Abstract
We have identified a clonal complex of Mycobacterium bovis present at high frequency in cattle in population samples from several sub-Saharan west-central African countries. This closely related group of bacteria is defined by a specific chromosomal deletion (RDAf1) and can be identified by the absence of spacer 30 in the standard spoligotype typing scheme. We have named this group of strains the African 1 (Af1) clonal complex and have defined the spoligotype signature of this clonal complex as being the same as the M. bovis BCG vaccine strain but with the deletion of spacer 30. Strains of the Af1 clonal complex were found at high frequency in population samples of M. bovis from cattle in Mali, Cameroon, Nigeria, and Chad, and using a combination of variable-number tandem repeat typing and spoligotyping, we show that the population of M. bovis in each of these countries is distinct, suggesting that the recent mixing of strains between countries is not common in this area of Africa. Strains with the Af1-specific deletion (RDAf1) were not identified in M. bovis isolates from Algeria, Burundi, Ethiopia, Madagascar, Mozambique, South Africa, Tanzania, and Uganda. Furthermore, the spoligotype signature of the Af1 clonal complex has not been identified in population samples of bovine tuberculosis from Europe, Iran, and South America. These observations suggest that the Af1 clonal complex is geographically localized, albeit to several African countries, and we suggest that the dominance of the clonal complex in this region is the result of an original introduction into cows naïve to bovine tuberculosis.
Mycobacterium bovis causes bovine tuberculosis (TB), an important disease of domesticated cattle that has a major economic and health impact throughout the world (61, 64, 65). The pathogen is a member of the Mycobacterium tuberculosis complex, which includes many species and subspecies that cause similar pathologies in a variety of mammalian hosts. The most notable member of the complex is M. tuberculosis, the most important bacterial pathogen of humans. In contrast to M. tuberculosis, which is largely host restricted to humans, M. bovis is primarily maintained in bovids, in particular, domesticated cattle, although the pathogen can frequently be recovered from other mammals, including humans (61). Bovine TB is found in cattle throughout the world and has been reported on every continent where cattle are farmed (3).
Bovine TB has been reduced or eliminated from domestic cattle in many developed countries by the application of a test-and-cull policy that removes infected cattle (3, 8, 16, 17, 61, 64, 65). However, in Africa, although bovine TB is known to be common in both cattle and wildlife, control policies have not been enforced in many countries due to cost implications, lack of capacity, and infrastructure limitations (8, 16, 17, 57). In 1998, Cosivi et al. reported of bovine TB, “Of all nations in Africa, only seven apply disease control measures as part of a test-and-slaughter policy and consider bovine TB a notifiable disease; the remaining 48 control the disease inadequately or not at all” (16). In the intervening years, the situation is not thought to have improved (8); however, preliminary surveys of bovine TB have been carried out in some African countries (4, 7, 12, 37, 44, 49, 53, 54, 56).
The most common epidemiological molecular-typing method applied to strains of M. bovis is spoligotyping. This method identifies polymorphism in the presence of spacer units in the direct-repeat (DR) region in strains of the M. tuberculosis complex (36, 67). The DR is composed of multiple, virtually identical 36-bp regions interspersed with unique DNA spacer sequences of similar size (direct variant repeat [DVR] units). Spacer sequences are unique to the DR region, and copies are not located elsewhere in the chromosome (68). The DR region may contain over 60 DVR units; however, 43 of the spacer units were selected from the spacer sequences of the M. tuberculosis reference strain H37Rv and M. bovis BCG strain P3 and are used in the standard application of spoligotyping to strains of the M. tuberculosis complex (29, 36). The DR region is polymorphic because of the loss (deletion) of single or multiple spacers, and each spoligotype pattern from strains of M. bovis is given an identifier (http://www.Mbovis.org).
Several studies of the DR regions in closely related strains of M. tuberculosis have concluded that the evolutionary trend for this region is primarily loss of single DVRs or multiple contiguous DVRs (22, 29, 68); duplication of DVR units or point mutations in spacer sequences were found to be rare. The loss of discrete units observed by Groenen et al. (29) led them to suggest that the mechanism for spacer loss was homologous recombination between repeat units. However, a study by Warren et al. (69) suggested that for strains of M. tuberculosis, insertion of IS6110 sequences into the DR region and recombination between adjacent IS6110 elements were more important mechanisms for the loss of spacer units.
The population structure of the M. tuberculosis group of organisms is apparently highly clonal, without any transfer and recombination of chromosomal sequences between strains (15, 30, 60, 61). In a strictly clonal population, the loss by deletion of unique chromosomal DNA cannot be replaced by recombination from another strain, and the deleted region will act as a molecular marker for the strain and all its descendants. Deletions of specific chromosomal regions (regions of difference [RDs] or large sequence polymorphisms) have been very successful at identifying phylogenetic relationships in the M. tuberculosis complex (11, 25, 26, 35, 48, 50, 61, 62, 66). However, because the loss of spoligotype spacer sequences is so frequent, identical spoligotype patterns can occur independently in unrelated lineages (homoplasy), and therefore, the deletion of spoligotype spacers may be an unreliable indicator of phylogenetic relationship (61, 69).
In samples of M. bovis strains from Cameroon, Nigeria, Chad, and Mali, spoligotyping was used to show that many of the strains had similar spoligotype patterns that lacked spacer 30, and it has been suggested that strains from these four countries are phylogenetically related (12, 18, 49, 53). We have extended the previous observations of spoligotype similarities between strains from these countries and confirmed the existence of a unique clonal complex of M. bovis, all descended from a single strain in which a specific deletion of chromosomal DNA occurred. We have named this clonal complex of M. bovis strains African 1 (Af1), and we show that this clonal complex is dominant in these four west-central African countries but rare in eastern and southern Africa. Extended genotyping, using variable-number tandem repeats (VNTR), of strains with the most common spoligotype patterns suggests that each of these four west-central African countries has a unique population structure. Evolutionary scenarios that may have led to the present day distribution of the Af1 clonal complex are discussed.
MATERIALS AND METHODS
Bacterial strains.
All of the strains analyzed in the study were isolated from cattle, and further information is provided in the supplemental material. Of 89 M. bovis strains isolated from Chadian cattle, 65 were isolated from animals sampled in the years 2000 to 2002 at the N′Djaména abattoir (18, 34); 24 additional strains originated from cattle sampled between July and November 2005 at the abattoir of Sarh in southern Chad, approximately 400 km from N′Djaména. Of 178 M. bovis strains isolated from Nigerian cattle, 15 strains isolated at the Bodija abattoir in Ibadan, Nigeria, in 2003 were previously published (12), while 163 strains isolated at the same abattoir between April and August 2004 are described in this study for the first time. The 75 strains from Cameroonian cattle were collected in 1989 and 1990 and in 1995 and 1996 from cattle in different abattoirs and were published previously (52); a representative subset of 17 strains was used for molecular analyses in this study. The 20 strains from Mali were isolated in March and April 2007 from cattle at the Bamako abattoir and have been previously described (49). All strains were characterized by spoligotyping, and the majority were subjected to VNTR typing, RD4 typing, and RDAf1 typing (see the supplemental material). Population samples of M. bovis from Madagascar (n = 8), South Africa (n = 11), Uganda (n = 13), Burundi (n = 10), Tanzania (n = 14), Ethiopia (n = 15), Mozambique (n = 20), and Algeria (n = 23) were also analyzed by spoligotyping and RDAf1 deletion typing (see the supplemental material). M. tuberculosis H37Rv and M. bovis AF2122/97 were included as reference strains in our experiments.
Spoligotyping and VNTR typing.
Strains were spoligotyped according to the method of Kamerbeek et al. (36) with minor modifications (12). VNTR typing targeted the six loci originally described by Frothingham and Meeker-O'Connell (24) according to the protocol described by Cadmus et al. (12). The presence of a 24-bp deletion frequently observed in one of the tandem repeats of the ETR-D locus is indicated by an asterisk, e.g., 4* (three 77-bp repeats and one 53-bp repeat). The ETR-F locus contains two sorts of tandem repeats of different lengths (79 bp and 55 bp). We display the repeat number of the 79-bp repeats followed by the repeat number of the 55-bp repeats separated by a period. All strains were VNTR typed at VLA, Weybridge, United Kingdom.
Microarray analysis.
For the microarray analysis, four isolates (no. 86, 111, 486, and 515) (see the supplemental material) were selected from the Chadian M. bovis collection, each representing a different spoligotype. The isolates lacked either spacer 30 (strains 111 and 486), spacers 20 to 22 (strain 515), or a combination of the two (strain 86). Approximately 1 to 4 μg of whole genomic DNA for each isolate was extracted as described previously (11). The array used in this study contained nonredundant protein-coding sequences from the two sequenced M. tuberculosis strains, H37Rv and CDC1551, and from the sequenced M. bovis strain, AF2122/97 (http://bugs.sghms.ac.uk). Random amplification, labeling of genomic DNA, and microarray hybridizations were performed as described previously (27). GeneSpring 5.0 was used for the data analysis, and a cutoff for the normalized test/control ratio of <0.5 was used to create gene deletion lists. Deletions found in regions associated with repetitive elements and insertion sequences, which are known to be prone to deletion events, were disregarded in this study.
Deletion typing.
The majority of the strains from Nigeria, Chad, Mali, and Cameroon identified as M. bovis by spoligotyping were additionally confirmed by the deletion of RD4 (11, 61). The presence or absence of RDAf1 was assessed by multiplex PCR with a set of three primers (RDAf1 primer set A): two primers targeting the flanking regions of RDAf1 (Mb0586c FW, 5′-ACTGGACCGGCAACGACCTGG, and Mb0590c Rev, 5′-CGGGTGACCGTGAACTGCGAC) and one primer hybridizing with the internal region of RDAf1 (Mb058xc Int Rev, 5′-CGGATCGCGGTGATCGTCGA). A 350-bp (RDAf1 intact) or a 531-bp (RDAf1 deleted) PCR product was identified by agarose gel electrophoresis. The RDAf1 primers and control strain supernatants are available on request. PCR mixtures contained (per reaction) 1 μl of supernatant of heat-killed mycobacterial cells, a final concentration of 1× HotStartTaq Master Mix (Qiagen), 1 μM of primer Mb0586c FW, 0.5 μM (each) of primer Mb0590c Rev and Mb058xc Int Rev, and sterile distilled water to a final volume of 20 μl. Thermal cycling was performed with an initial denaturation step of 15 min at 96°C and 30 cycles of 30 s at 96°C, 30 s at 65°C, and 1 min at 72°C, followed by a final elongation step of 10 min at 72°C. The PCR products were separated on a 1% agarose gel.
Nucleotide sequence accession numbers.
The RDAf1 deletion junctions of a sample of 20 strains of the Af1 clonal complex from different countries were sequenced using the primers Mb0586c FW and Mb0590c Rev described for RDAf1 deletion typing. The designations of the strains analyzed and the corresponding GenBank accession numbers are as follows: 11b, EU887538; 17b, EU887539; 24b, EU887540; 53b, EU887541; 950 gg mam P, EU887542; 806 rein P, EU887543; 57 HPS pm P, EU887544; 526 gg prescap P, EU887545; 208 gg prescap G, EU887547; 18 HPS pm G, EU887548; 81, EU887549; 45, EU887550; 50, EU887551; 52b, EU887552; C1-128, EU887553; C7-3438, EU887554; B15/05, EU887555; 55, EU887556; 86, EU887557; and 54, EU887558 (see the supplemental material).
RESULTS
Strains with spacer 30 absent.
It has been shown that many strains of M. bovis isolated from cattle in Mali, Chad, and Cameroon have a spoligotype pattern lacking spacer 30 (18, 49, 53). In a small sample of 15 strains from the Ibadan slaughterhouse in southwestern Nigeria, the spoligotype patterns also lacked spacer 30 (12). To supplement this observation, we spoligotyped another 163 strains from the same Nigerian source (Table 1). Only strains isolated from cattle were used in this analysis and throughout the study. All spoligotype patterns of strains from these four countries showed the loss of spacer 30, except for 2 of the 65 strains from Chad (SB1102) and 7 of the 20 strains from Mali (spoligotype patterns SB0134 and SB0991). The three most common spoligotype patterns from each of the four countries, representing over 65% of the strains from each of these countries, are shown in Table 2. In general, over 96% of the 338 strains sampled from these countries lacked spacer 30 in their spoligotype patterns (Table 1; see the supplemental material).
TABLE 1.
Frequencies of spoligotype patterns in four west-central African countries
| Country | Spoligotypea | Frequency
|
|
|---|---|---|---|
| No. | % | ||
| Chad | SB0944 | 26 | 40.0 |
| SB1025 | 13 | 20.0 | |
| SB0951 | 7 | 10.8 | |
| SB1027 | 4 | 6.2 | |
| SB0952 | 3 | 4.6 | |
| SB1098 | 2 | 3.1 | |
| SB1102b | 2 | 3.1 | |
| SB1103b | 2 | 3.1 | |
| SB1101 | 2 | 3.1 | |
| SB1099 | 1 | 1.5 | |
| SB1100 | 1 | 1.5 | |
| SB0328 | 1 | 1.5 | |
| SB1418 | 1 | 1.5 | |
| Total | 65 | ||
| Cameroon | SB0944 | 47 | 62.7 |
| SB1461 | 8 | 10.7 | |
| SB1460 | 6 | 8.0 | |
| SB0951 | 5 | 6.7 | |
| SB1459 | 2 | 2.7 | |
| SB0952 | 2 | 2.7 | |
| SB0955 | 2 | 2.7 | |
| SB1419 | 1 | 1.3 | |
| SB1462 | 1 | 1.3 | |
| SB1463 | 1 | 1.3 | |
| Total | 75 | ||
| Nigeria | SB0944 | 82 | 46.1 |
| SB1027 | 25 | 14.0 | |
| SB1025 | 8 | 4.5 | |
| SB1421 | 6 | 3.4 | |
| SB0328 | 5 | 2.8 | |
| SB1432 | 4 | 2.2 | |
| SB1444 | 4 | 2.2 | |
| SB0951 | 3 | 1.7 | |
| SB0952 | 3 | 1.7 | |
| SB1420 | 3 | 1.7 | |
| SB1439 | 3 | 1.7 | |
| SB1440 | 3 | 1.7 | |
| SB1099 | 2 | 1.1 | |
| SB1424 | 2 | 1.1 | |
| SB1428 | 2 | 1.1 | |
| SB1430 | 2 | 1.1 | |
| SB1435 | 2 | 1.1 | |
| SB1438 | 2 | 1.1 | |
| SB1445 | 2 | 1.1 | |
| SB1026 | 1 | 0.6 | |
| SB1422 | 1 | 0.6 | |
| SB1423 | 1 | 0.6 | |
| SB1425 | 1 | 0.6 | |
| SB1426 | 1 | 0.6 | |
| SB1427 | 1 | 0.6 | |
| SB1429 | 1 | 0.6 | |
| SB1431 | 1 | 0.6 | |
| SB1433 | 1 | 0.6 | |
| SB1434 | 1 | 0.6 | |
| SB1436 | 1 | 0.6 | |
| SB1437 | 1 | 0.6 | |
| SB1441 | 1 | 0.6 | |
| SB1442 | 1 | 0.6 | |
| SB1443 | 1 | 0.6 | |
| Total | 178 | ||
| Mali | SB0300 | 8 | 40.0 |
| SB0134b | 6 | 30.0 | |
| SB1410 | 2 | 10.0 | |
| SB0944 | 1 | 5.0 | |
| SB1411 | 1 | 5.0 | |
| SB1412 | 1 | 5.0 | |
| SB0991b | 1 | 5.0 | |
| Total | 20 | ||
International names for these spoligotype patterns were assigned by Mbovis.org (http://www.Mbovis.org).
Not a member of the Af1 clonal complex.
TABLE 2.
The three most common spoligotype patterns in each of four west-central African countries
| Country | Pattern designationa | Spoligotype patternb | Frequency (%) |
|---|---|---|---|
| Chad | SB0944 | 1101111101111110111111111111101111111100000 | 40 |
| SB1025 | 1101111101111110111111111111100011111100000 | 20 | |
| SB0951 | 1101111101111110111111111111101111110100000 | 11 | |
| Cameroon | SB0944 | 1101111101111110111111111111101111111100000 | 63 |
| SB1461 | 1101111100000000011111111111101111111100000 | 11 | |
| SB1460 | 1101111101110000001111111111101111111100000 | 8 | |
| Nigeria | SB0944 | 1101111101111110111111111111101111111100000 | 46 |
| SB1027 | 1101111101111110111111111100101111111100000 | 14 | |
| SB1025 | 1101111101111110111111111111100011111100000 | 4 | |
| Mali | SB0300 | 1101101101111110111111111111101111111100000 | 40 |
| SB0134c | 1100011101111110111111111111111111111100000 | 30 | |
| SB1410 | 1001101101111110111111111111101111111100000 | 10 |
International names for these spoligotype patterns were assigned by Mbovis.org (http://www.Mbovis.org).
The spoligotype pattern is shown as a series of ones and zeros, with one representing hybridization to the spacer and zero representing the absence of hybridization.
Not a member of the Af1 clonal complex.
Identification of a specific deletion, RDAf1.
The absence of specific spoligotype spacers can be characteristic of a clonal complex of M. bovis, a closely related group of strains all descended from a single common ancestor (61). However, the loss of spoligotype spacers can occur independently in unrelated lineages (homoplasy) and may misidentify members of a clonal complex (61, 69). Deletions of chromosomal DNA (RDs or large sequence polymorphisms) are less likely to generate homoplasies, provided that the deleted region is not flanked by repetitive sequences that generate identical deletions at high frequency (11).
To identify a suitable phylogenetically informative deletion, chromosomal DNA from two strains from Chad lacking spacer 30 in their spoligotype patterns were applied to an M. tuberculosis-M. bovis composite amplicon array and subjected to microarray analysis. As expected, the results of this analysis were compatible with the deletion of RD4, -7, -8, -9, -10, -12, and -13 (11); however, a previously unreported deletion of approximately 5.3 kb was identified in both strains and named RDAf1. In contrast, a strain from Chad with spoligotype spacer 30 present (SB1102) was intact in this region by microarray analysis. Sequencing of the RDAf1 region showed that 5,322 bp was deleted compared with the chromosomal sequences of M. bovis BCG (10) and M. bovis AF2122 (28). The RDAf1 deletion removed Mb0587c to Mb0589c and parts of Mb0586c and Mb0590c (corresponding to Rv0572c to Rv0574c and parts of Rv0571c and Rv0575c in M. tuberculosis H37Rv). The flanking regions of the deletion showed no similarity to insertion sequences or repetitive DNA and were not GC rich, suggesting that this region was not prone to generating homoplasies. We concluded that RDAf1 could be a suitable marker for a clonal complex of strains present at high frequency in Mali, Cameroon, Nigeria, and Chad.
Distribution of RDAf1 in west-central African strains.
To determine if the RDAf1 deletion could be used as a marker for an important clonal complex, we used PCR to survey the state of RDAf1 in all available strains from Chad and Mali and a representative sample of strains from Nigeria and Cameroon, a total of 239 strains (see the supplemental material). The PCR primers were located on either side of the RDAf1 region and were chosen to give easily visualized products of different sizes from strains with RDAf1 deleted and strains with RDAf1 intact.
From Nigeria, 148 strains, including the 15 strains published by Cadmus et al. in 2006 (12), were tested, as well as a sample of 17 strains representing the population of M. bovis from Cameroon. All 165 strains from Nigeria and Cameroon had the Af1 region deleted. In the sample of 20 strains from Mali, 7 strains were intact in the RDAf1 region, and these strains had spoligotype patterns SB0134 and SB0991; spacer 30 is present in both these spoligotype patterns. Among the 65 strains from Chad, 4 strains were intact in the RDAf1 region in contrast to the remaining 61 strains from that country; there were 2 strains with spacer 30 present (spoligotype pattern SB1102), but also 2 strains with both spacers 30 and 31 absent (spoligotype pattern SB1103) (Table 1). In general, of 250 strains surveyed from these four west-central African countries, 239 had the RDAf1 region deleted, and all 239 strains lacked spacer 30 in their spoligotype patterns (see the supplemental material).
To confirm that the RDAf1 deletions were identical by descent, we sequenced across the RDAf1 deletion boundaries in a total of 20 strains originating from different countries and showing distinct spoligotype patterns of the Af1 complex (see the supplemental material). In all 20 strains, the deletion boundaries of RDAf1 were identical, suggesting that these deletions are identical by descent in strains from these four countries.
We concluded that a single clonal complex of M. bovis strains, defined by the deletion of RDAf1 and marked by the loss of spacer 30, was present at high frequency in Mali, Chad, Cameroon, and Nigeria. We named this M. bovis clonal complex the Af1 clonal complex.
Af1 in other African countries.
Previously published population surveys of bovine TB strains from Burundi, Uganda, South Africa, and Madagascar showed that in general, spacer 30 was present in the spoligotype patterns of strains from these countries, suggesting that the Af1 clonal complex was not present at high frequency (44, 54, 56, 58). To confirm this observation, a sample of strains chosen to represent the spoligotype diversity of the population in each country was surveyed by PCR for the status of the RDAf1 region. For representative strains from Burundi (n = 10), Madagascar (n = 8), South Africa (n = 11), and Uganda (n = 13), the RDAf1 region was intact. In all of these strains, spacer 30 was present (see the supplemental material). We also surveyed the status of the RDAf1 region in previously unpublished collections of strains from Algeria (n = 23), Ethiopia (n = 15), Mozambique (n = 20), and Tanzania (n = 14). In 61 of the 72 strains, spacer 30 was present in the spoligotype pattern, and in all of the strains surveyed, the RDAf1 region was intact (see the supplemental material).
We concluded that the distribution and frequency of strains of the Af1 clonal complex were not uniform throughout Africa; Af1 strains were at high frequency in the four west-central African countries but were rare or absent in Algeria, Burundi, Ethiopia, Madagascar, Mozambique, South Africa, Tanzania, and Uganda (Fig. 1).
FIG. 1.
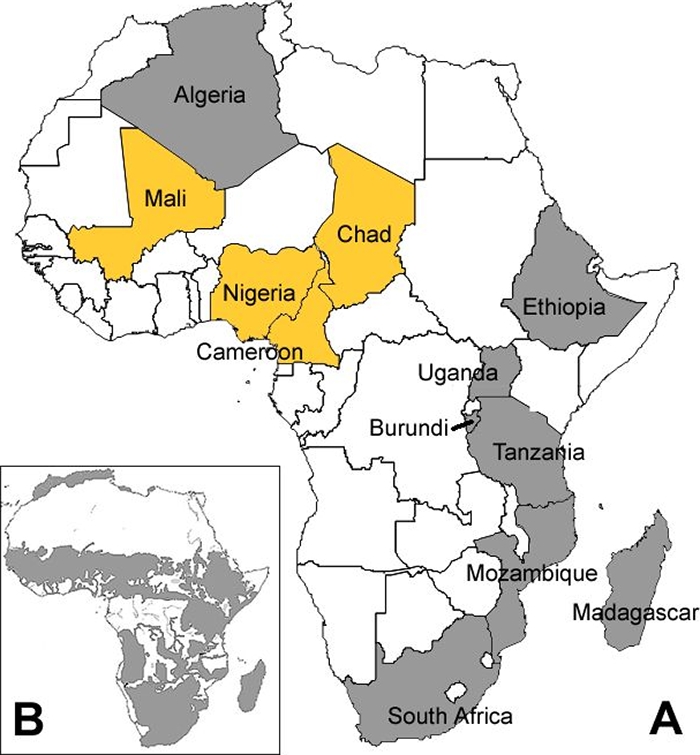
Localization of the Af1 clonal complex of M. bovis to west-central Africa. (A) The four west-central African countries where Af1 strains were found to be dominant are shown in yellow. Countries where Af1 strains were not found are shown in gray. (B) Cattle distribution on the continent of Africa (gray shaded area). (Modified from reference 32 with the author's permission.)
Geographical localization of genotypes in each country.
The population structures of the Af1 strains in Cameroon, Nigeria, and Chad are superficially similar; in each of these countries, over 90% of the strains included in this study are members of the Af1 clonal complex, and strains with spoligotype pattern SB0944 are most common in these three countries. However, strains from Mali show a distinct difference in population structure from those in the other three west-central African countries (Table 1), although some caution is needed in this interpretation because of the small number of isolates. In Mali, the majority of Af1 strains lack spacer 6 in the spoligotype patterns in addition to the loss of spacer 30 (SB0300 and related patterns); strains of the Af1 clonal complex lacking spacer 6 are rare or absent in Chad, Nigeria, or Cameroon. Furthermore, in Mali, members of the Af1 clonal complex make up only 60% of the sampled population, and a second group of strains with RDAf1 intact and spacer 30 present but lacking spacers 4 and 5 are also common (spoligotype patterns SB0134 and SB0991; 40% of strains) (Table 2) (49). This contrasts with the three other west-central African countries, where strains of the Af1 clonal complex are apparently ubiquitous (Table 1). These observations suggest that the population of M. bovis in Mali is markedly different from those in the other three west-central African countries analyzed here.
To further extend this observation, we VNTR typed (ETR-A to -F) all available strains with spoligotype pattern SB0944 from Cameroon, Nigeria, and Chad to give a genotype for these strains consisting of a combination of a spoligotype pattern and a six-locus VNTR pattern. Table 3 shows the genotypes of strains with spoligotype pattern SB0944 from Cameroon, Chad, and Nigeria.
TABLE 3.
Genotypes of strains with spoligotype pattern SB0944 from Chad, Nigeria, and Cameroon
| Country | Genotypea | Frequency (no.) |
|---|---|---|
| Chad | SB0944; 3 6 5 4* 3 3.1 | 2 |
| SB0944; 4 5 5 2 3 3.1 | 2 | |
| SB0944; 4 5 6 5* 3 3.1 | 2 | |
| SB0944; 4 7* 5 4* 3 3.1 | 2 | |
| SB0944; 4 8 5 4* 3 3.1 | 2 | |
| SB0944; 3 5 3 4* 3 3.1 | 1 | |
| SB0944; 3 5 5 4* 3 3.1 | 1 | |
| SB0944; 4 3 5 4* 3 3.1 | 1 | |
| SB0944; 4 5 6 4* 2 3.1 | 1 | |
| SB0944; 4 6 5 4* 3 3.1 | 1 | |
| SB0944; 4 7 5 4* 3 3.1 | 1 | |
| SB0944; 4 7 6 4* 3 3.1 | 1 | |
| SB0944; 4 8 5 3* 3 3.1 | 1 | |
| SB0944; 5 5 3 4* 3 3.1 | 1 | |
| Total | 19 | |
| Nigeria | SB0944; 5 5 3 4* 3 3.1 | 22 |
| SB0944; 5 5 5 4* 3 3.1 | 19 | |
| SB0944; 4 5 5 4* 3 3.1 | 2 | |
| SB0944; 5 4 5 4* 3 1.3 | 2 | |
| SB0944; 5 4 6 4* 3 3.1 | 2 | |
| SB0944; 5 5 3 4* 3 1.3 | 2 | |
| SB0944; 3 5 5 4* 3 3.1 | 1 | |
| SB0944; 4 4 3 4* 3 3.1 | 1 | |
| SB0944; 5 3 5 4* 3 3.1 | 1 | |
| SB0944; 5 4 5 4* 2 1.3 | 1 | |
| SB0944; 5 4 5 4* 3 3.1 | 1 | |
| SB0944; 5 5 4 4* 3 3.1 | 1 | |
| SB0944; 5 5 6 4* 3 3.1 | 1 | |
| SB0944; 5 6 5 4* 3 3.1 | 1 | |
| SB0944; 6 4 5 4* 3 3.1 | 1 | |
| SB0944; 6 4 7 4* 3 3.1 | 1 | |
| Total | 59 | |
| Cameroon | SB0944; 5 5 5 4* 3 2.1 | 3 |
| SB0944; 5 5 3 4* 3 3.1 | 2 | |
| SB0944; 5 3 4 4* 4 2.1 | 1 | |
| Total | 6 |
International name for the spoligotype pattern followed by a semicolon followed by the allele call for the ETR-A to -F loci. The two genotypes found in more than one country are marked by italics and boldface.
It is clear from Table 3 that the VNTR patterns of the strains with the most common spoligotype pattern in Nigeria and Chad (SB0944) differ significantly. Only 2 of the 16 genotypes found in Nigeria are also found in Chad. For Nigerian strains, allele 5 is common at the ETR-A locus (53 of 59 strains), whereas in Chad, only one of 19 strains had this allele. Furthermore, the most common genotype in Nigeria (SB0944; 5 5 3 4* 3 3.1), found in 22 of the 59 Nigerian strains, is rare in Chad. The small sample of strains from Cameroon with spoligotype pattern SB0944 also suggests that this population is unique compared to Nigeria and Chad. Four of the six strains genotyped from Cameroon have a 2.1 allele at the ETR-F locus, which is not found in the 78 strains with spoligotype pattern SB0944 from the other two countries from which we were able to generate a full genotype (Table 3).
The genotypes of strains with the second-most-common spoligotype pattern in Chad (SB1025) also suggest a difference in the population structure between Nigeria and Chad. Seven of 8 strains from Nigeria with spoligotype pattern SB1025 had allele 5 at both the ETR-A and ETR-B locus, whereas 8 of 10 strains from Chad had alleles 4 and 3 at the ETR-A and -B loci, respectively (see the supplemental material).
These observations suggest that each of the strains sampled from Mali, Chad, and Nigeria has a country-specific population structure. That is, given a strain blind from one of these countries, it would be possible, with reasonable accuracy, to identify the country of origin from the genotype. The limited data from Cameroon also suggest a country-specific population.
DISCUSSION
We have identified an epidemiologically important clonal complex of M. bovis dominant in Mali, Cameroon, Nigeria, and Chad and have named this clonal complex Af1. The clonal complex is epidemiologically important because it is at high frequency in these four countries; we do not yet know how phylogenetically distinct the clonal complex is from other strains of M. bovis. Members of the clonal complex are defined by a 5.3-kb deletion of chromosomal DNA, which we have named RDAf1. Sequencing of the RDAf1 region in many strains has shown that the deletion boundaries are identical, and in the absence of repetitive elements flanking RDAf1 and the apparent strict clonality of M. bovis, we conclude that this deletion is identical by descent in strains from the four countries. That is, RDAf1 was deleted from the most recent common ancestor of the clonal complex, and this region is therefore deleted in all descendants. All strains identified as members of the Af1 clonal complex by deletion analysis have spoligotypes that can be derived from spoligotype SB0944 by loss of spacers. We therefore assume that the most recent common ancestor (progenitor) of the Af1 clonal complex had spoligotype pattern SB0944. A definition and summary of the Af1 clonal complex is shown in Table 4.
TABLE 4.
Definition and summary of the Af1 clonal complex
| Parameter | Value |
|---|---|
| Definition | Presence of deletion RDAf1 (5.3 kb, between Mb0586c and Mb0590c) |
| Spoligotype marker | Absence of spacer 30 |
| Spoligotype signaturea | 1101111101111110111111111111101111111100000 (SB0944) |
| Distribution | At high frequency in sub-Saharan west-central Africa (Mali, Cameroon, Chad, and Nigeria) |
The spoligotype signature represents the assumed spoligotype pattern in the progenitor strain of the clonal complex and is shown as a series of ones and zeros, with 1 representing hybridization to the spacer and 0 representing absence of hybridization. The international name for this spoligotype pattern was assigned by Mbovis.org (http://www.Mbovis.org).
Strains of the Af1 clonal complex can be identified by the loss of spacer 30 in the spoligotype pattern, although this characteristic is not necessarily specific. It is theoretically possible for strains with the RDAf1 deletion to have spacer 30 present, although we have not yet identified such an isolate. Furthermore, because the loss of spacers in spoligotype patterns can be homoplastic (61, 69), strains that are not members of the Af1 clonal complex (with the RDAf1 region intact) can also lack spacer 30, for example, the strains with spoligotype pattern SB1103 from Chad.
We have surveyed for the presence of strains of the Af1 clonal complex in small samples of strains from other African countries and have shown that strains with RDAf1 deleted are not represented in our samples from Algeria, Burundi, Ethiopia, Madagascar, Mozambique, South Africa, Tanzania, and Uganda (Fig. 1). Although the number of strains sampled in each of these countries was small, the high frequency of Af1 in the four west-central African countries supports our conclusion that the Af1 clonal complex is not uniformly distributed throughout Africa but is dominant in sub-Saharan west-central Africa. Previously published large-scale spoligotype population surveys of M. bovis strains from Europe (France, Spain, United Kingdom, Italy, Germany, Belgium, Czech Republic, and Portugal) (2, 5, 6, 19, 20, 31, 33, 39, 55, 59, 61, 70), the Middle East (Iran) (63), and South and Central America (14, 45, 71) do not show a high frequency of strains with spacer 30 missing, suggesting that if Af1 strains are present, they are not at the high frequency seen in west-central Africa and supporting our suggestion of geographical localization of the Af1 clonal complex to this region of Africa.
National localization of Af1 genotypes.
The population structure of M. bovis in Mali is clearly different from the population structure in the three other west-central African countries (Table 1). We extended this observation to show that the genotypes (spoligotype plus VNTR type) of the most common types found in Chad and Nigeria also differ in both frequency and type. However, it could be suggested that these samples are not representative of the population structure of M. bovis in Chad and Nigeria; that is, for the short period when the strains were collected, a subset of the total M. bovis population present in each country was sampled. However, the strains from Chad were collected over a 3-year period and the two sets of Nigerian strains, although collected at the same abattoir, were sampled over a year apart. In both these samples from Nigeria, allele 5 was common at the ETR-A locus, in contrast to strains from Chad. A second small group of five strains with spoligotype pattern SB0944, collected at an abattoir in southern Chad at least 400 km from the collection site of the original Chadian strains in N′Djaména, and 3 years later, was also distinct from the Nigerian strains (unpublished data). Taken together, these data strongly suggest a distinct, country-specific population structure for M. bovis in each of these west-central African countries. This is notable, because it is known that there is extensive movement of cattle, both commercial and by transhumance, between Chad, Nigeria, and Cameroon; however, our results suggest there is not enough local transmission of extranational strains of M. bovis to obscure the country-specific population structure. It is common for livestock sold from Chad to either Cameroon or Nigeria to be directed to large city abattoirs and not resold to mix with the local populations. In a similar way, large cattle populations are sold in Mali for slaughtering in Abidjan (Ivory Coast) and do not mix with local cattle populations (46). The observation of a country-specific population structure is somewhat surprising, as these west-central African countries have a century-old, long-distance transhumant livestock production system and trade routes of cattle between the Sahelian countries and the large coastal cities, which were considered important determinants of the regional spread of bovine TB (23, 38, 42). More importantly, our results show that it would be worthwhile for each of these countries to establish simple genotype surveillance of M. bovis using spoligotyping and VNTR typing to monitor the import of extranational strains in a bovine TB control or eradication campaign.
Although our data do not address within-country geographic localization of M. bovis genotypes, as is commonly seen in Great Britain (61), we are aware that the previous population survey of M. bovis strains in Cameroon suggests a difference between the genotypes of strains isolated in the north and the south of the country (53). The samples of strains from Nigeria, Chad, and Mali were collected from a single abattoir in each country. It is known that these abattoirs process animals originating on a national and even, occasionally, an international scale. However, the absence of cattle-tracing systems in west-central Africa and the frequent trading of animals make the exact catchment area for each abattoir difficult to establish. These observations imply that it may be worthwhile to carry out more extensive studies of the geographical localization of M. bovis genotypes in each country, which, together with an in-depth knowledge of contemporary livestock trade and transhumance routes within Chad, Mali, and Nigeria, could lead to the development of useful epidemiological tools (genotype location maps) to identify key determinants of transmission and to aid in the control of bovine TB in these countries.
Evolution of the Af1 clonal complex.
The simplest explanation for the observed distribution and population structure of the Af1 clonal complex throughout these sub-Saharan west-central African countries is that a single strain or clonal complex of M. bovis spread between the four countries in cattle naïve to bovine TB. On the assumption that spacer sequences are lost and never regained, the progenitor strain would have had spacer 30 missing and would have carried the RDAf1 deletion, and this would account for the presence of strains descended from this progenitor Af1 strain in each of the countries (Table 4). Country-specific population structures could have evolved by drift, either during the spread of the Af1 clonal complex between countries (a series of founder events) or subsequently, as the population expanded in each country. In Mali, for example, the founding Af1 clone has lost spacer 6, and this subclone has become dominant, while in each of the other three west-central African countries differences in VNTR profiles have evolved. All spoligotype patterns of the Af1 clonal complex in these four countries can be derived from the spoligotype pattern of the progenitor strain by loss of spoligotype spacers. In support of the suggestion of a single introduction of bovine TB and subsequent spread through a population of cattle naïve to bovine TB, it is interesting that Alhaji, writing in 1976, reported that bovine TB first appeared in West Africa in the Cameroons in 1913 and that much of the bovine TB identified in Nigerian abattoirs in the 1940s came from cattle imported from Cameroon (1). If the Af1 clonal complex did originate in Cameroon, then we might expect that diversity would increase with distance as the clonal complex spread, and this may explain how the spoligotypes found in Mali, the most distant country from Cameroon surveyed here, are so different from the others.
The localization of the Af1 clonal complex to this region of west-central Africa may have been governed by geographical barriers. To the north of the Af1 range is the Sahara desert, and to the south are the forest areas in the Congo. These geographical features reduce both cattle density (Fig. 1B) and movement and therefore may have limited the spread of Af1 strains. There are no major geographical barriers between Chad, Mali, Cameroon, and Nigeria, and the cattle density throughout the region is fairly uniform (Fig. 1B). The absence of Af1 in East Africa, particularly in Ethiopia, can be explained by poor trade links between west-central and East Africa or the prior establishment of an M. bovis population in East African countries that may have limited the introduction of the Af1 clonal complex. We assume that other countries in this region, especially Niger, will also be dominated by strains of the Af1 clonal complex and that these countries will have country-specific population structures (Fig. 1). It is noteworthy that a strain of M. bovis with spacer 30 missing in the spoligotype pattern has been isolated from a human with pulmonary TB in Ghana (43).
It may seem unreasonable that a single strain with both a chromosomal deletion and the deletion of a spoligotype spacer spread between these countries. However, we can assume that the population of M. bovis that gave rise to the Af1 progenitor was very small (a founder population), and under those conditions in a clonal organism, the fixation of otherwise deleterious mutations may be quite common (61). Whether the progenitor of the Af1 clonal complex evolved in Africa or evolved elsewhere and was subsequently imported to Africa is unknown, although this may be resolved when the phylogenetic relationship of the Af1 clonal complex to strains from other countries is determined.
Strains of the Af1 clonal complex have virtually reached fixation in Nigeria, Cameroon, and Chad, and the suggestion of a single strain spreading throughout the region and subsequently establishing country-specific populations is, to a certain extent, dependent on the absence of M. bovis strains in these three countries prior to the spread of Af1. However, if each country had a prior population of M. bovis that was replaced by the Af1 clone, then it is difficult to explain, without invoking selection, how strains of this clonal complex went to fixation by drift, independently, in three countries. If we assume west-central African cattle were infected with a prior population of M. bovis, then the Af1 clone could have arisen with a selective advantage and spread throughout the region, going to fixation in Nigeria, Cameroon, and Chad and replacing the previous population.
It would be tempting to suggest that the RDAf1 deletion generated a phenotype that was selectively advantageous. For some M. bovis deletions, a phenotype has been described; for example, deletion RD4 induces a truncated form of a phenolic glycolipid (41), but any selective advantage of this phenotype is unknown. In general, attempts to assign selective advantages to the many other deletions in members of the M. tuberculosis complex have not been overly successful. Exceptions are the deletion of the esxAB genes and flanking regions seen in BCG (RD1), Mycobacterium microti (RDMic), and the Dassie bacillus (9, 40, 47) and the loss of an immunoregulatory locus from an epidemic strain of M. tuberculosis (51). The loss of the esxAB region in several independent deletion events is strong evidence for an unknown selective advantage for the deletion of this locus. However, before a selective advantage is assumed for any particular chromosomal deletion, it should be remembered that in a clonal organism, deletions that are not advantageous can easily go to fixation by hitchhiking with a selectively advantageous mutation (single nucleotide polymorphism or deletion) located elsewhere on the chromosome (61).
Homology searches in the databases at NCBI suggest that the five genes (Mb0586c to Mb0590c) affected by the RDAf1 deletion are conserved among several related Mycobacterium species, but none of the genes has yet been functionally characterized. However, Mb0589c (Rv0574c) shares homology with the genes for CapA from Bacillus anthracis and PgsAA from Bacillus subtilis; these proteins are proposed to be part of a complex for poly-gamma glutamate biosynthesis (13).
In Mali, two major clonal complexes of M. bovis have been described (49). Af1 strains make up 60% of the population, whereas a second clonal complex (spoligotype SB0134 and related patterns), marked by the loss of spacers 4 and 5 and the presence of spacer 30 in the spoligotype pattern, is also present at high frequency (49). This second clonal complex of strains in Mali, provisionally named Af5, has been shown to have spoligotype similarities to strains from Europe (49). Whether the Af1 clonal complex went to high frequency in Mali by drift or selection, the simplest explanation for the presence of the Af5 clonal complex is that it was introduced after Af1 had been established; however, we have no evidence to indicate the relative times of establishment of Af1 and Af5 or the phylogenetic relationship between them, except that strains of Af5 are not members of the Af1 clonal complex.
Global distribution of clonal complexes of M. bovis.
The local dominance of specific clonal complexes of M. bovis, identified initially by spoligotype signatures, is becoming a feature of the global population structure of this economically important pathogen (33). A clonal complex of limited diversity, provisionally called Eu1, dominates in the British Isles and their historic trading partners (61), and we also suspect that a second clonal complex of M. bovis (Af2) dominates in East African countries and a third (Af3) in Madagascar.
Identification of these clonal complexes will assist the epidemiological analysis of the international movement of M. bovis strains, not only in cattle, but also in humans (45). A strain of M. bovis isolated in 2005 from a human at the Midlands Regional Centre for Mycobacteriology, Birmingham, United Kingdom, had the commonest genotype seen in our Nigerian sample (SB0944; 5 5 3 4* 3 3.1) (21). This genotype is quite distinct from the genotypes currently isolated from cattle and humans in the British Isles, and we have shown that the strain has the RDAf1 region deleted and is therefore a member of the Af1 clonal complex (unpublished data). It is interesting that this patient was born in west-central Africa. Furthermore, a strain of M. bovis with spoligotype pattern SB1025, the second-most-common spoligotype identified in our sample from Chad, was isolated from a mother and daughter of Chadian origin in France in 2003. These strains also had the RDAf1 region deleted (unpublished data), and the infection was presumably acquired in Africa. These cases illustrate how the identification and analysis of M. bovis clonal complexes and genotypes can be used to suggest possible sources of infection.
Supplementary Material
Acknowledgments
We thank E. J. Vololonirina, M. Okker, and K. Gover for excellent technical assistance and I. Abubakar for his help.
This work was funded by the Swiss National Science Foundation (grant no. 320000-107559); the National Centre of Competence in Research (NCCR) North-South IP-4; Prionics AG (Zürich, Switzerland); ARC, MRC, and NRF in South Africa; the Institut Pasteur de Madagascar; and CHRU Arnaud de Villeneuve, Laboratoire de Bactériologie-Virologie, Montpellier, France, and by a Swedish International Development Cooperation Agency grant to Mozambique. Work in Mali is supported by the Swiss National Centre of Competence in Research (NCCR) North-South: Research Partnerships for Mitigating Syndromes of Global Change, by the Wellcome Trust Livestock for Life and Animal Health in the Developing World initiatives, by the University of Ibadan/MacArthur Foundation, and by the Department of Environment, Food and Rural Affairs, United Kingdom.
Footnotes
Published ahead of print on 9 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alhaji, I. 1976. Bovine tuberculosis: a general review with special reference to Nigeria. Vet. Bull. 46829-841. [Google Scholar]
- 2.Allix, C., K. Walravens, C. Saegerman, J. Godfroid, P. Supply, and M. Fauville-Dufaux. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 441951-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amanfu, W. 2006. The situation of tuberculosis and tuberculosis control in animals of economic interest. Tuberculosis 86330-335. [DOI] [PubMed] [Google Scholar]
- 4.Ameni, G., A. Aseffa, A. Sirak, H. Engers, D. B. Young, R. G. Hewinson, M. H. Vordermeier, and S. V. Gordon. 2007. Effect of skin testing and segregation on the prevalence of bovine tuberculosis, and molecular typing of Mycobacterium bovis, in Ethiopia. Vet. Rec. 161782-786. [PMC free article] [PubMed] [Google Scholar]
- 5.Aranaz, A., L. De Juan, N. Montero, C. Sanchez, M. Galka, C. Delso, J. Alvarez, B. Romero, J. Bezos, A. I. Vela, V. Briones, A. Mateos, and L. Dominguez. 2004. Bovine tuberculosis (Mycobacterium bovis) in wildlife in Spain. J. Clin. Microbiol. 422602-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aranaz, A., E. Liâebana, A. Mateos, L. Dominguez, D. Vidal, M. Domingo, O. Gonzolez, E. F. Rodriguez-Ferri, A. E. Bunschoten, J. D. Van Embden, and D. Cousins. 1996. Spacer oligonucleotide typing of Mycobacterium bovis strains from cattle and other animals: a tool for studying epidemiology of tuberculosis. J. Clin. Microbiol. 342734-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asiimwe, B. B., J. Asiimwe, G. Kallenius, F. K. Ashaba, S. Ghebremichael, M. Joloba, and T. Koivula. Molecular characterization of Mycobacterium bovis isolates from cattle carcasses at a city slaughterhouse in Uganda. Vet. Rec., in press. [DOI] [PubMed]
- 8.Ayele, W. Y., S. D. Neill, J. Zinsstag, M. G. Weiss, and I. Pavlik. 2004. Bovine tuberculosis: an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 8924-937. [PubMed] [Google Scholar]
- 9.Brodin, P., K. Eiglmeier, M. Marmiesse, A. Billault, T. Garnier, S. Niemann, S. T. Cole, and R. Brosch. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect. Immun. 705568-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. Dos Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 1045596-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadmus, S., S. Palmer, M. Okker, J. Dale, K. Gover, N. Smith, K. Jahans, R. G. Hewinson, and S. V. Gordon. 2006. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J. Clin. Microbiol. 4429-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candela, T., and A. Fouet. 2006. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 601091-1098. [DOI] [PubMed] [Google Scholar]
- 14.Cobos-Marin, L., J. Montes-Vargas, M. Zumarraga, A. Cataldi, M. I. Romano, I. Estrada-Garcia, and J. A. Gonzalez-y-Merchand. 2005. Spoligotype analysis of Mycobacterium bovis isolates from Northern Mexico. Can. J. Microbiol. 51996-1000. [DOI] [PubMed] [Google Scholar]
- 15.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 16.Cosivi, O., J. M. Grange, C. J. Daborn, M. C. Raviglione, T. Fujikura, D. Cousins, R. A. Robinson, H. F. Huchzermeyer, I. de Kantor, and F. X. Meslin. 1998. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 459-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosivi, O., F. X. Meslin, C. J. Daborn, and J. M. Grange. 1995. Epidemiology of Mycobacterium bovis infection in animals and humans, with particular reference to Africa. Rev. Sci. Technol. 14733-746. [DOI] [PubMed] [Google Scholar]
- 18.Diguimbaye-Djaibe, C., M. Hilty, R. Ngandolo, H. H. Mahamat, G. E. Pfyffer, F. Baggi, G. Hewinson, M. Tanner, J. Zinsstag, and E. Schelling. 2006. Mycobacterium bovis isolates from tuberculous lesions in Chadian zebu carcasses. Emerg. Infect. Dis. 12769-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte, E. L., M. Domingos, A. Amado, and A. Botelho. 2008. Spoligotype diversity of Mycobacterium bovis and Mycobacterium caprae animal isolates. Vet. Microbiol. 130415-421. [DOI] [PubMed] [Google Scholar]
- 20.Dvorská, L., M. Bartoš, G. Martin, W. Erler, and I. Pavlík. 2001. Strategies for differentiation, identification and typing of medically important species of mycobacteria by molecular methods. Vet. Med. (Prague) 46309-328. [Google Scholar]
- 21.Evans, J. T., E. G. Smith, A. Banerjee, R. M. Smith, J. Dale, J. A. Innes, D. Hunt, A. Tweddell, A. Wood, C. Anderson, R. G. Hewinson, N. H. Smith, P. M. Hawkey, and P. Sonnenberg. 2007. Cluster of human tuberculosis caused by Mycobacterium bovis: evidence for person-to-person transmission in the UK. Lancet 3691270-1276. [DOI] [PubMed] [Google Scholar]
- 22.Fang, Z., N. Morrison, B. Watt, C. Doig, and K. J. Forbes. 1998. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J. Bacteriol. 1802102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fokou, G., T. Haller, and J. Zinsstag. 2004. Identification of institutional factors affecting the well-being of sedentary and nomadic populations living in the Waza-Logone flood plain along the border between Cameroon and Chad. Med. Trop. 64464-468. [PubMed] [Google Scholar]
- 24.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 1441189-1196. [DOI] [PubMed] [Google Scholar]
- 25.Gagneux, S., K. Deriemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7328-337. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Pelayo, M. C., K. C. Caimi, J. K. Inwald, J. Hinds, F. Bigi, M. I. Romano, D. van Soolingen, R. G. Hewinson, A. Cataldi, and S. V. Gordon. 2004. Microarray analysis of Mycobacterium microti reveals deletion of genes encoding PE-PPE proteins and ESAT-6 family antigens. Tuberculosis 84159-166. [DOI] [PubMed] [Google Scholar]
- 28.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 1007877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 101057-1065. [DOI] [PubMed] [Google Scholar]
- 30.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 1621533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haddad, N., A. Ostyn, C. Karoui, M. Masselot, M. F. Thorel, S. L. Hughes, J. Inwald, R. G. Hewinson, and B. Durand. 2001. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 393623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanotte, O., D. G. Bradley, J. W. Ochieng, Y. Verjee, E. W. Hill, and J. E. Rege. 2002. African pastoralism: genetic imprints of origins and migrations. Science 296336-339. [DOI] [PubMed] [Google Scholar]
- 33.Hewinson, R. G., H. M. Vordermeier, N. H. Smith, and S. V. Gordon. 2006. Recent advances in our knowledge of Mycobacterium bovis: a feeling for the organism. Vet. Microbiol. 112127-139. [DOI] [PubMed] [Google Scholar]
- 34.Hilty, M., C. Diguimbaye, E. Schelling, F. Baggi, M. Tanner, and J. Zinsstag. 2005. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet. Microbiol. 109217-222. [DOI] [PubMed] [Google Scholar]
- 35.Huard, R. C., M. Fabre, P. de Haas, L. C. Lazzarini, D. van Soolingen, D. Cousins, and J. L. Ho. 2006. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J. Bacteriol. 1884271-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazwala, R. R., L. J. Kusiluka, K. Sinclair, J. M. Sharp, and C. J. Daborn. 2006. The molecular epidemiology of Mycobacterium bovis infections in Tanzania. Vet. Microbiol. 112201-210. [DOI] [PubMed] [Google Scholar]
- 38.Kilgour, V., and D. G. Godfrey. 1978. The influence of lorry transport on the Trypanosoma vivax infection rate in Nigerian trade cattle. Trop. Anim. Health Prod. 10145-148. [DOI] [PubMed] [Google Scholar]
- 39.Kubica, T., S. Rusch-Gerdes, and S. Niemann. 2003. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis-associated tuberculosis cases reported in Germany between 1999 and 2001. J. Clin. Microbiol. 413070-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1781274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malaga, W., P. Constant, D. Euphrasie, A. Cataldi, M. Daffe, J. M. Reyrat, and C. Guilhot. 2008. Deciphering the genetic bases of the structural diversity of phenolic glycolipids in strains of the Mycobacterium tuberculosis complex. J. Biol. Chem. 28315177-15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, A., P. Bonnet, D. Bourzat, R. Lancelot, and P. Souvenir. 1996. Importance of livestock production and its economic contribution to the countries of the Lake Chad Basin Commission, p. 79-96. In I. de Zborowski (ed.), Atlas d'élevage du Bassin du Lac Tchad. CITA, Wageningen, The Netherlands.
- 43.Meyer, C. G., G. Scarisbrick, S. Niemann, E. N. Browne, M. A. Chinbuah, J. Gyapong, I. Osei, E. Owusu-Dabo, T. Kubica, S. Rusch-Gerdes, T. Thye, and R. D. Horstmann. 2008. Pulmonary tuberculosis: virulence of Mycobacterium africanum and relevance in HIV co-infection. Tuberculosis 88482-489. [DOI] [PubMed] [Google Scholar]
- 44.Michel, A. L., T. M. Hlokwe, M. L. Coetzee, L. Mare, L. Connoway, V. P. Rutten, and K. Kremer. 2008. High Mycobacterium bovis genetic diversity in a low prevalence setting. Vet. Microbiol. 126151-159. [DOI] [PubMed] [Google Scholar]
- 45.Milian-Suazo, F., B. Harris, C. A. Diaz, C. Romero Torres, T. Stuber, G. A. Ojeda, A. M. Loredo, M. P. Soria, and J. B. Payeur. 2008. Molecular epidemiology of Mycobacterium bovis: usefulness in international trade. Prev Vet. Med. 87261-271. [DOI] [PubMed] [Google Scholar]
- 46.Mishra, G. S., and A. E. N′Depo. 1978. Cysticercus in animals slaughtered in the Port-Bouet abattoir (Abidjan). Rev. Elev. Med. Vet. Pays Trop. 31431-436. [DOI] [PubMed] [Google Scholar]
- 47.Mostowy, S., D. Cousins, and M. A. Behr. 2004. Genomic interrogation of the Dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J. Bacteriol. 186104-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mostowy, S., J. Inwald, S. Gordon, C. Martin, R. Warren, K. Kremer, D. Cousins, and M. A. Behr. 2005. Revisiting the evolution of Mycobacterium bovis. J. Bacteriol. 1876386-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller, B., B. Steiner, B. Bonfoh, A. Fane, N. H. Smith, and J. Zinsstag. 2008. Molecular characterisation of Mycobacterium bovis isolated from cattle slaughtered at the Bamako abattoir in Mali. BMC Vet. Res. 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narayanan, S., S. Gagneux, L. Hari, A. G. Tsolaki, S. Rajasekhar, P. R. Narayanan, P. M. Small, S. Holmes, and K. Deriemer. 2008. Genomic interrogation of ancestral Mycobacterium tuberculosis from south India. Infect. Genet. Evol. 8474-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newton, S. M., R. J. Smith, K. A. Wilkinson, M. P. Nicol, N. J. Garton, K. J. Staples, G. R. Stewart, J. R. Wain, A. R. Martineau, S. Fandrich, T. Smallie, B. Foxwell, A. Al-Obaidi, J. Shafi, K. Rajakumar, B. Kampmann, P. W. Andrew, L. Ziegler-Heitbrock, M. R. Barer, and R. J. Wilkinson. 2006. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc. Natl. Acad. Sci. USA 10315594-15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 412547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Njanpop-Lafourcade, B. M., J. Inwald, A. Ostyn, B. Durand, S. Hughes, M. F. Thorel, G. Hewinson, and N. Haddad. 2001. Molecular typing of Mycobacterium bovis isolates from Cameroon. J. Clin. Microbiol. 39222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oloya, J., R. Kazwala, A. Lund, J. Opuda-Asibo, B. Demelash, E. Skjerve, T. B. Johansen, and B. Djonne. 2007. Characterisation of mycobacteria isolated from slaughter cattle in pastoral regions of Uganda. BMC Microbiol. 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parra, A., J. Larrasa, A. Garcia, J. M. Alonso, and J. H. de Mendoza. 2005. Molecular epidemiology of bovine tuberculosis in wild animals in Spain: a first approach to risk factor analysis. Vet. Microbiol. 110293-300. [DOI] [PubMed] [Google Scholar]
- 56.Rasolofo Razanamparany, V., R. Quirin, A. Rapaoliarijaona, H. Rakotoaritahina, E. J. Vololonirina, T. Rasolonavalona, S. Ferdinand, C. Sola, N. Rastogi, H. Ramarokoto, and S. Chanteau. 2006. Usefulness of restriction fragment length polymorphism and spoligotyping for epidemiological studies of Mycobacterium bovis in Madagascar: description of new genotypes. Vet. Microbiol. 114115-122. [DOI] [PubMed] [Google Scholar]
- 57.Renwick, A. R., P. C. White, and R. G. Bengis. 2007. Bovine tuberculosis in southern African wildlife: a multi-species host-pathogen system. Epidemiol. Infect. 135529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rigouts, L., B. Maregeya, H. Traore, J. P. Collart, K. Fissette, and F. Portaels. 1996. Use of DNA restriction fragment typing in the differentiation of Mycobacterium tuberculosis complex isolates from animals and humans in Burundi. Tuber. Lung Dis. 77264-268. [DOI] [PubMed] [Google Scholar]
- 59.Serraino, A., G. Marchetti, V. Sanguinetti, M. C. Rossi, R. G. Zanoni, L. Catozzi, A. Bandera, W. Dini, W. Mignone, F. Franzetti, and A. Gori. 1999. Monitoring of transmission of tuberculosis between wild boars and cattle: genotypical analysis of strains by molecular epidemiology techniques. J. Clin. Microbiol. 372766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, N. H., J. Dale, J. Inwald, S. Palmer, S. V. Gordon, R. G. Hewinson, and J. M. Smith. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. USA 10015271-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, N. H., S. V. Gordon, R. de la Rua-Domenech, R. S. Clifton-Hadley, and R. G. Hewinson. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4670-681. [DOI] [PubMed] [Google Scholar]
- 62.Smith, N. H., K. Kremer, J. Inwald, J. Dale, J. R. Driscoll, S. V. Gordon, D. van Soolingen, R. G. Hewinson, and J. M. Smith. 2006. Ecotypes of the Mycobacterium tuberculosis complex. J. Theor. Biol. 239220-225. [DOI] [PubMed] [Google Scholar]
- 63.Tadayon, K., N. Mosavari, A. H. Shahmoradi, F. Sadeghi, A. Azarvandi, and K. Forbes. 2006. The epidemiology of Mycobacterium bovis in buffalo in Iran. J. Vet. Med. B 53(Suppl. 1)41-42. [Google Scholar]
- 64.Thoen, C., P. Lobue, and I. de Kantor. 2006. The importance of Mycobacterium bovis as a zoonosis. Vet. Microbiol. 112339-345. [DOI] [PubMed] [Google Scholar]
- 65.Thoen, C. O., J. Steele, and M. J. Gilsdorf. 2006. Mycobacterium bovis infection in animals and humans, 2nd ed. Blackwell Publishing Ltd., Oxford, United Kingdom.
- 66.Tsolaki, A. G., S. Gagneux, A. S. Pym, Y. O. Goguet de la Salmoniere, B. N. Kreiswirth, D. Van Soolingen, and P. M. Small. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 433185-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Zanden, A. G., A. H. Hoentjen, F. G. Heilmann, E. F. Weltevreden, L. M. Schouls, and J. D. van Embden. 1998. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol. Pathol. 51209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van Der Zeijst, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 1822393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 404457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanella, G., B. Durand, J. Hars, F. Moutou, B. Garin-Bastuji, A. Duvauchelle, M. Ferme, C. Karoui, and M. L. Boschiroli. 2008. Mycobacterium bovis in wildlife in France. J. Wildl. Dis. 4499-108. [DOI] [PubMed] [Google Scholar]
- 71.Zumarraga, M. J., C. Martin, S. Samper, A. Alito, O. Latini, F. Bigi, E. Roxo, M. E. Cicuta, F. Errico, M. C. Ramos, A. Cataldi, D. van Soolingen, and M. I. Romano. 1999. Usefulness of spoligotyping in molecular epidemiology of Mycobacterium bovis-related infections in South America. J. Clin. Microbiol. 37296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


