain revealed that GadE is indispensable for a functional GAD system and for survival of E. coli O157:H7 in a simulated gastric environment. Altogether, these data indicate that GadE is critical for the AR of E. coli O157:H7 and that it plays an important role in virulence by downregulating expression of LEE.
Escherichia coli O157:H7 is the prevalent variant of enterohemorrhagic E. coli (EHEC) associated with hemorrhagic enteritis and hemolytic uremic syndrome in humans in the United States (26, 51). E. coli O157:H7 has to survive a number of environmental stresses during transmission from cows to humans. Surviving acid stress is critical during transmission, as the typical human stomach pH ranges from 1.5 to 3.0 (40). E. coli strains exhibit more acid resistance (AR) than other enteric pathogens, and this AR is considered a virulence factor in E. coli O157:H7, as it contributes to the low infective dose (10, 40). E. coli has four distinct AR mechanisms, the oxidative (OXI) system, glutamate decarboxylase (GAD) system, arginine decarboxylase (ARG) system, and lysine decarboxylase system (18, 32), that are phenotypically distinct and provide protection against low pH dependent on the type of acidic environment encountered (42). In addition to the defined mechanisms, other factors of the general stress response, including the stress response sigma factor RpoS and the DNA binding protein Dps, also contribute to the AR of E. coli (13, 59).
The GAD system is the most effective system in protecting E. coli cells against low pH compared to other known AR mechanisms (10, 11, 27, 56). The GAD system has three components: two GAD isozymes, GadA and GadB, and the gamma-aminobutyric acid (GABA)-glutamate antiporter GadC (18, 31). gadA is a member of the acid fitness island (AFI), which is located at 78 min, whereas gadB and gadC form a separate operon located at 33 min in the E. coli K-12 chromosome (22, 33, 48). Environmental signals that induce the GAD system include entry into stationary phase and acidic pH (31).
Regulation of the GAD system is complex, involving multiple regulatory circuits that influence the expression of GAD components through the central activator, GadE (18). GadE, a LuxR family regulator, is transcribed as two transcripts of sizes 0.68 kb and 1.06 kb, and its secondary structure contains a potential helix-turn-helix DNA-binding domain (21, 31). However, a recent study demonstrated that GadE is possibly transcribed as three transcripts of sizes 0.9 kb, 1.1 kb, and 1.38 kb (J. W. Foster and A. Sayed, presented at the 108th General Meeting of American Society of Microbiology, Boston, MA, 1 to 5 June 2008). GadE binds to a conserved 20-bp GAD box sequence upstream of gadA and gadBC in E. coli K-12 and activates the transcription of these genes (9, 21, 31). Although the GAD system and GadE in E. coli K-12 are well characterized, they remain poorly defined in E. coli O157:H7.
A study conducted by our laboratory demonstrated that E. coli O157:H7 strains have a greater average level of survival under complex acidic conditions, such as a simulated gastric environment, compared to those of other serogroups of EHEC (5). O157:H7 also expresses higher transcript levels of gadA and gadB genes than other EHEC strains in minimal medium containing glucose (5). Also, we recently showed that gadA and gadB sequences remain divergent in E. coli O157:H7 compared to other E. coli strains (4). Taken together, these findings suggest that the regulation and function of the GAD system may be distinct in E. coli O157:H7.
Genome sequence comparisons revealed that the O157:H7 Sakai strain has approximately 1,650 O157-specific loci that are not present in E. coli K-12 (20). Through evolution, the O157 population has acquired many mobile elements such as lambdoid phages carrying virulence and fitness islands (43, 60). Some of the important virulence factors of E. coli O157:H7, including Shiga toxins and the locus of enterocyte effacement (LEE), are encoded by horizontally transferred phage elements (28). The LEE, which is encoded on an acquired pathogenicity island, encodes a type 3 secretion system that mediates intimate adherence of bacteria to the intestinal mucosa through formation of attaching and effacing lesions (34). Integrating these acquired elements into the chromosomal regulatory network is critical for a pathogen to be successful (1). An example of this is one of the GAD system regulators, GadX, which has been shown to influence the expression of the LEE (47). Hence, it is possible that a chromosomal regulator such as GadE has acquired additional functions, though the effects of the GadE regulator for E. coli O157:H7 on a genome-wide scale are still unknown. Comparisons of GadE amino acid sequences among pathogenic and nonpathogenic E. coli strains revealed no significant divergence. Recently, a study by Tatsuno et al. found that inactivation of gadE increases the expression of many LEE genes in E. coli O157:H7 (52). However, gadE inactivation did not affect the expression of ler, the LEE-encoded regulator, and hence the pathway through which this LEE downregulation occurs was not identified. Recently, Ler was found to negatively regulate expression of gadE and it was suggested that there is a reciprocal negative interaction between Ler and the GadE regulators (1).
The objectives of this study were to identify the genes regulated by GadE in E. coli O157:H7 and to gain insight into the mechanism underlying the negative regulation of the LEE by GadE. By comparing the whole-genome transcription profiles of E. coli O157:H7 Sakai and the isogenic ΔgadE strain, we found that gadE positively influences expression of the GAD system genes and other AFI genes, whereas it negatively impacts the expression of the LEE genes, including ler. Expression of gadE was markedly increased in the stationary phase, thereby affecting the expression of numerous genes in a growth phase-dependent manner. In addition, we demonstrate that a functional Ler is necessary for the downregulation of LEE by GadE. Characterization of the AR phenotype of the ΔgadE strain revealed that GadE is indispensable for a functional GAD system and for survival of E. coli O157:H7 in a simulated gastric environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are summarized in Table 1. All strains were stored at −70°C in LB broth containing 10% glycerol, inoculated into 10 ml of LB broth, and grown to an optical density at 600 nm (OD600) of ∼0.1 to recover cells. To minimize the confounding effect of the acidic pH that would develop in the stationary phase of growth in unbuffered medium, the strains were grown in morpholinepropanesulfonic acid (MOPS) minimal medium buffered to pH 7.4. Cells recovered in LB broth were then grown twice to the stationary phase in MOPS-buffered minimal medium before a final transfer at a 1:30 dilution into 100 ml of MOPS medium for RNA isolation and a model stomach assay (6).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotypea | Reference or source |
|---|---|---|
| Strains | ||
| TW08264 | E. coli O157:H7 RIMD0509952 (Sakai) wild type | 36 |
| TW15901 | TW08264 harboring pKM208 plasmid | This study |
| TW15902 | ΔgadE | This study |
| TW15903 | ΔgadE Δler::Km | This study |
| TW15904 | Δler::Km | This study |
| TW15905 | ΔgadE/pCR2.1gadE | This study |
| Plasmids | ||
| pKM208 | Red recombinase expression plasmid, Apr | 16 |
| pKD4 | Template plasmid for lambda Red recombination system, Kmr | 16 |
| pCP20 | Flp recombinase expression plasmid, Cmr | 16 |
| pCR2.1 | Cloning vector | Invitrogen |
| pCR2.1gadE | gadE cloned into pCR2.1 | This study |
Ap, ampicillin; Cm, chloramphenicol.
Genetic manipulations.
E. coli O157:H7 Sakai ΔgadE and Δler strains were constructed by the modified one-step gene inactivation method for EHEC developed by Datsenko and Wanner and by Murphy et al. (16, 37). Briefly, recombinant PCR products containing a kanamycin (Km) resistance marker flanked by 45- to 50-bp sequences homologous to the upstream and downstream regions of target genes were generated from plasmid pKD4 (16) by use of the primers listed in Table 2. PCR products were electroporated into Red recombinase-producing E. coli O157:H7 (TW15901) as described previously (37), and the transformants were identified on LB agar plates with 25 μg of Km/ml at 37°C. The Km resistance marker was removed from the ΔgadE strain by introducing plasmid pCP20, which encodes Flp recombinase (16). Subsequently, the ΔgadE Δler double mutant was constructed by one-step inactivation of ler in the ΔgadE strain by use of the method described above.
TABLE 2.
Oligonucleotide primers used for one-step inactivation
| Primera | Sequence (5′ to 3′)b |
|---|---|
| gadE-H-F | ATCAATTCCCTGTCAGAGATCAAAAAAGTAGGCAATAAACCCTTCAAGGTgtgtaggctggagctgcttc |
| gadE-H-R | CTCGTCATGCCAGCCATCAATTTCAGTTGCTTATGTCCTGACTAAAAATAcatatgaatatcctccttag |
| ler-H-F* | TTTCATCTTCCAGCTCAGTTATCGTTATCATTTAATTATTTCATGgtgtaggctggagctgcttc |
| ler-H-R* | GTTGGTCCTTCCTGATAAGGTCGCTAATAGCTTAAAATATTAAAGcatatgaatatcctccttag |
The homology regions for the ler primers are from Iyoda and Watanabe (23).
Priming sites for pKD4 are in lowercase letters.
For complementation of the ΔgadE strain, DNA fragments of 2,630 bp containing the Sakai gadE coding region and additional flanking regions of gadE were amplified from E. coli O157:H7 Sakai chromosomal DNA by use of TaKaRa LA Taq polymerase (Takara Bio USA, Madison, WI). The resulting PCR products were cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) to make the pCR2.1gadE plasmid, which was transformed into the ΔgadE strain, creating the ΔgadE/pCR2.1gadE strain.
RNA isolation and cDNA labeling.
For RNA isolation, the wild-type Sakai and ΔgadE strains were grown to the early exponential phase (2.25 h [OD600 ∼ 0.25]) and stationary phase (5.5 h [OD600 ∼ 1.5]) in MOPS medium as described above. RNA was isolated from five independent cultures by use of hot acid-phenol-chloroform extraction. At each growth phase, 5 ml of culture was mixed with an equal volume of hot acid-phenol-chloroform (Ambion, Austin, TX) (pH 4.5 with isoamyl alcohol; 125:25:1) and incubated at 65°C with periodic shaking for 10 min. The samples were centrifuged at 3,220 × g for 20 min, and the supernatant was subjected to further extractions with phenol-chloroform and chloroform-isoamyl alcohol (6). RNA was precipitated overnight at −70°C in 2.5 volumes of 100% ethanol and 1/10 volume of 3 M sodium acetate (pH 5.2). RNA purification and DNase treatment of RNA samples were done with an RNeasy kit (Qiagen, Valencia, CA), and RNA quality was assessed on a formaldehyde-agarose gel.
Six micrograms of RNA was used for reverse transcription reactions containing 3 μg of random primers (Invitrogen), 1× first-strand buffer (Invitrogen), 10 mM dithiothreitol, 400 U of Superscript II (Invitrogen), 0.5 mM (each) dATP, dCTP, and dGTP, 0.3 mM dTTP, and 0.2 mM aminoallyl dUTP (6). After incubation at 42°C overnight, the cDNAs were purified with Qiagen PCR cleanup columns with phosphate wash buffer (5 mM K2HPO4 [pH 8.0], 80% ethyl alcohol) and phosphate elution buffer (4 mM K2HPO4 [pH 8.5]) and were coupled with either Cy3 or Cy5 dyes (Amersham Biosciences, Piscataway, NJ) as described previously (6).
cDNA hybridizations.
Hybridizations were performed according to a loop design, which included between-strain (wild type versus mutant at the same growth phase) and within-strain (same strain at two growth phases) comparisons. Five biological replicates were included for each comparison, resulting in 20 arrays. As described previously (6), the cDNAs were hybridized onto microarray slides printed with 6,088 open reading frames (ORFs), including 110 ORFs from the pO157 plasmid, representing E. coli strains K-12, EDL933, and Sakai. Arrays were scanned with an Axon 4000b scanner (Molecular Devices, Sunnyvale, CA) followed by image analysis using GenePix 6.0 (Molecular Devices) (6).
Data analysis.
The microarray data were analyzed using R (version 2.2.1) and the MAANOVA (version 0.98.8) package. Raw intensity values from replicate probes were averaged and log2 transformed after normalization by the pin-tip LOWESS method. The normalized intensity values were fitted to a mixed-model analysis of variance (ANOVA) by considering array and biological replicates as random factors and dye, strain, and growth phase as fixed factors (14). The linear model tested was Y (intensity) = array plus dye plus strain (wild type or mutant) plus growth phase (exponential or stationary) plus strain × growth phase plus sample (biological replicate) plus error. Each main effect had two levels: (i) mutant and wild type for the strain and (ii) exponential and stationary phases for the growth phase. The design included between- and within-strain comparisons using five biological replicates. Significant differences in expression due to strain, growth phase, and strain × growth phase were determined using the Fs test in MAANOVA, which uses a shrinkage estimator for gene-specific variance components that makes no assumption about the variances across genes (15), with 500 random permutations to estimate the P values. ANOVA has been used with mixed linear models to analyze microarray experiments with repeated calculations in which transcript levels of the strains at two different growth phases are measured (3, 24, 29). The q-value package in R was used for determining the false discovery rate (FDR) (49).
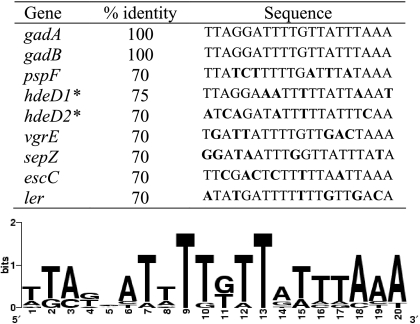
Overrepresentation of gene sets with a common biological function in the wild-type or mutant strain was determined using preranked gene set enrichment analysis (GSEA) and the GSEA program (version 2.0) (Broad Institute, Massachusetts Institute of Technology) (50). The gene sets were designated based on The Institute for Genomic Research annotation for the Sakai genome (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=ntec03). Additionally, two gene sets, the LEE gene set and the AFI and GAD gene set, were included in the analysis (17, 19, 33, 54). The pattern search tool in coliBASE (http://xbase.bham.ac.uk/colibase/pattern.pl?id=1073) was used to identify GAD box (9, 32) sequences (5′-TTAGGATTTTGTTATTTAAA-3′) in the putative promoter regions of genes differentially regulated between the wild type and ΔgadE strain. A cutoff of 70% similarity to the query sequence was set to apply higher stringency, because experimental confirmation of GadE binding was not conducted. A sequence logo for the consensus sequence was created at http://weblogo.berkeley.edu/logo.cgi.
Quantitative real-time PCR.
RNA isolations were conducted at both the exponential and stationary phases of growth. For assays with the ΔgadE/pCR2.1gadE, ΔgadEΔler, and Δler strains, RNA was isolated only at the exponential phase. TaqMan assays (5) were used for quantifying the expression of gadA, gadB, and ler, with mdh used as a reference for normalization. For the remaining genes, SYBR green chemistry was used for measuring expression levels. Primers were designed using the Primer3 server (44) based on the published reference genome sequence of E. coli O157:H7 strain Sakai (see Table S3 in the supplemental material). cDNA synthesis was conducted using an iScript Select cDNA synthesis kit (Bio-Rad, Hercules, CA) and 1 μg of total RNA according to the manufacturer's instructions. After reverse transcription, fivefold serial cDNA dilutions were used for quantitative PCR (Q-PCR) assays containing 12.5 μl of 2× iQ SYBR green SuperMix (Bio-Rad), 0.63 μl of each primer (10 μM stock), 9.24 μl of water, and 2 μl of cDNA, with cycle conditions of 95°C for 2 min followed by 40 cycles of 10 s at 95°C and then 20 s at the specific annealing temperature (6). The expression levels of the 16S rRNA gene were used for normalization of data, and the relative expression levels were quantified using Pfaffl's method (41). The results presented are averages of the results from at least three biological replicate experiments ± standard errors of the means (SEM).
Expression studies using EG minimal medium.
Wild-type and ΔgadE cells were grown in EG minimal medium (E minimal medium containing 0.4% glucose) at pH 7.0 and pH 5.0 (30) to the late exponential phase (OD600 ∼ 0.5). RNA extractions were conducted using a modified hot-phenol extraction protocol (7) that utilized 5% acidic phenol in ethanol. cDNA synthesis and Q-PCR methods are described above.
AR mechanism assays.
AR mechanism assays for the GAD, ARG, and OXI systems were conducted as described previously (27, 30). Briefly, for the GAD system, strains grown in LB broth with 0.4% glucose were challenged at pH 2.0 in a test environment (EG plus glutamate) and in a control environment (EG), whereas for the ARG system, after growth in BHI broth with 0.6% glucose, strains were analyzed in test (EG plus arginine) and control (EG) environments at pH 2.5. For testing of the OXI system, strains were grown in LBMES (pH 5.0) (Luna-Bertani broth buffered with morpholineethanesulfonic acid) and EG (pH 7.0) and challenged at pH 2.5 in EG. Samples were withdrawn at specific time points (30-min or 1-h intervals) and plated onto LB agar plates by use of an Autoplate 4000 spiral plater (Spiral Biotech, Bethesda, MD). Colonies were counted after overnight incubation at 37°C by use of a Q-Count system (Spiral Biotech). Assays were conducted for at least two biological replicate experiments, each with two technical replicate experiments. Levels of CFU per milliliter from technical replicates were averaged and converted to log10 CFU per milliliter. The data reported represent averages ± SEM for the results of at least three experiments.
Model stomach assay.
The model stomach system (MSS) (25) was prepared as described previously (5). Gerber Turkey Rice Dinner baby food (30 g) was mixed with 120 ml of synthetic gastric fluid (pH 1.75), yielding a final acidity of pH 2.5. Contents of the MSS were left for 30 s, sampled, diluted, and plated onto LB agar plates every 30 min for 1.5 h to enumerate viable cells. CFU per milliliter from duplicate plates were averaged and converted to log10 CFU per milliliter. The data reported represent the average results ± SEM for three experiments.
Microarray data accession.
Microarray data are available at NCBI GEO (accession number GSE13132; http://www.ncbi.nlm.nih.gov/geo).
RESULTS
Identification of genes regulated by GadE.
Inactivation of gadE did not cause a significant difference in the growth rate of E. coli O157:H7; the generation time of the wild-type strain was 43.7 min, and that of the ΔgadE strain was 44.1 min. A two-factor ANOVA with two main effects (strain and growth phase) and the interaction effect (strain × growth phase) was used to determine the impact of gadE inactivation on the transcriptome of E. coli O157:H7 at both the exponential and stationary phases. Genes with FDR < 0.1 for strain effect and FDR < 0.05 for interaction effect were considered to be regulated by GadE. Significant strain effects were identified for 60 genes, indicating differential expression between the wild type and ΔgadE strain (FDR < 0.1) (see Table S1 in the supplemental material). Of these, 58 genes had higher transcript levels in the ΔgadE strain, demonstrating that GadE has a negative influence on their transcription, and 2 genes had lower transcript levels in the ΔgadE strain, indicating that GadE has a positive influence on their transcription. Of the two genes with lower transcript levels in the ΔgadE strain, ECs3904 had significantly greater transcript levels in the exponential phase than in the stationary phase, and ECs2294 had greater levels in stationary phase. Among the 58 genes with higher transcript levels in the ΔgadE strain, the transcript levels of 33 were significantly higher in the exponential phase, including those of LEE genes tir, espF, and cesT, and 25 were higher in the stationary phase, including those of genes vgrE, ilvG, and treR (see Table S1 in the supplemental material). A significant interaction effect, which indicates that inactivation of gadE affects expression of genes differently at each growth phase, was identified for 122 genes, including the AFI and GAD genes gadA, gadB, and gadC (FDR < 0.05) (see Table S2 in the supplemental material). GSEA identified significant enrichment of the AFI and GAD genes (FDR < 0.05) in the wild type and of the LEE genes (FDR < 0.05) in the ΔgadE strain. In summary, the array data demonstrated that inactivation of gadE had an effect on several other genes, including members of the LEE pathogenicity island, in addition to the GAD and AFI genes.
Interaction effects of gadE inactivation and growth phase.
Expression of gadE was 84.2- ± 13.4-fold higher in the stationary phase than in the exponential phase for the wild type, as measured by Q-PCR. Since the expression of gadE is dependent on the growth phase, it affected the expression of 122 genes in a growth phase-dependent manner, leading to a significant interaction effect (FDR < 0.05) (see Table S2 in the supplemental material). Genes with a significant interaction effect included several belonging to the AFI and GAD system, as well as a number involved in energy metabolism and encoding transcriptional regulators. In the exponential phase, GadE exhibited a positive effect on the transcript levels of genes involved in energy metabolism such as cyoDC, sdhCDAB, and sucAB, while in the stationary phase, GadE exhibited a negative effect on transcript levels of these genes (see Table S2 in the supplemental material). Transcript levels of genes such as rpoS, lysR, and pspF that encode sigma factors and transcriptional regulators were slightly elevated in the ΔgadE strain in the exponential phase but were 1.3- to 2.9-fold higher in the wild type than in the ΔgadE strain in the stationary phase, indicating that these genes are positively regulated by GadE at the stationary phase (see Table S2 in the supplemental material).
Putative GadE binding site upstream of GadE-regulated genes.
To determine whether differentially expressed genes identified by the microarray analysis are regulated directly by GadE, a pattern search of the E. coli O157:H7 Sakai genome was conducted in coliBASE to identify potential GAD boxes upstream of these genes. A conserved GAD box sequence described previously in studies of E. coli K-12 (9, 32) was used for the pattern search. Typically, one GAD box is observed upstream of the GadE-regulated gadA and gadBC genes in E. coli K-12 (9). Sequences with ≥70% similarity to the query sequence were considered putative binding sites of GadE. Matching sequences were detected upstream of eight genes that were significantly differentially expressed in the ΔgadE strain. As expected, GAD box sequences preceding gadA and gadB showed 100% similarity to the conserved sequence in E. coli K-12. There were two putative GadE binding regions identified upstream of hdeD. Matching sequences were also identified in the upstream regions of vgrE and of three LEE genes (sepZ, escC, and ler) (Fig. 1).
FIG. 1.
Alignment of putative GAD box sequences. Sequences upstream of eight gadE-regulated genes with greater than 70% identity to the conserved GAD box sequence were identified by the pattern search tool in coliBASE. Letters in boldface in the table represent unmatched bases. Asterisks indicate the two matching sequences found upstream of hdeD.
Expression of AFI and GAD genes in the ΔgadE strain.
Inactivation of gadE resulted in a decrease in expression of many of the AFI and GAD genes, and the magnitude of decrease was dependent on the growth phase. Six AFI and GAD genes, including gadA, gadBC, and hdeBAD, had a significant interaction effect (Table 3). These results were verified with Q-PCR. For gadA, gadB, gadC, hdeA, and hdeB, microarray data at the exponential phase revealed higher expression in the mutant whereas Q-PCR detected higher expression in the wild type. This discrepancy could be due to the negligible expression of these genes at the exponential phase in minimal medium at neutral pH (31). It has been shown that microarrays are less sensitive than Q-PCR in detecting changes in expression when the transcript levels are low (8). At the stationary phase, Q-PCR supported the microarray results for these five genes, though a greater difference was detected in transcript levels compared to the microarray results (Table 3). This underestimation by microarrays of expression level changes has been reported in many of the previous studies (12, 61). The increase in expression of gadA and gadB in the wild type compared to that seen with the ΔgadE strain was approximately 10 to 20 times higher at the stationary phase than at the exponential phase. Similarly, the increase in expression of hdeAB in the wild type was 6 to 12 times higher at the stationary phase than at the exponential phase (Table 3). In summary, both the microarray and Q-PCR data demonstrated that the difference between the wild type and ΔgadE strain in expression of AFI and GAD genes was minimal at the exponential phase whereas at the stationary phase there was a marked decrease in expression of gadABC and hdeAB in the mutant. Moreover, in the wild type, the expression of AFI and GAD genes increased markedly from the exponential to the stationary phase, whereas in the ΔgadE strain, their expression decreased minimally or remained unchanged as the cells entered the stationary phase (see Fig. S1 in the supplemental material). This demonstrates that inactivation of gadE abrogates the growth-phase regulation of AFI and GAD genes in E. coli O157:H7.
TABLE 3.
Effect of gadE inactivation on expression of AFI and GAD genes in exponential and stationary phase
| IDa | Geneb | Function | Exponential-phase expression ratio (wild type/ΔgadE strain)
|
Stationary-phase expression ratio (wild type/ΔgadE strain)
|
||
|---|---|---|---|---|---|---|
| Microarray | Q-PCRc | Microarray | Q-PCRc | |||
| ECs4389 | hdeB | Periplasmic chaperone | 0.6 | 5.2 ± 2.0 | 2.8 | 31.4 ± 1.5 |
| ECs4390 | hdeA | Protection from organic acid metabolites | 0.3 | 3.2 ± 0.3 | 11.1 | 39.3 ± 3.0 |
| ECs4391 | hdeD | Acid resistance at high cell densities | 0.6 | 2.5 | ||
| ECs4397 | gadA | Glutamate decarboxylase isozyme | 0.4 | 2.2 ± 0.8 | 6.4 | 46.3 ± 13.7 |
| ECs2097 | gadC* | Glutamate-GABA antiporter | 0.5 | 2.3 ± 0.5 | 4.1 | 16 ± 0.9 |
| ECs2098 | gadB* | Glutamate decarboxylase isozyme | 0.4 | 2.2 ± 1.5 | 8.0 | 22.5 ± 1.67 |
Locus identification numbers (ID) for O157:H7 strain Sakai (GenBank accession no. BA000007).
Genes marked with an asterisk are GAD system genes not encoded in the AFI. Genes in boldface have putative GAD boxes upstream of the sequence.
Ratio ± SEM as determined by Q-PCR.
Six AFI genes, including gadX and gadW, two AraC-like regulators of the GAD system, did not show differential expression between the mutant and the wild type. A significant increase in the expression of yhiU, a multidrug resistance-related efflux pump gene, was observed in the mutant (see Table S1 in the supplemental material). As is consistent with previous observations (6), all of the AFI and GAD genes had significantly higher expression at the stationary phase than at the exponential phase. GSEA confirmed the enrichment of the gene set representing the AFI and GAD genes in the wild type.
GadE represses expression of LEE.
Inactivation of gadE significantly elevated the expression of 19 LEE genes independently of the growth phase; among those 19 genes, 8 showed a ≥1.35-fold increase in expression in the mutant (Table 4). Changes in expression levels between the mutant and the wild type detected by microarray and Q-PCR were highly correlated for the LEE genes (Table 4). Contrary to previous findings (52), ler, the LEE-encoded regulator, was upregulated 2.2-fold in the mutant as determined by Q-PCR (1.35-fold in the microarray). Other LEE genes such as tir and espD were upregulated 1.8- and 1.6-fold, respectively (the microarray detected 1.4-fold upregulation for both) (Table 4). Most of the LEE genes with significantly different expression between the ΔgadE strain and the wild type also had higher expression in the exponential phase compared to the stationary phase. Six LEE genes, including sepZ (espZ), had higher expression at the stationary phase, and sepZ showed a 1.4-fold (1.36-fold in the microarray) increase in expression in the mutant. Two non-LEE-encoded effectors, nleG2-2 and nleG2-3, also had a significant increase in expression in the mutant at the stationary phase. In the array data, eae, espA, and espB were not significantly upregulated, whereas Q-PCR identified an increase in expression of 1.3- to 1.6-fold in the mutant, in similarity to the results seen with other significant LEE genes (Table 4). Significant enrichment of the LEE gene set in the ΔgadE strain was detected by GSEA. To further confirm the negative influence of GadE on LEE expression, exponential-phase transcript levels of select LEE genes were measured in the ΔgadE/pCR2.1gadE complement strain, which overexpresses gadE. Expression of ler decreased 6-fold in the complement, whereas that of espD and sepZ decreased 9.9- and 9.5-fold, respectively. Altogether, these data demonstrate that GadE is a repressor of LEE genes, including ler, in E. coli O157:H7.
TABLE 4.
LEE genes significantly upregulated in the ΔgadE strain (FDR < 0.1)
| IDa | Geneb | Function | Microarray expression ratio (ΔgadE strain/wild type) | Q-PCR expression ratio (ΔgadE strain/wild type)c | Microarray expression ratio (stationary/exponential)d |
|---|---|---|---|---|---|
| ECs4550 | espF | Type III secretion system, secreted effector | 1.30 | 1.34 ± 0.08 | 0.6 |
| ECs4553 | cesD2 | Type III secretion system, chaperone | 1.36 | 0.5 | |
| ECs4554 | espB* | Type III secretion system, secreted translocator | 1.20 | 1.38 ± 0.09 | |
| ECs4555 | espD | Type III secretion system, secreted translocator | 1.38 | 1.63 ± 0.04 | |
| ECs4555 | espA* | Type III secretion system, secreted translocator | 1.10 | 1.58 ± 0.2 | |
| ECs4558 | escD | Type III secretion system, structural protein | 1.18 | 1.3 | |
| ECs4558 | eae* | Gamma intimin | 1.20 | 1.3 ± 0.1 | |
| ECs4560 | cesT | Type III secretion system, chaperone | 1.38 | 0.3 | |
| ECs4561 | tir | Translocated intimin receptor protein | 1.39 | 1.84 ± 0.06 | 0.5 |
| ECs4562 | map | Type III secretion system, secreted effector | 1.26 | ||
| ECs4563 | cesF | Type III secretion system, chaperone | 1.31 | 1.6 | |
| ECs4564 | espH | Type III secretion system, secreted effector | 1.33 | 3.1 | |
| ECs4565 | sepQ | Type III secretion system, structural protein | 1.27 | 2.9 | |
| ECs4567 | orf15 | orf of unknown function | 1.19 | 0.9 | |
| ECs4571 | sepZ | Type III secretion system, secreted effector | 1.36 | 1.44 ± 0.06 | 6.4 |
| ECs4572 | rorf8 | orf of unknown function | 1.26 | 3.0 | |
| ECs4575 | escC | Type III secretion system, structural protein | 1.21 | 0.3 | |
| ECs4584 | orf5 | orf of unknown function | 1.34 | 0.5 | |
| ECs4585 | orf4 | orf of unknown function | 1.41 | 0.6 | |
| ECs4586 | orf3 | orf of unknown function | 1.37 | 0.6 | |
| ECs4587 | cesAB | Type III secretion system, chaperone | 1.35 | 0.6 | |
| ECs4588 | ler | Type III secretion system, regulator | 1.35 | 2.23 ± 0.26 | 0.8 |
| ECs4590 | espG | Type III secretion system, secreted effector | 1.27 | 0.6 | |
| ECs1994 | nleG2-2 | Non-LEE-encoded effector | 1.26 | 2.5 | |
| ECs2156 | nleG2-3 | Non-LEE-encoded effector | 1.28 | 2.9 |
Locus identification numbers (ID) for E. coli O157:H7 strain Sakai (GenBank accession no. BA000007).
Underlined gene designations represent non-LEE-encoded effectors; genes designated with asterisks did not give significant microarray results but were detected as upregulated by Q-PCR. Genes indicated with boldface have putative GAD boxes upstream of the sequence.
Data represent exponential-phase change ± SEM as determined by Q-PCR.
Expression ratios between two growth phases = 2[log2 (stat) − log2(exp)], as determined by microarray. Ratios are reported only for genes with a significant growth phase effect (FDR < 0.05).
Repression of LEE by GadE is mediated through Ler.
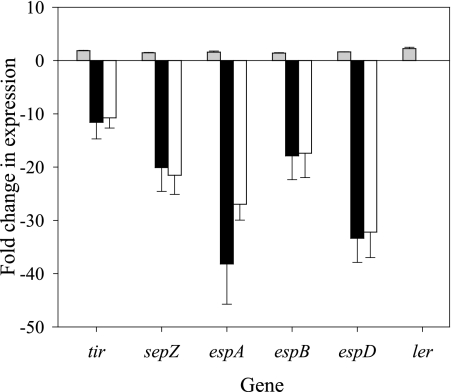
Expression data from ΔgadE strains and gadE-overexpressing strains demonstrated that GadE negatively regulates the expression of ler. Moreover, a pattern search in coliBASE to find GAD box sequences in the LEE island region revealed a putative GAD box upstream of ler (−199 to −180 bp) with six mismatches (70% identity). To determine whether repression of LEE genes by GadE is mediated by Ler, we inactivated ler in both the ΔgadE and wild-type strains and compared the expression results for select LEE genes. If GadE downregulates LEE expression independently of ler, then an increase in expression of LEE genes in the ΔgadE Δler double mutant similar to that seen with the ΔgadE strain is expected. In this study, however, expression of tir, sepZ, espA, espB, and espD decreased by 11.6-, 20.1-, 38.2-, 17.9- and 33.4-fold, respectively, in the ΔgadE Δler strain (Fig. 2). A similar decrease in LEE expression was observed for the Δler strain. These data demonstrate that the positive effect of gadE inactivation on LEE expression is reversed by ler inactivation, suggesting that ler is essential for the repression of LEE by GadE.
FIG. 2.
Exponential-phase expression of the LEE genes tir, sepZ, espA, espB, espD, and ler in the ΔgadE (gray bars), ΔgadE Δler::Km (black bars), and Δler::Km (white bars) strains compared to the wild-type results. Results shown represent average changes in expression measured by Q-PCR for at least three biological replicate experiments. Bars represent SEM.
Effect of acidic pH on expression of LEE genes.
To determine the influence of acidic pH on expression of the LEE genes, wild-type and ΔgadE cultures were grown to the exponential phase (OD600 ∼ 0.5) in EG minimal medium adjusted to pH 7.0 (control) and pH 5.0 (moderately acidic) for comparing gadE and LEE expression levels. Growth in pH 5.0 EG resulted in a 34-fold increase in the wild-type expression of gadE. Expression of three LEE genes, ler, espD, and sepZ, was downregulated in pH 5.0 EG compared to pH 7.0 EG for the wild type (Table 5). To determine whether the downregulation of LEE genes in response to moderate acidity was directed exclusively by GadE, the expression of LEE genes in the ΔgadE strain grown in pH 5.0 EG was assessed. There were 3.9- and 6.5-fold decreases in expression of espD and sepZ, respectively, in the ΔgadE strain; however, this decrease was lower than that seen with the wild type, where expression of espD and sepZ decreased by 7.5- and 9.4-fold, respectively. Interestingly, the pattern of ler expression at acidic pH was different from that seen with the other two LEE genes tested. In the wild type, there was a 6-fold decrease in ler expression, whereas in the ΔgadE strain, ler expression increased 4.5-fold at pH 5.0 (Table 5). Together, these observations indicate that downregulation of ler, the major LEE regulator, is mediated through GadE in response to moderate acid stress, whereas repression of other LEE genes under the same conditions involves additional GadE/Ler-independent factors.
TABLE 5.
Effect of pH 5.0 on expression of GAD regulators and LEE genes
| Gene | Fold change in expression (pH 5.0/pH 7.0)a
|
|
|---|---|---|
| Wild type | ΔgadE strain | |
| gadE | 34.4 ± 6.9 | No expression |
| gadX | 10.4 ± 2.5 | 4.3 ± 1.0 |
| evgA | 2.8 ± 0.3 | 3.4 ± 0.7 |
| ler | 0.16 ± 0.0 | 4.5 ± 0.4 |
| espD | 0.16 ± 0.06 | 0.26 ± 0.02 |
| sepZ | 0.12 ± 0.04 | 0.17 ± 0.05 |
Ratios greater than 1 indicate increases in expression, and ratios less than 1 indicate decreases in expression.
Two regulators that may affect the expression of LEE genes at acidic pH, independently of GadE and Ler, are GadX and EvgA, which have a negative effect on LEE expression in enteropathogenic E. coli (38, 47). Hence, we measured the expression of these regulators in pH 7.0 EG and pH 5.0 EG in the wild type and the ΔgadE strain and observed a strong induction of both genes in both strains at pH 5.0. The gadX gene exhibited more than twofold-higher induction in the wild type, whereas evgA induction results for the wild type and the ΔgadE strain were similar (Table 5).
Functional GadE is necessary for optimal performance of the three principal AR mechanisms in E. coli O157:H7.
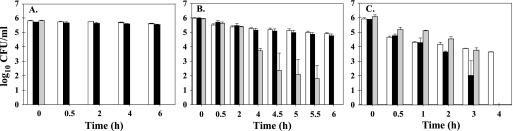
To functionally confirm the microarray data, which revealed a marked decrease in the expression of GAD and AFI genes in the ΔgadE strain, we conducted AR mechanism assays for the GAD, ARG, and OXI systems. The ARG and OXI systems were included in the study because GadE has been shown to influence their function in E. coli K-12 (31). The ΔgadE strain could not survive in the test environment for the GAD system (pH 2.0 with glutamate) for even 30 min, indicating a nonfunctional GAD system (Fig. 3A). The wild type and complement showed log reductions of 0.20 ± 0.08 and 0.17 ± 0.02 CFU/ml, respectively, after 6 h of exposure to the test environment. In the ARG system test environment (pH 2.5 with arginine), survival of the ΔgadE strain was similar to the results seen with the wild type and the complement for up to 2 h. However, at 4 h there was reduction in viable cell numbers and the mutant showed a high variation in cell numbers up to 5.5 h, and by 6 h, no viable mutants were recovered. The wild type and complement showed log reductions of 1.07 ± 0.08 and 1.22 ± 0.16 CFU/ml, respectively, after 6 h (Fig. 3B). The OXI system was less effective than the ARG system in protecting all three strains. The mutant survived for only 3 h at pH 2.5, and the log reduction after 4 h was 2.27 ± 0.1 CFU/ml for the wild type (Fig. 3C). Interestingly, the complement also did not survive after 3 h, indicating that gadE in trans does not reconstitute the phenotype for the OXI system. It is possible that flooding the cell with multiple copies of gadE, as performed in the complement experiments, adversely affected the functioning of the OXI system. These findings demonstrate that inactivation of gadE abolished the functioning of the GAD system and rendered the ARG and OXI systems less effective in protecting the cells against low pH.
FIG. 3.
AR mechanism assays. Survival of the wild-type (white bars) and ΔgadE (gray bars) and ΔgadE/pCR2.1gadE (black bars) strains in the three AR systems. (A) Survival in the GAD system test system at pH 2.0. (B) Survival in the ARG test system at pH 2.5. (C) Survival in the OXI test system at pH 2.5. The results represent average CFU per milliliter from three experiments for each AR system. Bars represent SEM.
Survival of the ΔgadE strain in a simulated gastric environment.
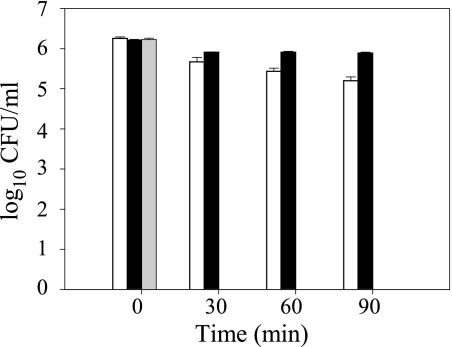
Since the three principal AR systems were defective in protecting the ΔgadE strain from acidic stress in defined minimal test conditions, the ability of the ΔgadE strain to survive in a complex acidic environment was assessed using the MSS (pH 2.5). The wild-type and complemented cells showed average log reductions of 1.05 ± 0.06 and 0.32 ± 0.04 CFU/ml, respectively, after 1.5 h in the MSS (Fig. 4). Viable cells could not be recovered from the MSS inoculated with the ΔgadE strain, which indicates that functional GadE is necessary for survival in the simulated gastric environment.
FIG. 4.
Survival of the wild-type (white bars) and ΔgadE (gray bars) and ΔgadE/pCR2.1gadE (black bars) strains in the MSS. The average log CFU per milliliter of the results from three experiments is plotted for each time point. Bars represent SEM.
DISCUSSION
Although the upstream regulatory circuits and downstream effects of GadE in nonpathogenic E. coli strains have been examined (18, 31), little is known about GadE and its role in AR and virulence among pathogenic E. coli strains. Here, the role of the GadE regulator in the AR and virulence of E. coli O157:H7 was investigated by constructing an isogenic ΔgadE strain and comparing its expression profiles with that of the wild-type strain. Our findings demonstrate that besides being a positive regulator of GAD and many AFI genes, GadE acts as a negative regulator of the LEE pathogenicity island, an important factor in the virulence of E. coli O157:H7. GadE, along with additional regulators, is involved in the downregulation of LEE expression at moderately acidic pH. In addition, the characterization of AR phenotypes of the ΔgadE strain revealed that GadE is indispensable for a functional GAD system and plays a vital role in the survival of E. coli O157:H7 in a simulated gastric environment.
In this study, the microarray data demonstrated that inactivation of gadE in E. coli O157:H7 altered expression of 60 genes independently of growth phase and of 122 genes in a growth phase-dependent manner. The genes with altered expression included both AR and virulence genes, indicating that the regulatory function of GadE is not restricted to AR but has a more global effect on the transcriptome of E. coli O157:H7. Overexpression of gadE in nonpathogenic E. coli was shown to affect the expression of ∼40 genes, including GAD genes (21). Most of these genes, however, differed from those identified following inactivation of gadE in O157:H7, suggesting that apart from its effect on the GAD system, GadE has additional regulatory functions in E. coli O157:H7.
Expression of gadA, gadB, and gadC was not completely abolished in E. coli O157:H7 ΔgadE, which was a result similar to that seen with E. coli K-12, where minimal expression of GadAB proteins in the ΔgadE strain was observed (31). Expression of GAD system components in the absence of GadE could be induced by the GadX regulator, which has been shown to bind to and activate gadA and gadBC transcription directly under in vitro conditions but not during in vivo growth (46, 55, 57). Another interesting observation was that at the stationary phase, the magnitude of increase in expression of gadA and gadB in the wild type was different from that seen with the ΔgadE strain, indicating that the inactivation of gadE affects these duplicated genes in distinct ways. This corroborates the recent finding that the sequences of gadA and gadB are divergent in E. coli O157:H7, in contrast to the results seen with other E. coli strains, where gene conversion events between gadA and gadB have led to genetic homogenization (4). In contrast to the wild-type results, no increase in the expression of GAD genes in the ΔgadE strain as the cells entered the stationary phase was observed, demonstrating that GadE is required for the growth phase regulation of GAD genes.
This study demonstrates that gadE inactivation has differential effects on the expression of AFI genes in E. coli O157:H7. In nonpathogenic E. coli strains, gadE induces the expression of AFI genes such as hdeB, hdeA, hdeD, gadX, and yhiF in addition to gadA (21, 33, 45). In this study, expression of gadA and hdeBAD in the ΔgadE strain showed growth phase-dependent downregulation. However, gadX and yhiF were not differentially expressed in the ΔgadE strain, indicating that at pH 7.0, loss of gadE does not influence the expression of these two genes in E. coli O157:H7. These differences between O157:H7 and nonpathogenic strains could also be due to the differences in growth conditions, since the strains were grown in rich or minimal media at acidic pH in most of the previous studies.
The relationship between AR and virulence of pathogenic E. coli, particularly the interactions between the GAD system and LEE, remains poorly defined. A few studies in the past have shown that some of the GAD system regulators negatively affect LEE expression (11, 38, 47, 53). In E. coli O157:H7 Sakai, gadE inactivation was found to increase the expression of LEE-encoded espB, espD, and tir genes but not that of ler (52). Hence, Mellies et al. considered that GadE-mediated downregulation of LEE was independent of Ler and that the pathway through which GadE affects LEE was undetermined (35). Moreover, the extent to which GadE inhibits the transcription of LEE genes has not been demonstrated quantitatively before. The data presented here demonstrate that GadE has a global effect on LEE genes: GadE influences the expression of at least 19 LEE-encoded genes belonging to all five LEE operons and of 2 non-LEE-encoded effectors. These data also provide insight about the mechanism underlying GadE-mediated LEE downregulation. In contrast to a previous study (52), there was a significant increase in the expression of ler in the ΔgadE strain which may have been due to differences in growth medium used. The previous study used Dulbecco's modified Eagle's medium containing glycerol (52), whereas in this study MOPS minimal medium was used for growing the cells. This discrepancy could also be due to differences in the sensitivity of the assays used (Northern blotting versus Q-PCR and microarray). The negative correlation between expression of gadE and that of ler was marked in the gadE-overexpressing strain, in which ler expression was substantially downregulated. Furthermore, the identification of a putative GAD box sequence upstream of ler with 70% identity to the conserved GAD box sequence provides additional evidence of direct regulation, as GadE has been shown to bind to box sequences with as low as 60% identity to the conserved sequence (32). Additionally, inactivation of ler in the ΔgadE strain led to a marked decrease in LEE expression, confirming that ler is essential for the upregulation of LEE in the ΔgadE strain. Taken together, these findings illustrate that GadE indirectly downregulates LEE expression, most likely through downregulation of Ler. However, additional putative GAD boxes were observed upstream of two other LEE genes, sepZ and escC; therefore, it is possible that GadE directly regulates these LEE genes independently of Ler.
Because GadE negatively influences LEE expression, we hypothesized that environmental conditions that induce gadE may downregulate the expression of LEE. Two conditions that lead to induction of gadE are entry into the stationary phase and acidic pH (18). Stationary-phase expression of LEE genes has been described previously (6). Similarly, the influence of several environmental factors, such as temperature, bicarbonate ion concentration, and membrane stress, on the expression of LEE has been investigated (2, 39, 53, 58). However, the effect of pH on LEE expression and the factors regulating that effect in EHEC remain largely unknown. Our experiments demonstrated that exposure to moderately acidic pH strongly induces gadE and has a substantial negative effect on LEE expression in E. coli O157:H7. This inhibitory effect could be more profound in extreme acidic conditions such as the gastric environment. To determine whether the acidic pH-induced downregulation of LEE is exclusively regulated by GadE, we measured the expression of LEE genes in the ΔgadE strain following growth at pH 5.0. Except for ler, a partial downregulation at acidic pH was observed in the expression of the LEE genes in the ΔgadE strain, indicating that GadE is not the only regulator responsible for the pH-induced downregulation of LEE. This partial downregulation of LEE is not mediated through Ler, as expression of ler was increased in the ΔgadE strain at pH 5.0. To increase our understanding of this phenomenon, we analyzed the expression of other AR regulators that could act on LEE independently of GadE and found that gadX and evgA were also strongly induced at acidic pH in both the wild-type and ΔgadE strains. GadX has been shown to negatively regulate the expression of LEE genes through Per, the plasmid-encoded regulator, in enteropathogenic E. coli (47). However, the effect of GadX on expression of LEE in EHEC has not been determined. It is possible that GadX regulates LEE through an unknown regulator in EHEC. The decrease in LEE gene expression in an acidic environment in the ΔgadE strain is likely also to be mediated by EvgA. Previously, EvgA had been shown to repress LEE, independently of Ler, by activating ydeO and ydeP (38). Because YdeO is a positive regulator of gadE, the partial decrease in LEE expression in the ΔgadE strain may occur through YdeP. Collectively, these experiments suggest that GadE, GadX, and EvgA may cooperatively repress the expression of LEE genes at acidic pH and that GadE is the sole regulator responsible for the changes in expression of ler in the acidic conditions used in this study.
The AR of the ΔgadE strain also was characterized by assessing survival at pH 2.0 and 2.5 in the minimal AR mechanism environments and in the complex acidic conditions of the MSS. The ΔgadE strain failed to survive the acid challenge at pH 2.0 in medium that included glutamate, indicating a lack of a functional GAD system. Inactivation of gadE in E. coli O157:H7 negatively impacted the protective ability of the ARG and OXI AR systems. The survival of the ΔgadE strain was tested in the MSS, which evaluates the ability of bacterial strains to survive in a gastric environment after ingestion of food (25). The GadE central regulator, and thus a functional GAD system, is a critical component for survival, as inactivation of gadE abrogated the ability of E. coli O157:H7 to survive in the MSS. Hence, GadE most likely plays a protective role during the passage of O157 through the gastric environment. This assumption is supported by a previous study by Price et al. (42) which demonstrated that gadC is required for the survival of E. coli O157:H7 in calves.
In summary, this study shows that GadE is an important regulator that modulates the expression of AR and virulence genes in E. coli O157:H7 in response to environmental conditions similar to those that are found in various food matrices and the human gastrointestinal tract. GadE has acquired additional functions in E. coli O157:H7 and acts as a link between AR and virulence: it activates the GAD system of AR and at the same time downregulates the expression of LEE genes, which are important for the adhesion of the organism to intestinal mucosa and development of attaching and effacing lesions. Consequently, we propose that, during passage through the human stomach, GadE protects E. coli O157:H7 by inducing the GAD system and aids in energy conservation by inhibiting the unnecessary expression of the LEE genes. As the organism reaches the intestine, environmental changes, including alkaline pH and a high NaHCO3 concentration, induce Ler, the LEE regulator, which negatively regulates expression of gadE (1), leading to inhibition of the GAD system.
Supplementary Material
Acknowledgments
This work was supported by Food Safety National Research Initiative 2005-35201-16362 from the U.S. Department of Agriculture and in part by funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), and the Department of Health and Human Services via the Food and Waterborne Diseases Integrated Research Network (NIH research contract N01-AI-30058).
We thank Vanessa Sperandio, University of Texas South Western Medical Center, for helpful suggestions for creating genetic complements and Galeb Abu-Ali, James T. Riordan, and Shannon Manning for critical review of the manuscript.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abe, H., A. Miyahara, T. Oshima, K. Tashiro, Y. Ogura, S. Kuhara, N. Ogasawara, T. Hayashi, and T. Tobe. 2008. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 1525-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, H., I. Tatsuno, T. Tobe, A. Okutani, and C. Sasakawa. 2002. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 703500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alignan, M., T. Hewezi, M. Petitprez, G. Dechamp-Guillaume, and L. Gentzbittel. 2006. A cDNA microarray approach to decipher sunflower (Helianthus annuus) responses to the necrotrophic fungus Phoma macdonaldii. New Phytol. 170523-536. [DOI] [PubMed] [Google Scholar]
- 4.Bergholz, T. M., C. L. Tarr, L. M. Christensen, D. J. Betting, and T. S. Whittam. 2007. Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. Mol. Biol. Evol. 242323-2333. [DOI] [PubMed] [Google Scholar]
- 5.Bergholz, T. M., and T. S. Whittam. 2007. Variation in acid resistance among enterohaemorrhagic Escherichia coli in a simulated gastric environment. J. Appl. Microbiol. 102352-362. [DOI] [PubMed] [Google Scholar]
- 6.Bergholz, T. M., L. M. Wick, W. Qi, J. T. Riordan, L. M. Ouellette, and T. S. Whittam. 2007. Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol. 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhagwat, A. A., R. P. Phadke, D. Wheeler, S. Kalantre, M. Gudipati, and M. Bhagwat. 2003. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J. Microbiol. Methods 55399-409. [DOI] [PubMed] [Google Scholar]
- 8.Canales, R. D., Y. Luo, J. C. Willey, B. Austermiller, C. C. Barbacioru, C. Boysen, K. Hunkapiller, R. V. Jensen, C. R. Knight, K. Y. Lee, Y. Ma, B. Maqsodi, A. Papallo, E. H. Peters, K. Poulter, P. L. Ruppel, R. R. Samaha, L. Shi, W. Yang, L. Zhang, and F. M. Goodsaid. 2006. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 241115-1122. [DOI] [PubMed] [Google Scholar]
- 9.Castanie-Cornet, M.-P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147709-715. [DOI] [PubMed] [Google Scholar]
- 10.Castanie-Cornet, M.-P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 1813525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castanié-Cornet, M.-P., H. Treffandier, A. Francez-Charlot, C. Gutierrez, and K. Cam. 2007. The glutamate-dependent acid resistance system in Escherichia coli: essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology 153238-246. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., J. Lozach, E. W. Garcia, B. Barnes, S. Luo, I. Mikoulitch, L. Zhou, G. Schroth, and J.-B. Fan. 25 June 2008. Highly sensitive and specific microRNA expression profiling using BeadArray technology. Nucleic Acids Res. 36e87. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, S. H., D. J. Baumler, and C. W. Kaspar. 2000. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 663911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, X., and G. A. Churchill. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui, X., J. T. Hwang, J. Qiu, N. J. Blades, and G. A. Churchill. 2005. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 659-75. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 1013597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2898-907. [DOI] [PubMed] [Google Scholar]
- 19.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 732573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C.-G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 21.Hommais, F., E. Krin, J.-Y. Coppee, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 15061-72. [DOI] [PubMed] [Google Scholar]
- 22.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J.-P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 4020-36. [DOI] [PubMed] [Google Scholar]
- 23.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 1502357-2571. [DOI] [PubMed] [Google Scholar]
- 24.Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel, and G. Gibson. 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29389-395. [DOI] [PubMed] [Google Scholar]
- 25.Just, J. R., and M. A. Daeschel. 2003. Antimicrobial effects of wine on Escherichia coli O157:H7 and Salmonella typhimurium in a model stomach system. J. Food Sci. 68285-290. [Google Scholar]
- 26.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295405-418. [DOI] [PubMed] [Google Scholar]
- 27.Large, T. M., S. T. Walk, and T. S. Whittam. 2005. Variation in acid resistance among Shiga toxin-producing clones of pathogenic Escherichia coli. Appl. Environ. Microbiol. 712493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law, D. 2000. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J. Appl. Microbiol. 88729-745. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., C. Wood, T. Getchell, M. Getchell, and A. Stromberg. 2004. Analysis of oligonucleotide array experiments with repeated measures using mixed models. BMC Bioinformatics 5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J., M. Smith, K. Chapin, H. Baik, G. Bennett, and J. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 623094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 491309-1320. [DOI] [PubMed] [Google Scholar]
- 32.Ma, Z., N. Masuda, and J. W. Foster. 2004. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 1867378-7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mates, A. K., A. K. Sayed, and J. W. Foster. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 1892759-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDaniel, T., K. Jarvis, M. Donnenberg, and J. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 921664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mellies, J. L., A. M. S. Barron, and A. M. Carmona. 2007. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 754199-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michino, H., K. Araki, S. Minami, S. Takaya, N. Sakai, M. Miyazaki, A. Ono, and H. Yanagawa. 1999. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150787-796. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, K., and K. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nadler, C., Y. Shifrin, S. Nov, S. Kobi, and I. Rosenshine. 2006. Characterization of enteropathogenic Escherichia coli mutants that fail to disrupt host cell spreading and attachment to substratum. Infect. Immun. 74839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi, N., H. Abe, Y. Ogura, T. Hayashi, K. Tashiro, S. Kuhara, N. Sugimoto, and T. Tobe. 2006. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 61194-205. [DOI] [PubMed] [Google Scholar]
- 40.Peterson, W. L., P. A. Mackowiak, C. C. Barnett, M. Marling-Cason, and M. L. Haley. 1989. The human gastric bactericidal barrier: mechanisms of action, relative antibacterial activity, and dietary influences. J. Infect. Dis. 159979-983. [DOI] [PubMed] [Google Scholar]
- 41.Pfaffl, M. W. 1 May 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, S. B., J. C. Wright, F. J. DeGraves, M.-P. Castanie-Cornet, and J. W. Foster. 2004. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl. Environ. Microbiol. 704792-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 40664-67. [DOI] [PubMed] [Google Scholar]
- 44.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132365-386. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz, C., L. M. McMurry, and S. B. Levy. 2008. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J. Bacteriol. 1901290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayed, A. K., C. Odom, and J. W. Foster. 2007. The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology 1532584-2592. [DOI] [PubMed] [Google Scholar]
- 47.Shin, S., M.-P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 411133-1150. [DOI] [PubMed] [Google Scholar]
- 48.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 1745820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarr, P. I., C. A. Gordon, and W. L. Chandler. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 3651073-1086. [DOI] [PubMed] [Google Scholar]
- 52.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 712598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobe, T., H. Ando, H. Ishikawa, H. Abe, K. Tashiro, T. Hayashi, S. Kuhara, and N. Sugimoto. 2005. Dual regulatory pathways integrating the RcsC-RcsD-RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity. Mol. Microbiol. 58320-333. [DOI] [PubMed] [Google Scholar]
- 54.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 10314941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tramonti, A., M. De Canio, I. Delany, V. Scarlato, and D. De Biase. 2006. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 1888118-8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 1846551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 1853190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 1482735-2744. [DOI] [PubMed] [Google Scholar]
- 59.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 1871591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 1871783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 15 May 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30e48. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.