Abstract
Streptococcus agalactiae (group B Streptococcus [GBS]) remains a leading cause of invasive infections in neonates and has emerged as a pathogen of the immunocompromised and elderly populations. The virulence mechanisms of GBS are relatively understudied and are still poorly understood. Previous evidence indicated that the GBS cspA gene is necessary for full virulence and the cleavage of fibrinogen. The predicted cspA product displays homology to members of the extracellular cell envelope protease family. CXC chemokines, many of which can recruit neutrophils to sites of infection, are important signaling peptides of the immune system. In this study, we purified CspA and demonstrated that it readily cleaved the CXC chemokines GRO-α, GRO-β, GRO-γ, neutrophil-activating peptide 2 (NAP-2), and granulocyte chemotactic protein 2 (GCP-2) but did not cleave interleukin-8. CspA did not cleave a panel of other test substrates, suggesting that it possesses a certain degree of specificity. CXC chemokines also underwent cleavage by whole GBS cells in a cspA-dependent manner. CspA abolished the abilities of three representative CXC chemokines, GRO-γ, NAP-2, and GCP-2, to attract and activate neutrophils. Genetic and biochemical evidence indicated that CspA is a serine protease with S575 at its active site. D180 was also implicated as part of the signature serine protease catalytic triad, and both S575 and D180 were required for both N-terminal and C-terminal autocatalytic processing of CspA.
Streptococcus agalactiae (group B Streptococcus [GBS]) is one of the most common causes of invasive infections in human neonates. The principle clinical manifestations of GBS infections in neonates include pneumonia, sepsis, and meningitis. Recently, GBS has emerged as an increasingly common cause of infections in elderly or immunocompromised nonpregnant adults (1, 18). A common theme underlying GBS pathogenesis involves the ability of the organism to evade phagocytic cells, a key host defense mechanism against the bacterium. Early studies demonstrated a delay in the influx of neutrophils to infection sites (22); this delay may give GBS an opportunity to replicate to high densities and subsequently overwhelm the host defense.
Several virulence factors from streptococci belong to the multidomain cell envelope protease (CEP) family, a diverse family of extracellular proteases that also includes caseinases from lactococcal species (4, 8, 13, 14, 24, 25). The prototype of streptococcal CEPs is the C5a peptidase, which specifically cleaves the neutrophil chemotactic factor C5a (2-4). The crystal structure of the GBS C5a peptidase has been reported, shedding new light on the structure and function of this important CEP (4). A novel CEP (SpyCEP, also known as ScpC) produced by Streptococcus pyogenes (group A Streptococcus [GAS]) is an important virulence factor that has the ability to proteolyse many human and murine CXC chemokines, including interleukin-8 (IL-8) (8, 14, 27, 29). This serine protease allows GAS to evade the immune system by disrupting the abilities of chemokines to stimulate the activation and chemotaxis of neutrophils (8) and diminishing the formation of neutrophil extracellular traps (29). In relation to noninvasive isolates, invasive GAS isolates produce high levels of SpyCEP/ScpC, and this protease has been implicated in necrotizing fasciitis (8). A Streptococcus iniae homolog of SpyCEP/ScpC (CepI) has recently been identified; it also cleaves IL-8 and contributes to virulence (29).
Harris et al. described a putative GBS CEP encoded by the cspA gene (13). The inactivation of cspA decreased GBS virulence in a neonatal rat model of sepsis and diminished the capacity of GBS to resist opsonophagocytic killing by neutrophils. The cspA mutant, in contrast to the wild-type (wt) strain, was unable to cleave fibrinogen. This study provided strong evidence that cspA encodes a protease that can cleave fibrinogen.
Here, we have purified CspA and examined its biochemical properties. Our findings revealed that in addition to cleaving fibrinogen, CspA cleaves and inactivates a number of CXC chemokines that act on neutrophils. We have also identified the putative catalytic residues of CspA and assessed their role in the processing of the protease.
MATERIALS AND METHODS
Chemicals, growth media, and peptide reagents.
Chemical reagents were purchased from Sigma-Aldrich, unless otherwise noted. Recombinant human chemokines were obtained from Peprotech. Escherichia coli was grown in Luria-Bertani broth (Becton and Dickinson). GBS was grown in Todd-Hewitt broth; Lactococcus lactis was grown in M17 medium (Becton and Dickinson) for routine purposes and in M9CAYEE (10) for protein production (23).
Cloning methodology.
The cspA gene was previously cloned and expressed in L. lactis strain MG1363 (see Table 1 for a description of strains); the cspA allele utilized in the expression system is engineered to lack the region encoding the putative cell wall anchor in order to facilitate the isolation of the encoded protein from culture supernatants (23). Mutated cspA alleles were constructed with the QuikChange site-directed mutagenesis kit as recommended by the manufacturer (Stratagene). Plasmid pJB101 (23) (see Table 1 for a description of plasmids) was used as a template for PCR with the oligonucleotides 5′GATATGATGAGTGGGACAGCTATGGCTTCTCCCCATGTCGCTGG3′ and 5′CCAGCGACATGGGGAGAAGCCATAGCTGTCCCACTCATCATATC3′ to generate a cspA allele encoding the S575A variant (pJB103) and the oligonucleotides 5′GGAACTGTTGTAGCAATTATTGCCTCAGGACTAGATACCAATCAC3′ and 5′GTGATTGGTATCTAGTCCTGAGGCAATAATTGCTACAACAGTTCC3′ to generate a cspA allele encoding the D180A variant (pJB104). LA Taq polymerase (Takara) was utilized in the reactions. The E. coli CopyCutter strain (Epicentre) was transformed with pJB103 and pJB104, resulting in E. coli strains JDB1 and JDB2, respectively. The pJB103 and pJB104 inserts were sequenced to ensure that the desired mutations were present and that no spurious mutations were introduced during PCR amplification. All DNA sequencing was performed at the Arizona State University sequencing facility. These cspA-bearing inserts were liberated by treatment with NcoI and SphI and ligated to NcoI/SphI-cut pMSP3545, generating pJB105 (S575A variant) and pJB106 (D180A variant). L. lactis strain MG1363 (11) was transformed with pJB105 and pJB106, resulting in strains DSL103 (S575A variant; pJB105) and DSL104 (D180A variant; pJB106). pJB107 was constructed by digesting pJB103 with HindIII and ligating the liberated 1.4-kb insert to similarly cut pHY304, a derivative of pVE6007 (15) with an erythromycin resistance gene.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| COH1 | wt clinical isolate | 17 |
| DS107 | COH1 with cspA(S575A) allele | This work |
| DS108 | COH1 harboring pJB107 | This work |
| TOH121 | COH1 bearing cspA::erm allele | 13 |
| L. lactis | ||
| MG1363 | L. lactis host for pMSP3545 | 11 |
| DSL102 | MG1363 harboring pJB102 (wt CspA) | 23 |
| DSL103 | MG1363 harboring pJB105 (S575A CspA) | This work |
| DSL104 | MG1363 harboring pJB106 (D180A CspA) | This work |
| E. coli | ||
| CopyCutter | Used for maintenance of cspA-bearing plasmids | Epicentre |
| JDB1 | CopyCutter harboring pJB103 | This work |
| JDB2 | CopyCutter harboring pJB104 | This work |
| Plasmids | ||
| pCR-XL-TOPO | Vector for cloning PCR products | Stratagene |
| pJB101 | pCR-XL-TOPO bearing wt cspA allele | 23 |
| pJB103 | pCR-XL-TOPO bearing cspA(S575A) allele | This work |
| pJB104 | pCR-XL-TOPO bearing cspA(D180A) allele | This work |
| pMSP3545 | Vector for nisin-inducible gene expression | 5 |
| pJB102 | pMSP3545 with wt cspA allele | 23 |
| pJB105 | pMSP3545 bearing cspA(S575A) allele | This work |
| pJB106 | pMSP3545 bearing cspA(D180A) allele | This work |
| pHY304 | Temp-sensitive vector used for allelic replacements; derivative of pVE6007 (15); Ermr | Craig Rubens |
| pJB107 | pHY304 with cspA(S575A) allele | This work |
Allelic exchange mutagenesis for GBS cspA.
Allelic exchange was conducted in a manner similar to that used previously to generate nonpolar mutations in the GBS capsule operon (6). The wt cspA chromosomal allele was replaced with the allele encoding the S575A variant of CspA. Plasmid pJB107 was introduced into GBS strain COH1 by electroporation, and the resulting strain (DS108) was cultured at 30°C in the presence of erythromycin. Cells harboring pJB107 were shifted to 37°C in the presence of erythromycin to select for the chromosomal integration of pJB107. Integration was confirmed by the isolation of chromosomal DNA and subsequent PCR. A single isolate with the integrated plasmid was chosen and passaged five times in the absence of erythromycin at 30°C. The resulting strain was then cured of the plasmid by passage at 37°C. Colonies sensitive to erythromycin were then tested for their abilities to degrade human fibrinogen. A colony that lacked the ability to degrade fibrinogen was identified; chromosomal DNA was isolated from this colony and used as a template for PCR, and the presence of the desired mutation was verified by DNA sequencing. The resulting strain was designated DS107.
Heterologous expression of CspA and variants.
CspA production in L. lactis has been described previously and utilizes a nisin-inducible expression system and a cspA allele bearing a deletion of the region that encodes the putative cell wall anchor. Briefly, 100 μg/ml nisin was added to DSL102, DSL103, and DSL104 cultures to induce the production of the wt and variant CspA proteins. L. lactis secreted CspA into the supernatants, which were harvested and concentrated.
N-terminal sequencing and MALDI-TOF.
Lactococcal culture supernatants from strains DSL102, DSL103, and DSL104 were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroblotted onto polyvinylidene difluoride membranes using 1× CAPS [3-(cyclohexylamino)-1-propanesulfonic acid]-10% methanol buffer, and stained with Coomassie blue. The bands corresponding to CspA were excised, and N-terminal sequencing was performed at the Columbia University protein core facility. In preparation for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of CspA, SDS-PAGE analyses of samples were conducted; the gels were stained with Coomassie blue, and the CspA-containing gel slices were excised and sent to the Columbia University protein core facility. The masses of peptides identified by MALDI-TOF were compared to peptide masses predicted from the published CspA sequence for identification.
Purification of CspA.
CspA-containing culture supernatant was concentrated 500-fold using a stirred ultrafiltration cell (Amicon) and a regenerated cellulose membrane with a 50-kDa cutoff. This concentrated medium was then diluted 10-fold in 25 mM MOPS (morpholinepropanesulfonic acid; pH 7.4)-10% glycerol (buffer A). Chromatography was performed using a ceramic hydroxyapatite type I 80-μm-particle-size column (2.5 cm in diameter by 7.6 cm in height; Bio-Rad) equilibrated with buffer A and coupled to a BioLogic LP low-pressure liquid chromatography system (Bio-Rad). The ceramic hydroxyapatite column was then washed with 25 mM MOPS (pH 7.4)-10% glycerol-1 M NaCl. CspA was eluted from the column with a 30-column-volume linear gradient of 0 to 100% 500 mM phosphate in buffer A. CspA eluted at approximately 310 mM phosphate. The separation of CspA from a peak of UV-absorbing, nonproteinaceous material was apparent based on inspection of the chromatogram. CspA-containing fractions were identified using fibrinogen cleavage activity assays and SDS-PAGE, pooled, and concentrated with a 50-kDa-cutoff centrifugal ultrafiltration device (Amicon). This material was diluted 10-fold in 20 mM Tris (pH 7.4)-10% glycerol (buffer C). Chromatography was performed using a Mono Q 4.6/100PE anion exchange column (GE Healthcare) coupled to an AKTApurifier high-performance liquid chromatography system (GE Healthcare). CspA was eluted from the column with a 40-column-volume linear gradient of 0 to 1 M NaCl in buffer C. CspA-containing fractions were identified using activity assays and SDS-PAGE. Again, the separation of CspA from media containing nonproteinaceous, UV-absorbing material was apparent from the examination of the chromatogram, SDS-PAGE, and UV absorbance during the chromatography. CspA-containing fractions were pooled and concentrated 10-fold, giving a final volume of 1 ml. One unit of CspA was defined as the smallest amount of CspA having the activity required to cleave 1 pmol of the fibrinogen α subunit in 1 min. The yield of the procedure was 0.33 mg of CspA from 1 liter of supernatant harvested from an L. lactis culture (final optical density at 600 nm [OD600] of 1.5). The final specific activity of purified CspA was equivalent to the cleavage of 92.1 mol of the fibrinogen α subunit per μg of protein. The purification resulted in a 3.34-fold overall increase in CspA specific activity and 26% recovery of CspA activity. UV-absorbing material from culture medium supernatants that was not observable on SDS-PAGE gels was also eliminated during the purification process (data not shown).
Protein, proteolysis, and peptidase assays.
Total protein was assayed using the Bio-Rad dye-binding assay. For proteolysis/peptidase assays using purified CspA, 54 U of purified CspA was added to 2 μg of recombinant human chemokine (Peprotech) in a total volume of 10 μl. This mixture was incubated for 1 h at 37°C. The entire reaction mixture was then analyzed by Tris-Tricine-SDS-PAGE (13.5 or 20% acrylamide) (21). For the proteolysis of fibrinogen, 7 μg of purified human fibrinogen (Enzyme Research Laboratories) was mixed with 14 U of purified protease in a total volume of 10 μl. This mixture was incubated at 37°C for 3 min and then analyzed by SDS-PAGE (10% acrylamide).
For proteolysis/peptidase assays using whole GBS cells, a 50-ml sample from a GBS culture was cultivated in 80% Todd-Hewitt broth-20% fetal bovine serum (HyClone) to an OD600 of 0.3. Cells were then harvested by centrifugation, washed twice in phosphate-buffered saline (PBS; Gibco), and resuspended in 50 μl of PBS. Five micrograms of recombinant human chemokine (Peprotech) and 50 μl of a GBS suspension in PBS were incubated for 16 h at 37°C with mixing. GBS cells were then removed by centrifugation. Twenty microliters of supernatant was analyzed by Tris-Tricine-PAGE (13.5% acrylamide) and Coomassie blue staining. The proteolysis of fibrinogen was conducted as described previously (23).
Proteolysis/peptidase assays with CspA-containing lactococcal media were conducted as described previously (23), except that in place of fibrinogen, 2 μl of concentrated medium was incubated with 2 μg of chemokine for 1 h. Tris-Tricine-PAGE (13.5% acrylamide) was conducted as described above. Fibrinogen and CspA self-proteolysis assays were conducted as described previously (23).
Isolation of human neutrophils.
Blood was collected from healthy donors after informed consent as approved by the instructional review board at Louisiana State University Health Sciences Center in Shreveport. The freshly collected blood was added to a heparin-dextran-NaCl solution. Erythrocytes were allowed to sediment for 1 h at 25°C. The neutrophil-rich top layer was extracted and placed on ice. Cells were then centrifuged at low speed and resuspended in a cold hypotonic solution for 30 s to lyse any remaining erythrocytes. The solution was then immediately made isotonic. Cells were harvested by centrifugation again, washed twice, and resuspended in Eagle's minimal essential medium (Cambrex). The number of recovered neutrophils was determined by staining a sample of recovered Eagle's minimal essential medium with trypan blue and counting with a hemacytometer.
Chemotaxis assays.
Recombinant human CXC chemokines (Peprotech) were either treated with purified CspA or mock treated and added to the bottom well of a QCM 3-μm cell migration assay kit (Chemicon International). Aliquots of freshly isolated human neutrophils (105) were added to the top wells. The assay plates were incubated for 1 h at 37°C and 5% CO2 in a humidified cell culture incubator (Forma). Neutrophils in the bottom chamber were lysed by adding lysis-fluorescence buffer as directed by the manufacturer (Chemicon International). Fluorescence was quantified with a 96-well plate reader using a 480- and 520-nm filter set (BMG Labtech). The number of migrated neutrophils was determined by using a standard curve. Three technical replicates (three wells) for each chemokine were examined, and each experiment was performed at least three times.
Neutrophil activation assays.
Neutrophil activation was quantified by elastase release assays. Cytochalasin B (Sigma-Aldrich) was added to freshly isolated human neutrophils to a concentration of 5 μg/ml, and the mixture was incubated at 25°C for 10 min. The neutrophils were then added to a mixture of either mock-treated or CspA-treated CXC chemokines and the colorimetric elastase substrate N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide (Sigma-Aldrich). Absorbance at 405 nm was monitored by a 96-well plate reader (BMG Labtech). Three technical replicates for each chemokine were performed, and each experiment was repeated at least three times.
RESULTS
Purification of CspA.
We recently reported the development of a system for expressing recombinant CspA in a functional form by using L. lactis. The system utilizes a recombinant allele of cspA lacking the region that encodes the putative cell wall anchor and employs a nisin-inducible promoter (23). Concentrated culture supernatant harvested from the strain expressing the recombinant cspA allele was free of contaminants visible on Coomassie blue-stained SDS-PAGE gels (23) (Fig. 1A) and was used as starting material for the purification of CspA.
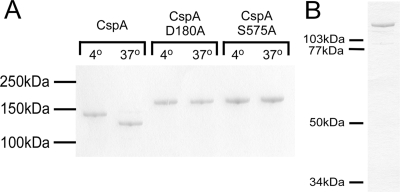
FIG. 1.
(A) Putative catalytic residues are necessary for autocatalytic cleavage of CspA. Equivalent amounts of media from lactococcal strains expressing wt CspA, D180A CspA, and S575A CspA were incubated at 4 or 37°C for 5 h as described previously (23). The reaction mixtures were subjected to SDS-PAGE and subsequent Coomassie blue staining. (B) Purified CspA. Heterologously expressed CspA was purified from L. lactis medium supernatants by hydroxyapatite chromatography followed by ion exchange chromatography.
Contaminants not visible on SDS gels (i.e., medium components) were removed via hydroxyapatite and anion exchange chromatography. The recovery of catalytic activity was assessed by fibrinogen cleavage assays. CspA with a 3.3-fold increase in specific activity was obtained, and 26% of activity was recovered. The purified protein was free of detectable contaminants (Fig. 1B). As described below, purified CspA exhibited the same catalytic activities as did whole GBS cells and CspA-enriched L. lactis media.
Residues in the predicted active site of CspA are necessary for catalytic activity.
The catalytic triad of serine proteases (His, Asp, and Ser), a distinguishing characteristic of proteins in the CEP family, was identified by aligning the predicted amino acid sequences of CspA and other members of the CEP family (24). We used site-directed mutagenesis to engineer recombinant variants of CspA. S575A and D180A variants of CspA and wt CspA were expressed in parallel and recovered from L. lactis growth medium supernatants. Figure 1A depicts a Coomassie blue-stained SDS-PAGE gel with the harvested L. lactis growth media; single bands from the harvested media are apparent, indicating that each protein was intact.
The catalytic activities of the proteins were examined by incubating equivalent amounts of concentrated growth media from the three strains with fibrinogen. The cleavage of fibrinogen was evaluated by SDS-PAGE (Fig. 2A). When the proteins were expressed in L. lactis, wt CspA (in the media and in purified form) cleaved fibrinogen but the variant proteins did not. These results indicate that S575 and D180 are essential for the full activity of CspA.
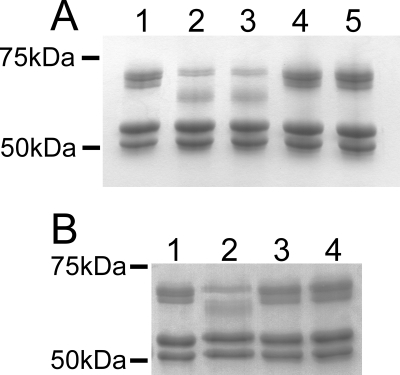
FIG. 2.
The fibrinolytic activity of CspA is dependent on the D180 and S575 residues of the CspA protein. (A) Fibrinogen cleavage by purified CspA and supernatants of L. lactis expressing CspA. Seven micrograms of purified human fibrinogen (Enzyme Research Laboratories) was incubated in PBS alone for 3 min (lane 1), with 14 U of purified CspA (lane 2), or with equivalent volumes of concentrated L. lactis medium supernatants from strains expressing wt CspA (lane 3), D180A CspA (lane 4), or S575A CspA (lane 5). (B) Fibrinogen cleavage by whole cells of GBS. Fibrinogen was incubated with PBS (lane 1), whole cells of wt GBS (lane 2), an isogenic mutant bearing a cspA::Ermr mutation (13) (lane 3), or the strain bearing the cspA(S575A) mutation (lane 4).
The experiments described above demonstrated that the S575 and D180 residues of heterologously expressed CspA are required for the cleavage of fibrinogen. An important question to address was whether the putative catalytic serine residue (S575) is necessary for CspA activity in the context of GBS cells. As CspA is a surface-localized protein, the activity of this enzyme can be readily examined without disrupting cells (13). Allelic exchange mutagenesis was performed, replacing the wt chromosomal cspA gene with an allele encoding the S575A CspA variant. The fibrinolytic activity of the resulting GBS strain (DS107) was compared with that of the wt. The mutant failed to cleave fibrinogen, while the wt strain displayed fibrinolytic activity (Fig. 2B).
N- and C-terminal processing of CspA requires its putative active site residues.
The N terminus of heterologously expressed CspA recovered from L. lactis supernatants is an asparagine residue at position 152 (23), and the propeptide (PRO) domains of other CEPs end at similar positions (e.g., between ∼101 and 192) (24). Therefore, we hypothesized that CspA has a PRO domain and that the protease autocatalyzes the removal of its PRO domain. The catalytically inactive CspA variant proteins were electrophoresed and analyzed via N-terminal sequencing and MALDI-TOF mass spectrometry. These analyses revealed that the N-terminal sequences of both the CspA S575A and D180A variants were 45DSVINK. In contrast, as previously reported (23), the N terminus of the wt CspA was 152NIDSNK. Taken together, these results indicate that during processing, CspA cleaves itself between Q151 and N152 and that this activity depends on the S575 and D180 putative catalytic site residues.
Next, we examined the C-terminal processing of CspA. The incubation of heterologously expressed wt CspA at 37°C resulted in a shift to a lower apparent molecular mass (Fig. 1A). Previously, we used MALDI-TOF analysis to demonstrate that this shift results from cleavage near the C terminus (23). This shift was not observed when CspA was incubated at 4 or 25°C, suggesting that the event is catalytic in nature. We hypothesized that this shift was due to CspA autocatalysis. We incubated heterologously expressed wt CspA, S575A CspA, and D180A CspA at 4 and 37°C for 4 h and analyzed the resulting reaction mixture via SDS-PAGE (Fig. 1A). After incubation at 4°C, all proteins migrated to positions that were consistent with their predicted molecular masses. The variants migrated to positions corresponding to slightly higher molecular masses than the wt protein, consistent with the presence of an uncleaved N-terminal PRO sequence (see above). The incubation of the variant proteins at 37°C had no effect on the positions to which they migrated on gels. In contrast, after incubation at 37°C, wt CspA shifted to a lower apparent molecular mass, as we have reported previously (23). Taken together, these results indicate that the cleavage of the C terminus of CspA is an autocatalytic event that is dependent on S575 and D180.
Previous studies using whole cells of wt GBS and an isogenic cspA mutant (13) indicated that a very limited range of proteins are cleaved by CspA, suggesting a certain degree of substrate specificity. We examined purified CspA for the ability to cleave an additional panel of potential substrates (Table 2). The molecular masses of the test substrates ranged from 4.3 kDa for peptides to 75 kDa for proteins. By incubating the test substrates with purified CspA, conducting SDS-PAGE, and examining the stained gels for shifts in the migration positions of test substrates, we found that only fibrinogen and certain CXC chemokines (see below) were cleaved by CspA. It remains a possibility, however, that some other test molecules were cleaved at the extreme C or N terminus, resulting in a change in molecular mass that could not be detected by our SDS-PAGE analysis.
TABLE 2.
Proteins and peptides not cleaved by CspA
| Protein or peptidea | Molecular mass (kDa) |
|---|---|
| IGF-1 | 7.7 |
| MIP-1α | 7.8 |
| MCP-1 | 8.6 |
| RANTES | 7.8 |
| Beta-defensin 2 | 4.3 |
| Beta-defensin 3 | 5.1 |
| Beta-defensin 1 | 3.9 |
| ICAM-1 | 50 |
| HCC-1 | 8.4 |
| Complement C3b | 75 |
| Ubiquitin | 10.7 |
| Lectin (from Pseudomonas aeruginosa) | 13.5 |
| Cytochrome c | 11 |
IGF-1, insulin-like growth factor 1; ICAM-1, intercellular adhesion molecule 1.
CXC chemokines are cleaved by CspA.
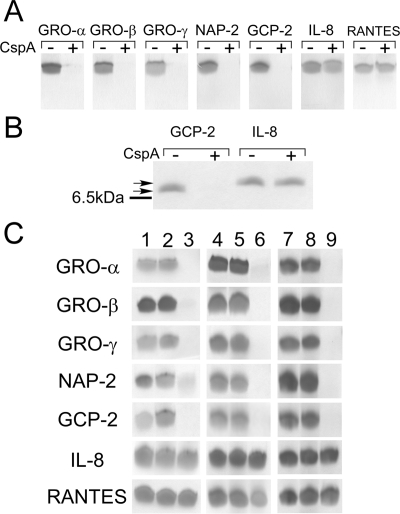
Next, we found that CXC chemokines but not CC chemokines are cleaved by purified CspA. Purified CspA was incubated with the following CXC and CC chemokines: GRO-α, GRO-β, GRO-γ, neutrophil-activating peptide 2 (NAP-2), granulocyte chemotactic protein 2 (GCP-2), IL-8, RANTES (regulated on activation, normal T cell expressed and secreted), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), and hemofiltrate CC chemokine 1 (HCC-1). The CC chemokines RANTES (Fig. 3A) and MCP-1, MIP-1α, and HCC-1 (data not shown) were not cleaved by CspA. However, the CXC chemokines GRO-α, GRO-β, GRO-γ, NAP-2, and GCP-2 were cleaved by CspA. In fact, inspection of the Tris-Tricine-SDS gels revealed that no Coomassie blue-staining material was detectable, consistent with the CXC chemokines' being cleaved into fragments too small to observe on the gels. Interestingly, incubation of the CXC chemokine IL-8 with CspA resulted in no mobility shift (Fig. 3A and B). Preliminary MALDI-TOF analysis of CspA-treated IL-8 indicated that the chemokine was intact (J. Bryan and D. Shelver, unpublished observations). Taken together, these results indicate that CspA, in addition to having proteolytic activity toward fibrinogen, cleaves many of the CXC chemokines but does not cleave CC chemokines.
FIG. 3.
Chemokine cleavage by CspA. Chemokines were incubated with purified CspA, whole GBS cells, or media from cspA-expressing L. lactis cultures. The reaction products were then analyzed by Tris-Tricine-SDS-PAGE. (A) Chemokine cleavage by purified CspA. Two micrograms of each recombinant chemokine (Peprotech) was incubated for 1 h at 37°C with PBS (−) or 13.7 U of CspA (+). Reaction products were analyzed using Tris-Tricine-SDS-PAGE (13.5% acrylamide). (B) The same experiment described in the legend to panel A was performed, except that a 20% acrylamide-Tris-Tricine gel system was utilized. The line to the left of the gel indicates the migration position of the bovine aprotinin standard (6.5 kDa), the lower arrow indicates the migration position of mock-treated GCP-2 (7.9 kDa), and the upper arrow indicates the migration position of mock-treated IL-8 (8.4 kDa). (C) Chemokine cleavage by wt CspA and catalytic site variants of CspA. Lanes 1 to 3, cleavage of chemokines by whole GBS cells. Chemokines were untreated (lane 1) or incubated with logarithmic-phase (OD600 = 0.3), concentrated, PBS-washed whole cells of GBS strain DS107 (expressing S575A CspA) (lane 2) or with identically prepared whole cells of wt GBS strain COH1 (lane 3). Lanes 4 to 9, chemokine cleavage by medium supernatants from lactococcal strains expressing the D180A and S575A CspA variants. Chemokines were incubated for 1 h at 37°C with PBS (lanes 4 and 7) or with equivalent volumes of concentrated L. lactis medium supernatants from strains expressing D180A CspA (lane 5), S575A CspA (lane 8), or wt CspA (lanes 6 and 9).
GBS cleaves CXC chemokines, and cleavage depends on CspA residues S575 and D180.
We then examined whether the putative active site residues identified above are necessary for CspA-mediated chemokine cleavage. We tested medium supernatants containing L. lactis-expressed wt and active site variants of CspA for the ability to cleave chemokines. In separate experiments, we evaluated the effect of the analogous GBS cspA(S575A) chromosomal mutation on GBS whole cell-mediated chemokine cleavage. The same panel of chemokines (GRO-α, GRO-β, GRO-γ, NAP-2, GCP-2, IL-8, and RANTES) tested as described above with purified wt CspA was evaluated in these experiments. After incubation with either wt GBS cells or L. lactis-expressed wt CspA, bands corresponding to GRO-α, GRO-β, GRO-γ, NAP-2, and GCP-2 disappeared from gels (Fig. 3) while those corresponding to IL-8 and RANTES remained and migrated at the same apparent molecular masses as those corresponding to the untreated chemokines. In contrast, CspA active site variants, in the contexts of both heterologously expressed CspA and whole GBS cells, failed to cleave GRO-α, GRO-β, GRO-γ, NAP-2, and GCP-2. Taken together, these results indicate that CspA is responsible for chemokine cleavage by GBS and that S575 and D180 are necessary for CspA-mediated proteolysis of CXC chemokines. The results obtained with whole GBS cells and heterologously expressed CspA were in concordance.
CspA eliminates the ability of CXC chemokines to induce chemotaxis by human neutrophils in vitro.
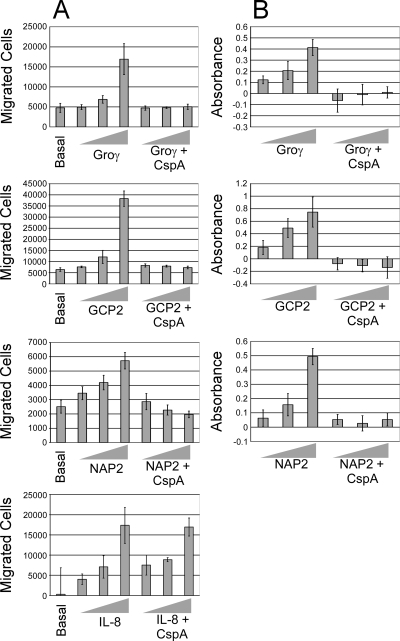
We hypothesized that the proteolysis of CXC chemokines would abolish their biological activities. One important function of certain CXC chemokines is to attract neutrophils to infection sites. NAP-2, GCP-2, and GRO-γ were chosen as representative CspA substrates for neutrophil chemotaxis assays. IL-8 was also tested, since SDS-PAGE analysis indicated that CspA did not appear to cleave this chemokine. The chemotaxis of neutrophils toward mock- or CspA-treated chemokines was examined using commercial Boyden-type chamber kits (Chemicon). Our results, depicted in Fig. 4A, showed that neutrophils migrated by chemotaxis toward mock-treated chemokines but that treatment with purified CspA abolished the neutrophil-attracting activities of NAP-2, GCP-2, and GRO-γ. In contrast, the treatment of IL-8 with CspA did not affect neutrophil chemotaxis, consistent with our finding that IL-8 was not cleaved by CspA.
FIG. 4.
Effects of CspA-treated chemokines on neutrophil migration and activation. (A) Assays of the chemotaxis of freshly isolated human neutrophils toward increasing concentrations (0 [basal], 10, 100, and 1,000 nM) of CspA-treated or mock-treated GRO-γ, GCP-2, NAP-2, and IL-8 were conducted using a commercial Boyden-type chamber fitted with a porous membrane (3-μm pore size) according to the recommendations of the manufacturer (Chemicon). Cells present in the lower chamber at the conclusion of the assay were quantitated per the manufacturer's instructions. The experiment was repeated at least three times, and results from a representative experiment are shown. The shaded bars represent the mean numbers of cells in three replicate wells in the lower chamber, and the error bars represent the standard deviations of the means. (B) Elastase release from human neutrophils was used as a measure of neutrophil activation after incubation with CspA-treated or mock-treated GRO-γ, GCP-2, and NAP-2 (at 10, 100, and 1,000 nM). Released elastase was assayed using the elastase substrate N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide by monitoring at 405 nm. All values were normalized to those for untreated cells. The shaded bars represent the means of results for at least three replicates. Error bars represent the standard deviations of the means. The experiment was repeated at least three times, and results from a representative experiment are depicted.
CspA inhibits neutrophil activation by GRO-γ, GCP-2, and NAP-2.
In addition to attracting neutrophils, CXC chemokines activate the bactericidal activity of neutrophils. Since CspA cleaves many CXC chemokines, we hypothesized that the treatment of chemokines with CspA would inhibit the neutrophil-activating abilities of chemokines. Neutrophil activation was assessed via elastase release assays. To determine if CspA rendered GRO-γ, GCP-2, and NAP-2 unable to activate neutrophils, the chemokines were either mock treated or treated with CspA. Elastase secretion increased in a dose-dependent manner with GRO-γ, GCP-2, and NAP-2 stimulation. However, CspA-treated GRO-γ, GCP-2, and NAP-2 failed to stimulate the secretion of elastase (Fig. 4B). Thus, in addition to abolishing the neutrophil-attracting activities of GRO-γ, GCP-2, and NAP-2, CspA eliminates the abilities of these chemokines to activate neutrophils.
DISCUSSION
In this study, we identified the catalytic site residues of CspA and examined the involvement of the residues in the self-processing of the protease and the cleavage of exogenous substrates. We also identified CXC chemokines as substrates of CspA and determined the effects of CspA-mediated chemokine cleavage on the function of neutrophils. The CspA purification scheme developed in this study will facilitate further studies on the structure and function of CspA and the relationship of this protein to GBS pathogenesis.
Identification of the CspA SS, PRO domain, and catalytic residues.
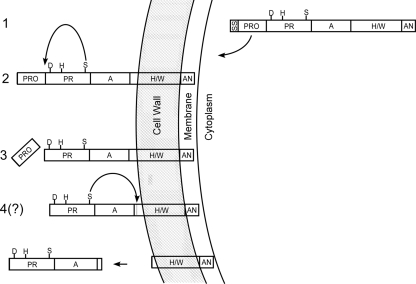
Residues 1 to 44 appear to constitute the predicted CspA signal sequence (SS) (23, 24); our findings provide further support for this prediction, as the SS was removed in an active site-independent manner. We propose that residues 45 to 151 constitute the CspA PRO domain. PRO domains of CEPs are believed to preclude the catalysis of exogenous substrates before removal. A previous bioinformatics study predicted a CspA PRO domain (24), but the assignment was tentative as the sequence does not resemble other CEP PRO sequences. Together with the results from our previous study (23), our present findings support the existence of a CspA PRO domain, as the removal of the putative PRO sequence occurred in an active site-dependent manner, consistent with the mechanism utilized by other CEPs. A model for the processing of CspA is presented in Fig. 5.
FIG. 5.
Model for CspA export, maturation, and processing. Depicted are the cell wall, membrane, and cytoplasm of GBS. Putative assignment and nomenclature of CspA domains were predicted in silico in conjunction with comparisons of other CEPs by Siezen (24). The predicted SS and PRO domain and the protease domain (PR) with residues (S575, D180, and H246 [S, D, and H]) constituting the putative serine protease catalytic site are indicated, as are the domains A (domain of unknown function), H/W (helical/cell wall-spanning region), and AN (cell wall anchor bearing the LPXTG motif). After the synthesis of the immature protein (1), the SS is recognized by the host, resulting in protein translocation across the membrane and SS removal. Subsequently, the protein is covalently anchored to the cell wall (2) via the AN domain, presumably by a host sortase-dependent mechanism (16), and CspA catalyzes the removal (arrow) of its N-terminal PRO domain via its serine active site (3). Finally, evidence from this and previous work (23) suggests that recombinant CspA autocatalyzes its own C-terminal cleavage (arrow), which may allow the release of the protease from the cell (4).
Our results indicate that CspA is a serine protease with D180 and S575 as catalytic site residues. Studies using sequence alignments (24) and protease inhibitors (23) suggested that CspA is a serine protease. Our present work provides strong experimental support for these predictions.
C-terminal autocatalytic cleavage of CspA.
The C-terminal cleavage of L. lactis-expressed CspA occurred in an active site-dependent manner, and it may be speculated that this activity can release the protein from GBS. PrtP, a CEP of L. lactis, has the ability to autocatalytically release itself from the cell (12). SpyCEP/ScpC localizes either to the cell surface or to culture supernatants, depending upon the strain background (27), but its release mechanism has not been determined. C-terminal proteolysis has been proposed as a release mechanism (8); alternative release mechanisms involving the cell-anchoring system have also been suggested previously (8, 27). The complementary findings from the studies of SpyCEP/ScpC and CspA make it tempting to speculate that autocatalysis may function for both proteins. The experiments we performed demonstrating C-terminal autocatalytic cleavage utilized L. lactis-expressed CspA lacking its putative cell wall anchor region. Due to technical barriers, including the lack of a suitable antibody, we have not yet determined if full-length, native GBS CspA undergoes autocatalysis and if this process leads to release from the cell in an active form.
Chemokine cleavage and its role in GBS pathogenesis.
The cleavage of chemokines by GBS has not been reported previously. A critical time window in which the neonate has the opportunity to clear GBS may exist, and delays in neutrophil recruitment may favor infection. In addition to numerous other factors, S. pyogenes avoids phagocytes by utilizing two distinct CEPs, a chemokine-cleaving CEP (SpyCEP/ScpC) and a C5a-cleaving CEP (ScpA). GBS may utilize CspA in addition to its C5a protease to avoid or delay neutrophil attraction and activation.
Human cells respond to pathogens, including GBS (7), by secreting a variety of neutrophil-attracting CXC chemokines. It remains unclear why the immune system utilizes such a large number of chemokines that act on neutrophils. A recent review article (20) noted that chemokines may be a “language” the immune system uses to orchestrate a response to different infections. The specific chemokines utilized and their deployment patterns may depend upon the location and stage of the infection, among other factors. A study utilizing an IL-8-expressing mouse transgene showed that even in the presence of elevated IL-8 levels in plasma, the local recruitment of neutrophils is increased due to other chemokines (19). The activity of CspA observed in our study may suggest that GBS faces situations in which the elimination of chemokines other than IL-8 is important.
Substrate recognition and cleavage by CspA.
CspA appears to have a limited substrate range, based on results from this work and the work of Harris et al. (13). CXC chemokines have very similar three-dimensional structures but share limited amino acid identity. Thus, it is possible that a common structural motif present on the chemokines mediates binding or entry to the CspA active site. Substrate recognition may also trigger a conformational change in the protease that allows the catalytic site to gain access to the substrate. A structure-based recognition mechanism for the chemokine-cleaving protease SpyCEP/ScpC has been suggested previously (8), and a conformational change induced by binding to integrins has been discussed as a potential mechanism that facilitates the access of the C5a peptidase catalytic site to its substrate (4).
Chemokine substrates of CspA appear to be cleaved into small fragments; results from preliminary MALDI-TOF analysis are consistent with this observation (Bryan and Shelver, unpublished). Though its cleavage site preferences have not yet been defined, CspA may cleave substrates between a variety of amino acids. In the models described above, substrate recognition would be relatively specific, but once accessible to the catalytic site, substrates would undergo rather nonspecific cleavage. Surprisingly, CspA did not appear to cleave IL-8, even though the IL-8 structure is highly similar to that of other CspA-cleaved CXC chemokines such as GRO-β (28). Minor structural differences or specific amino acids in IL-8 may preclude its recognition by and entry to the CspA active site. It is unclear why CspA can cleave fibrinogen in addition to chemokines. The Aα subunit of fibrinogen appears to be cleaved by CspA (13). A portion of this subunit may adopt a structure that is recognized by CspA while the remainder of this large molecule is excluded from the active site.
CspA and other streptococcal chemokine-cleaving CEPs.
Three streptococcal CEPs cleave CXC chemokines: CspA of GBS and SpyCEP/ScpC of GAS (8, 14, 27, 29) and CepI of S. iniae (29). However, there appear to be a number of differences among the proteases. The inactivation of cspA reduces virulence in a GBS systemic infection model (13), while the inactivation of the gene encoding SpyCEP/ScpC (cepA) increases virulence in a GAS systemic infection model (26).
Though CspA and SpyCEP/ScpC have some substrates, such as GCP-2 and GRO-α, in common (27), CspA did not appear to cleave IL-8 under the conditions we employed. Also, the two proteases seem to cleave recognized substrates in different manners. CspA appears to cleave its chemokine substrates into small fragments. In contrast, GAS SpyCEP/ScpC cleaves its chemokine substrates at a single location near the C terminus (e.g., between Q59 and R60 in IL-8 and at the analogous position between Q60 and K61 in MIP-2 [8]), even with extended incubation (14). However, the above comparisons are based on results obtained from separate studies; conclusions concerning the differences must be taken in this context and are thus preliminary. A future side-by-side comparative study of the two purified proteases would be valuable to comprehensively evaluate their biochemical differences and similarities.
Levels of identity shared by the streptococcal CEPs and the chromosomal locations of the chemokine protease-encoding genes may indicate that CspA is distinct from SpyCEP/ScpC and CepI. The two proteases that cleave IL-8 (SpyCEP/ScpC and S. iniae CepI [29]) share a significantly higher level of amino acid identity with each other than either does with CspA. CepI and SpyCEP/ScpC share 55% identity, while CspA shares only 33 and 32% identity with SpyCEP/ScpC and CepI, respectively. In contrast, the GBS and GAS C5a peptidases, which appear to be true functional homologs, share a much higher level of identity (∼95%) with each other. The C5a peptidases are functionally distinct from the chemokine-cleaving proteases; these peptidases also belong to the CEP family and are ∼30% identical to CspA, SpyCEP/ScpC, and CepI. Thus, it is not entirely surprising that CspA appears to have different biochemical properties than the other chemokine-cleaving streptococcal proteases; even minor differences in the sequences of CEPs can result in divergent cleavage site preferences and substrate specificities (9). The S. iniae cepI gene and the GAS cepA gene appear to be at conserved chromosomal locations, based on the observation that a number of genes flanking cepA share high levels of identity with genes flanking cepI (29). Though genes that are highly similar to the cepA- and cepI-flanking genes colocalize on the GBS chromosome, no CEP-encoding gene is present in this region. Instead, cspA is located at a completely different region of the GBS chromosome and is flanked by genes distinct from those that flank cepA and cepI.
In summary, we have purified CspA, demonstrated that it is a novel serine protease, and identified its catalytic site residues. The processing of the protease was shown to be dependent on the active site. It was revealed that although CspA does not cleave a large range of substrates, it inactivates a number of CXC chemokines. The necessity of cspA for full GBS virulence and the ability of CspA to inactivate chemokines suggest a mechanism by which CspA may contribute to pathogenesis.
Acknowledgments
We thank Ken Peterson and David McGee for review of the manuscript, Andrew Yurochko, Craig Rubens, and Theresa Harris for helpful discussions and for providing strains, and Jung-Eun Lee for technical assistance.
This work was supported by NIH grants K22AI56271 and R21AI073818 to D.W.S.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Balter, S., C. G. Whitney, and A. Schuchat. 2000. Epidemiology of group B streptococcal infections, p. 154-162. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 2.Bohnsack, J. F., K. Widjaja, S. Ghazizadeh, C. E. Rubens, D. R. Hillyard, C. J. Parker, K. H. Albertine, and H. R. Hill. 1997. A role for C5 and C5a-ase in the acute neutrophil response to group B streptococcal infections. J. Infect. Dis. 175847-855. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack, J. F., X. N. Zhou, P. A. Williams, P. P. Cleary, C. J. Parker, and H. R. Hill. 1991. Purification of the proteinase from group B streptococci that inactivates human C5a. Biochim. Biophys. Acta 1079222-228. [DOI] [PubMed] [Google Scholar]
- 4.Brown, C. K., Z. Y. Gu, Y. V. Matsuka, S. S. Purushothaman, L. A. Winter, P. P. Cleary, S. B. Olmstead, D. H. Ohlendorf, and C. A. Earhart. 2005. Structure of the streptococcal cell wall C5a peptidase. Proc. Natl. Acad. Sci. USA 10218391-18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44183-190. [DOI] [PubMed] [Google Scholar]
- 6.Cieslewicz, M. J., D. L. Kaspar, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276139-146. [DOI] [PubMed] [Google Scholar]
- 7.Doran, K. S., G. Y. Liu, and V. Nizet. 2003. Group B streptococcal beta-hemolysin/cytolysin activates neutrophil signaling pathways in brain endothelium and contributes to development of meningitis. J. Clin. Investig. 112736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, R. J., G. W. Taylor, M. Ferguson, S. Murray, N. Rendell, A. Wrigley, Z. Bai, J. Boyle, S. J. Finney, A. Jones, H. H. Russel, C. Turner, J. Cohen, L. Faulkner, and S. Sriskandan. 2005. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J. Infect. Dis. 192783-790. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Espla, M. D., P. Garault, V. Monnet, and F. Rul. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 664772-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 633539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 1541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haandrikman, A. J., R. Meesters, H. Laan, W. N. Konings, J. Kok, and G. Venema. 1991. Processing of the lactococcal extracellular serine protease. Appl. Environ. Microbiol. 571899-18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, T. O., D. W. Shelver, J. F. Bohnsack, and C. E. Rubens. 2003. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Investig. 11161-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hidalgo-Grass, C., I. Mishalian, M. Dan-Goor, I. Belotserkovsky, Y. Eran, V. Nizet, A. Peled, and E. Hanski. 2006. A streptocococcal protease that degrades CXC chemokines and impairs bacterial clearance from infected tissues. EMBO J. 254628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 1745633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, T. R., C. E. Rubens, and C. B. Wilson. 1988. Lung antibacterial defense mechanisms in infant and adult rats: implications for the pathogenesis of group B streptococcal infections in the neonatal lung. J. Infect. Dis. 15791-100. [DOI] [PubMed] [Google Scholar]
- 18.Phares, C. R., R. Lynfield, M. M. Farley, J. M. Mahle-Boctani, L. H. Harrison, S. Petit, A. S. Craig, W. Schaffner, S. M. Zansky, K. Gershman, K. R. Stefonek, B. A. Albanese, E. R. Zell, A. Schuchat, and S. J. Schrag. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 2992056-2065. [DOI] [PubMed] [Google Scholar]
- 19.Remick, D. G., L. B. Green, D. E. Newcomb, S. J. Garg, G. L. Bolgos, and D. R. Call. 2001. CXC chemokine redundancy ensures local neutrophil recruitment during acute inflammation. Am. J. Pathol. 1591149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rot, A., and U. H. von Andrian. 2004. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 22891-928. [DOI] [PubMed] [Google Scholar]
- 21.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 22.Schuit, K. E., and R. DeBiasio. 1980. Kinetics of phagocyte response to group B streptococcal infections in newborn rats. Infect. Immun. 28319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shelver, D., and J. D. Bryan. 2008. Expression of the Streptococcus agalactiae virulence-associated protease CspA in a soluble, active form utilizing the Gram-positive host, Lactococcus lactis. J. Biotechnol. 136129-134. [DOI] [PubMed] [Google Scholar]
- 24.Siezen, R. J. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie van Leeuwenhoek 76139-155. [PubMed] [Google Scholar]
- 25.Siezen, R. J., B. Renckens, and J. Boekhorst. 2007. Evolution of prokaryotic subtilases: genome-wide analysis reveals novel subfamilies with different catalytic residues. Proteins 67681-694. [DOI] [PubMed] [Google Scholar]
- 26.Sjölinder, H., L. Lövkvist, L. Plant, L., J. Eriksson, H. Aro, A. Jones, and A. B. Jonsson. 2008. The ScpC protease of Streptococcus pyogenes affects the outcome of sepsis in a murine model. Infect. Immun. 763959-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumby, P., S. Zhang, A. R. Whitney, F. Falugi, G. Grandi, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2008. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect. Immun. 76978-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yian, Y. Q., K. O. Johanson, and P. McDevitt. 1999. Nuclear magnetic resonance solution structure of truncated human GROβ [5-73] and its structural comparison with CXC chemokine family members GROα and IL-8. J. Mol. Biol. 2941065-1072. [DOI] [PubMed] [Google Scholar]
- 29.Zinkernagal, A. S., A. M. Timmer, M. A. Pence, J. B. Locke, J. T. Buchanan, C. E. Turner, I. Mishalian, S. Sriskandan, E. Hanski, and V. Nizet. 2008. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe 14170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]