Abstract
FimH is an adhesive subunit of type 1 fimbriae expressed by different enterobacterial species. The enteric bacterium Klebsiella pneumoniae is an environmental organism that is also a frequent cause of sepsis, urinary tract infection (UTI), and liver abscess. Type 1 fimbriae have been shown to be critical for the ability of K. pneumoniae to cause UTI in a murine model. We show here that the K. pneumoniae fimH gene is found in 90% of strains from various environmental and clinical sources. The fimH alleles exhibit relatively low nucleotide and structural diversity but are prone to frequent horizontal-transfer events between different bacterial clones. Addition of the fimH locus to multiple-locus sequence typing significantly improved the resolution of the clonal structure of pathogenic strains, including the K1 encapsulated liver isolates. In addition, the K. pneumoniae FimH protein is targeted by adaptive point mutations, though not to the same extent as FimH from uropathogenic Escherichia coli or TonB from the same K. pneumoniae strains. Such adaptive mutations include a single amino acid deletion from the signal peptide that might affect the length of the fimbrial rod by affecting FimH translocation into the periplasm. Another FimH mutation (S62A) occurred in the course of endemic circulation of a nosocomial uropathogenic clone of K. pneumoniae. This mutation is identical to one found in a highly virulent uropathogenic strain of E. coli, suggesting that the FimH mutations are pathoadaptive in nature. Considering the abundance of type 1 fimbriae in Enterobacteriaceae, our present finding that fimH genes are subject to adaptive microevolution substantiates the importance of type 1 fimbria-mediated adhesion in K. pneumoniae.
Klebsiella pneumoniae is recognized as an important opportunistic pathogen that frequently causes urinary tract infections (UTI), septicemia, or pneumonia, particularly in immunocompromised individuals (25). K. pneumoniae is responsible for up to 10% of all nosocomial bacterial infections (12, 35). In recent years, a high incidence of community-acquired K. pneumoniae pyogenic liver abscess with a high mortality rate has been reported, especially from Taiwan, but also from other Asian countries, Europe, and North America (6, 8, 19, 27, 44). Furthermore, 15% to 30% of K. pneumoniae isolates are resistant to broad-spectrum cephalosporins via plasmid-encoded extended-spectrum β-lactamases (5).
In contrast to many other bacterial pathogens, K. pneumoniae is ubiquitous in nature. Its nonclinical habitats include environmental locations, such as vegetation, soil, and surface waters, as well as transient commensal colonization of mucosal surfaces in humans and other animals (1). Several studies have reported K. pneumoniae isolates of environmental origin to be nearly identical to clinical isolates with respect to several phenotypic properties (16, 22, 23, 25, 30). It has been suggested that environmental isolates of K. pneumoniae may be as virulent as clinical isolates (24, 39).
Several virulence factors have been identified in K. pneumoniae (25, 38). The prominent polysaccharide capsule expressed by most isolates, together with the lipopolysaccharide layer, protects the bacteria against phagocytosis and the bactericidal activity of serum. Fimbrial adhesins expressed by the bacteria are protein structures able to recognize molecular receptors and to facilitate adherence to specific tissue surfaces in the host. K. pneumoniae produces two major fimbrial adhesion organelles, type 1 and type 3 fimbriae (9). Type 1 fimbriae have mannose-sensitive hemagglutinins, while type 3 fimbriae have mannose-resistant hemagglutinins (21).
Type 1 fimbriae are the most common adhesive organelle in Enterobacteriaceae and have been most extensively studied in Escherichia coli. The type 1 fimbrial structures of K. pneumoniae are homologous to those of E. coli with regard to genetic composition and regulation (37). Type 1 fimbriae and the adhesive subunit FimH, in particular, play an important role in UTI caused by both K. pneumoniae and E. coli (3, 15, 17, 30, 37). Analysis of E. coli fimH variation at the population level has revealed that the FimH adhesin in urinary E. coli isolates accumulates amino acid replacements that increase its tropism toward the uroepithelium and various components of basement membranes (14, 26, 31, 33, 46). Most of the replacements increase the monomannose binding capability of FimH under low shear by altering allosteric catch bond properties of the protein (40). The natural FimH mutants were shown to provide an advantage in colonization of the urinary tract in a mouse model (32) and correlate with the overall extraintestinal virulence of E. coli (11). Thus, FimH mutations are pathoadaptive in nature. No such population-wide analysis has been performed for K. pneumoniae fimH.
Population genetic analysis involves comparison of the nucleotide and structural variability of the locus of interest across multiple bacterial strains of different clonalities and geographic origins. The clonal structure of the strains can be determined by multiple-locus sequence typing (MLST), in which 400- to 500-bp sequences of multiple genetically unlinked loci are determined in order to define the phylogenetic relationship of the strains and the extent of interclonal gene recombination (horizontal gene transfer). MLST has been used to reveal the epidemiological relationship of ceftazidime- and ciprofloxacin-resistant K. pneumoniae isolates of nosocomial origin (4). In addition, the analysis of gene variability enables the determination of the type of selection processes acting on loci of interest, with possible identification of mutational changes of functional significance that could enhance the organism's ability to cause disease, i.e., that could be of a pathoadaptive nature.
In this study, the population dynamics of the K. pneumoniae FimH adhesin were determined by analysis of fimH allelic diversity in strains of environmental and various clinical origins in the context of K. pneumoniae clonal structure based on the allelic diversity of three loci—tonB, mdh and fumC—commonly used for MLST.
MATERIALS AND METHODS
Bacterial isolates.
The collection under study consisted of 65 K. pneumoniae isolates collected from 1990 through 2006 (Table 1). The environmental isolates (n = 10) originated from various streams and lakes and the Baltic Sea in the area of Schleswig-Holstein, Germany (24). The liver isolates (n = 12) originated from patients in Canada and Seattle, WA, with liver abscess. The urinary tract isolates (n = 21) originated from various hospitals, foster homes, and general practitioners in Denmark (gifts of Niels Frimodt-Møller, Statens Serum Institut [SSI]), apart from the UTI isolate K. pneumoniae C3091, which came from Washington, DC (18). The bloodstream isolates (n = 20) originated from patients suffering from bacteremia in hospitals in the Copenhagen area in Denmark between 1990 and 1992 (10). Also included in the study for comparison was the reference strain MGH78578 (ATCC 700721; GenBank) and K. pneumoniae strain 342 (7).
TABLE 1.
Strains used in the study
| Strain | Isolation sourcea | Dateb | Place of isolationc | Capsule serotyped | fimHe | Reference or sourcef |
|---|---|---|---|---|---|---|
| C3091 | U | 1997 | DC, USA | 16 | + | 18 |
| 3824 | L | 9/10/2004 | Sea, USA | 1 | + | UW |
| 3857 | L | 5/13/2005 | Sea, USA | NT | NO | UW |
| 3858 | L | 5/13/2005 | Sea, USA | 1 | + | UW |
| 3859 | L | 5/13/2005 | Sea, USA | 1 | + | UW |
| 3860 | L | 5/13/2005 | Sea, USA | 1 | + | UW |
| 3861 | L | 5/13/2005 | Sea, USA | 2 | + | UW |
| 3928 | L | 11/15/2005 | CAN | 2 | + | UW |
| 3950 | L | 12/2/2005 | CAN | 1 | + | UW |
| 3951 | L | 12/2/2005 | CAN | 1 | + | UW |
| 4041 | L | 6/14/2006 | CAN | 1 | + | UW |
| 4121 | L | 12/9/2006 | CAN | 1 | + | UW |
| 4133 | L | 12/22/2006 | CAN | 1 | + | UW |
| sp3 | B | 1990-1992 | Cph, DK | 48 | + | 10 |
| sp7 | B | 1990-1992 | Cph, DK | 27 | + | 10 |
| sp10 | B | 1990-1992 | Cph, DK | 43 | + | 10 |
| sp13 | B | 1990-1992 | Cph, DK | 24 | + | 10 |
| sp14 | B | 1990-1992 | Cph, DK | 31 | + | 10 |
| sp15 | B | 1990-1992 | Cph, DK | 74 | + | 10 |
| sp19 | B | 1990-1992 | Cph, DK | 6 | + | 10 |
| sp20 | B | 1990-1992 | Cph, DK | 3 | + | 10 |
| sp22 | B | 1990-1992 | Cph, DK | 29 | + | 10 |
| sp25 | B | 1990-1992 | Cph, DK | 21 | NO | 10 |
| sp28 | B | 1990-1992 | Cph, DK | 68 | NO | 10 |
| sp29 | B | 1990-1992 | Cph, DK | 1 | + | 10 |
| sp30 | B | 1990-1992 | Cph, DK | 52 | + | 10 |
| sp31 | B | 1990-1992 | Cph, DK | 61 | + | 10 |
| sp32 | B | 1990-1992 | Cph, DK | 26 | NO | 10 |
| sp33 | B | 1990-1992 | Cph, DK | 31 | NO | 10 |
| sp34 | B | 1990-1992 | Cph, DK | 18 | + | 10 |
| sp37 | B | 1990-1992 | Cph, DK | 62 | + | 10 |
| sp39 | B | 1990-1992 | Cph, DK | 31 | + | 10 |
| sp41 | B | 1990-1992 | Cph, DK | 64 | + | 10 |
| cas119 | W | 1997-1998 | DE | NA | + | 24 |
| cas120 | W | 1997-1998 | DE | 23 | + | 24 |
| cas121 | W | 1997-1998 | DE | 60 | + | 24 |
| cas122 | W | 1997-1998 | DE | NA | + | 24 |
| cas123 | W | 1997-1998 | DE | NA | + | 24 |
| cas124 | W | 1997-1998 | DE | 19 | NO | 24 |
| cas125 | W | 1997-1998 | DE | NA | + | 24 |
| cas126 | W | 1997-1998 | DE | 1 | + | 24 |
| cas127 | W | 1997-1998 | DE | 40 | + | 24 |
| cas128 | W | 1997-1998 | DE | NA | + | 24 |
| cas663 | U | 1/24/2006 | Nyk, DK | NA | + | SSI |
| cas664 | U | 1/30/2006 | Cph, DK | NA | + | SSI |
| cas665 | U | 2/1/2006 | Cph, DK | NA | + | SSI |
| cas666 | U | 2006 | Cph, DK | NA | + | SSI |
| cas667 | U | 2/14/2006 | Cph, DK | NA | NO | SSI |
| cas668 | U | 2006 | DK | NA | + | SSI |
| cas669 | U | 2/28/2006 | Vib, DK | NA | + | SSI |
| cas670 | U | 3/2/2006 | DK | NA | + | SSI |
| cas671 | U | 3/10/2006 | DK | NA | + | SSI |
| cas672 | U | 3/14/2006 | Cph, DK | NA | + | SSI |
| cas673 | U | 3/27/2006 | Aar, DK | NA | + | SSI |
| cas674 | U | 3/28/2006 | DK | NA | + | SSI |
| cas675 | U | 3/17/2006 | Cph, DK | NA | + | SSI |
| cas676 | U | 3/22/2006 | Nyk, DK | NA | + | SSI |
| cas677 | U | 2006 | DK | NA | + | SSI |
| cas678 | U | 4/3/2006 | Cph, DK | NA | + | SSI |
| cas679 | U | 4/4/2006 | Cph, DK | NA | + | SSI |
| cas680 | U | 5/6/2006 | Cph, DK | NA | + | SSI |
| cas681 | U | 5/4/2006 | Nyk, DK | NA | + | SSI |
| cas682 | U | 2006 | DK | NA | NO | SSI |
| Kp342 | P | 2000 | USA | 33 | + | 7 |
| MGH78578 | S | 1994 | USA | 52 | + | (G) |
L, Liver; U, urinary tract, B, blood; W, water; P, plant; S, sputum.
Dates are in one of the following formats: year(s) or month/day/year.
Aar, Aarhus; Cph, Copenhagen; Nyk, Nykøbing Falster; Vib, Viborg; DK, Denmark; DE, Schleswig-Holstein, Germany; Sea, Seattle; CAN, Canada.
NT, not typeable; NA, not applicable.
+, present; NO, absent.
UW, University of Washington strain collection; SSI, Statens Serom Institut; (G), ATCC 700721, GenBank.
Isolates were identified as K. pneumoniae by standard biochemical and MINIBACT E assays (developed by Nissen B., Statens Serum Institut, no. 905 and 906). Serotyping was performed by the International Escherichia coli and Klebsiella Reference Centre (WHO), SSI, Denmark, as described by Ørskov and Ørskov (20).
Detection of the magA (wzy) gene was carried out as previously described (6) with the magA-specific primer pairs 5′ GGTGCTCTTTACATCATTGC plus 5′ GCAATGGCCATTTGCGTTAG and 5′ CGCCGCAAATACGAGAAGTG plus 5′ GCAATCGAAGTGAAGAGTGC, used to amplify 1,282-bp and 540-bp fragments of the magA gene (GenBank AB355924), respectively.
Sequence analysis.
Sequences of the type 1 fimbrial adhesin (fimH) subunit were obtained from 54 isolates. Three housekeeping locus fragments (fumC, mdh, and tonB) were also sequenced for these 54 isolates. The primers used for amplification were fimH forward (5′ CACGCAAGGCACCATTC) and reverse (5′ GCTCAGAATCAACATCGGTAAC), fumC forward (5′ TCCACAGTCGCCAACGCTTC) and reverse (5′ GCATCGCAGTGAAAAAGACTC), and mdh forward (5′ ATGAAAGTTGCAGTCCTTGGCGCCGCCGGTGG) and reverse (5′ TTGACGAAGTCTTCGCCGAGCGCGATATCTTTCT).
Primers for tonB were obtained from the K. pneumoniae MLST website (http://pubmlst.org/kpneumoniae/) and were as follows: forward, 5′ CTTTATACCTCGGTACATCAGGTT, and reverse, 5′ ATTCGCCGGCTGRGCRGAGAG.
Electropherograms for all singleton mutations were visually inspected for consistency between strands, and any ambiguous nucleotides were resolved by resequencing.
Nucleotide sequences were aligned using ClustalW and ClustalX (http://www.ebi.ac.uk/Tools/clustalw/index.html); phylogenetic trees were built using ClustalX 1.83 (41) and were viewed using Tree View (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Nucleotide sequence accession numbers.
All sequences obtained in this study were deposited in GenBank under accession numbers FJ483550 through FJ483756.
RESULTS
Allelic diversity of fimH.
The fimH gene could be amplified for 57 of 65 (87.5%) isolates, including 19 out of 21 urinary tract isolates, 16 out of 20 bloodstream isolates, 11 out of 12 liver isolates, and 9 out of 10 environmental isolates (Table 1). Reliable sequence data were obtained for 54 isolates. Sequencing of fimH revealed a total of 18 unique alleles, with a pairwise nucleotide diversity (π) of 1.9% ± 0.3% and a >20-fold-higher rate of synonymous mutations (dS) over nonsynonymous mutations (dN) (Table 2). The levels and patterns of diversity of fimH did not differ significantly between isolates of different origins (not shown).
TABLE 2.
Overall nucleotide diversity and rates of synonymous and nonsynonymous variations in mdh, fumC, tonB, and fimH genes (or gene fragments) from 54 strains of K. pneumoniae
| Gene | Lengtha (bp) | No. of isolates | No. of alleles | π/nucleotide | dS | dN | dN/dS |
|---|---|---|---|---|---|---|---|
| mdh | 450 | 54 | 11 | 0.042 ± 0.007 | 0.172 | 0.006 | 0.035 |
| fumC | 465 | 54 | 13 | 0.046 ± 0.005 | 0.226 | 0.017 | 0.075 |
| tonB | 399 | 54 | 24 | 0.050 ± 0.006 | 0.188 | 0.014 | 0.075 |
| fimH | 903 | 54 | 19 | 0.019 ± 0.003 | 0.069 | 0.003 | 0.044 |
The length is indicated for regions used for the sequence analysis.
Internal regions (399 to 465 bp in length) of three additional genes were amplified: mdh, encoding malate dehydrogenase (total length, 939 bp); fumC, for fumarase (total length, 1,401 bp); and tonB, for the outer membrane porin protein (total length, 747 bp). The pairwise nucleotide variabilities of mdh (4.2% ± 0.7%), fumC (4.6% ± 0.5%), and tonB (5.0% ± 0.6%) were more than twofold higher than that of fimH. Most of the increase in π of non-fimH loci was due to dS values higher than those for fimH. However, the dS was significantly (15- to 50-fold) higher than the dN for all four loci.
Clonal congruence of fimH alleles.
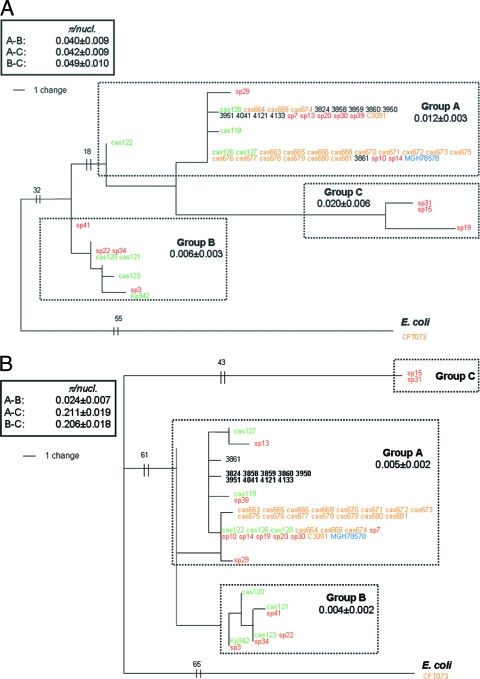
Based on the DNA sequences of four loci, phylogenetic trees were constructed as phylograms rooted with a homologous sequence of E. coli. According to trees formed by fumC, mdh, and tonB, K. pneumoniae isolates split into three distinct phylogenetic clades (groups): A, B, and C (Fig. 1A, B, C, and D). The groups formed by different loci were highly congruent with each other, with a single exception: the fumC allele of strain sp19 shifted from group C to group A. The nucleotide diversity within the groups was between 0.5% and 2.0%, which is at the level of within-species diversity found in many bacteria. Diversity between the groups (measured using the nodes that were phylogenetically closest to each other) was significantly higher, corresponding to the diversity seen between (sub)species. Thus, the A, B, and C groups represent distinct clonal groups of K. pneumoniae, possibly at the level of subspecies, with very limited recombination between groups in the fumC, mdh, and tonB regions.
FIG. 1.
Phylogenetic trees of mdh (A), fumC (B), tonB (C), and fimH (D) genes of K. pneumoniae, rooted with the corresponding gene of E. coli CFT073. Internal fragments of 450 bp, 465 bp, and 399 bp were analyzed for mdh, fumC, and tonB genes, respectively. Different colors are used for strains from different origins of isolation: orange for UTI, red for sepsis, green for environment, black for liver, and blue for sputum. Each phylogenetic group is blocked by a dotted rectangle showing the average nucleotide diversity (π) within the group. The π values in solid rectangles denote between-group (A-B, A-C, or B-C) values for the phylogenetically closest pair of strains. In fimH (D), the asterisks along the branches denote the deletion of codon 12 (i.e., CTG in group A or TTG in group B) in the corresponding strains.
In contrast, there were only two distinct groups formed on the fimH tree, as group C merged with group A and alleles from group A strains cas119, cas122, and cas128 formed a distinct group with strains from group B. Thus, the clonal structure of K. pneumoniae based on fimH alleles is less diverse than that seen with three other genes, providing evidence for relatively frequent recombination (horizontal transfer) between groups.
Clonal diversity of K. pneumoniae isolates of different origins.
Different gene sequences from the same isolates were concatenated with each other, and a single MLST tree was built (Fig. 2). A total of 34 unique MLST clones were identified, with distinct differences in the clonal distributions of strains from different sources.
FIG. 2.
Phylogenetic tree, constructed by concatenation of the 54 isolates, showing a total of 34 unique MLST alleles. Different colors are used for strains from different origins of isolation: orange for UTI, red for sepsis, green for environment, black for liver, and blue for sputum.
Environmental isolates.
Environmental isolates were represented across different tree clades, indicating extensive clonal diversity in the K. pneumoniae isolates of environmental origin. The least diverse locus in the environmental isolates was fimH; four isolates from group B that differed in other loci had identical fimH alleles (Fig. 1D), supporting frequent horizontal transfer of this gene both between and within the clonal groups.
Sepsis isolates.
Twenty clinical isolates of bloodstream origin (sepsis isolates) were also highly diverse and carried 15 unique MLST types. While most sepsis isolates were unique by MLST, one (isolate sp3) had an MLST profile identical to that of an environmental isolate (reference strain Kp342). Horizontal transfer of fimH was implied by the fact that five isolates with identical mdh and fumC alleles (sp7, sp13, sp20, sp30, and sp39) had distinct, phylogenetically unlinked fimH alleles. Interestingly, tonB was also variable among these five isolates, indicating that tonB and fimH alike are subject to frequent horizontal transfer (by independent recombination events) during clone diversification.
Liver isolates.
K. pneumoniae liver isolates exhibited significantly less diversity than environmental and sepsis isolates in all alleles, with eight isolates forming a single clone within clonal group A. Only one liver isolate, 3861, had a completely different profile in all four alleles, but this strain also belonged to phylogenetic group A. Another liver isolate, 3859, had tonB, mdh, and fumC sequences identical to those of the main clone but a completely different fimH allele, providing additional support for horizontal transfer of the last gene. The presence of magA is indicative of the K1 serotype (36). Serotyping and detection of the magA gene revealed that 9 out of 11 liver isolates belonged to the K1 serotype while the remaining two (3861 and the partially sequenced strain 3928) belonged to the K2 serotype (Table 1).
Urinary isolates.
Finally, the 19 K. pneumoniae isolates of urinary origin also displayed relatively low diversity. All isolates belonged to phylogenetic group A, with 13 isolates having identical MLST profiles. One isolate, cas673, had fumC and mdh identical to those of the large 13-isolate clone but distinct tonB and fimH alleles, again consistent with relatively frequent horizontal transfer of the last two genes. This was further supported by another group of four urinary isolates—cas664, cas669, cas674, and C3091—with identical mdh and fumC sequences and variation in tonB and fimH. Yet another isolate, cas665, had all loci identical to those of the large 13-isolate clone but had a single-nucleotide difference in fimH, suggesting the recent acquisition of a fimH point mutation.
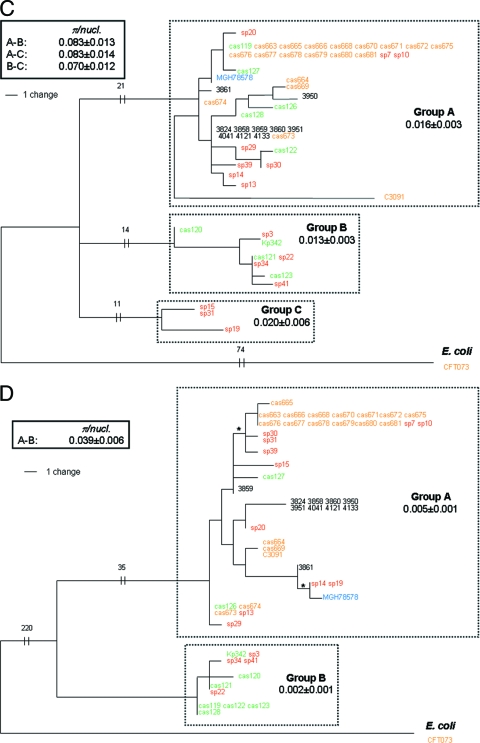
Structural variability of FimH.
The structural variability of the FimH protein within K. pneumoniae isolates was analyzed by constructing a phylogenetic tree of nascent FimH protein variants (Fig. 3D) by collapsing silent changes along the branches of a DNA-based fimH tree (Fig. 1D). In this way, independent structural mutations in the same position (“hot-spot” mutations) could be detected. The protein tree of nascent FimH contained seven protein variant nodes formed by nine independently occurring mutations, including eight amino acid replacements and one deletion of a single amino acid. When K. pneumoniae FimH was aligned with FimH of E. coli (∼85% homologous at the structural level), three of these mutations occurred in the leader peptide, five in the lectin domain, and two in the pilin domain regions defined in E. coli FimH (Fig. 4). The only repeated mutation was a single amino acid deletion, L(−12), in the leader peptide. The DNA phylogeny of fimH (Fig. 1D) suggests that two deletions were acquired on independent occasions. It was shown previously that E. coli FimH is targeted by a variety of point mutations occurring under positive selection for pathoadaptive functional changes. Among the K. pneumoniae FimH variants, one mutation, Ser to Ala in position 62 of the mature peptide, has been found to occur in FimH variants of E. coli (34, 47) belonging to a highly pathogenic clonal group of extraintestinal strains (13). None of the other mutations found in this study has, to our knowledge, been detected in E. coli.
FIG. 3.
Protein phylograms of mdh (A), fumC (B), tonB (C) and fimH (D) genes of K. pneumoniae derived from the corresponding DNA trees by collapsing the branches carrying only synonymous variations. Structural mutations are shown along the branches. Structural mutations [or L(−12) deletion in panel A for FimH] that occurred multiple times across independent lineages of DNA phylogeny are termed hot-spot mutations and are shown as two groups (as in Fig. 1D). Different colors are used for strains from different origins of isolation: orange for UTI, red for sepsis, green for environment, black for liver, and blue for sputum.
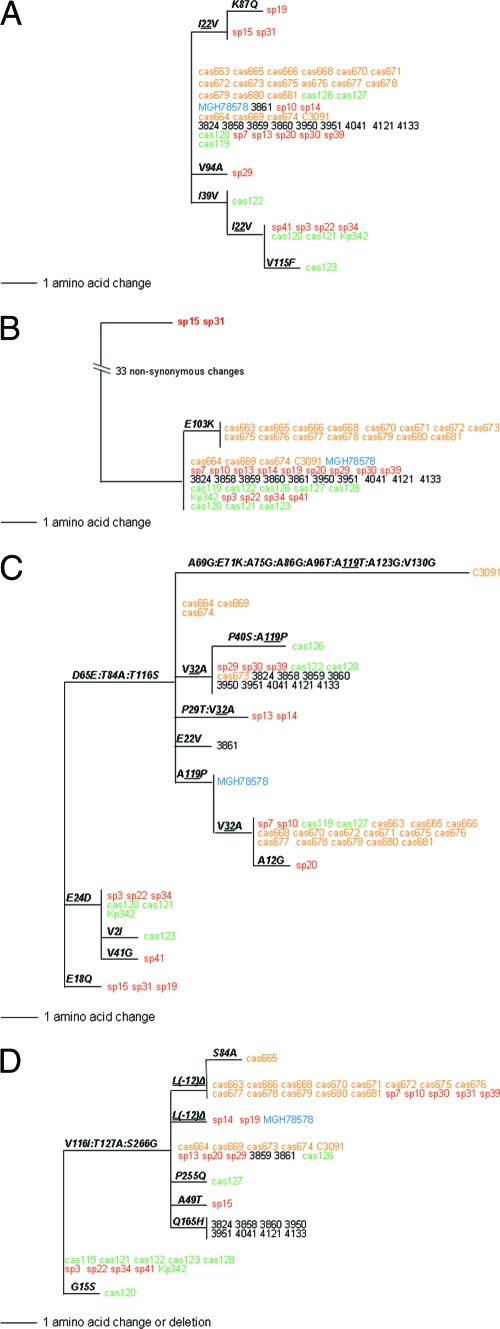
FIG. 4.
Amino acid polymorphisms in the FimH proteins of K. pneumoniae structural variants and E. coli CFT073, showing domain borders of the E. coli FimH protein, which are as follows: in amino acid positions 1 to 21, the leader peptide; in positions 22 to 177, the lectin domain; in positions 178 to 180, the linker chain; and in positions 181 to 300, the pilin domain. The dots correspond to identical amino acids relative to the consensus FimH of K. pneumoniae (represented by cas664). The numbers above the sequences indicate corresponding positions of the structural mutations. The numbers in parentheses correspond to the number of isolates carrying the FimH variant.
The structural diversity of the Mdh and FumC protein regions was also relatively low (Fig. 3A and B), with no mutational hot spots. In contrast, the structural variability of the tonB region was significantly higher than in FimH (Fig. 3C and D), with several hot-spot mutations, likely reflecting positive selection for the structural variability of TonB, as described previously in E. coli (43).
DISCUSSION
In this study, our main observations were that K. pneumoniae fimH (i) has relatively low nucleotide diversity, (ii) moves horizontally across clonal groups (subspecies), (iii) contributes substantially to the diversification of K. pneumoniae clones, and (iv) has acquired a pathoadaptive mutation during endemic circulation of a large clone that causes nosocomial UTI.
At the population level, fimH was analyzed in a group of 65 K. pneumoniae isolates of diverse origins. The fimH locus could be amplified from almost 90% of the isolates, indicating the ubiquitous nature of type 1 fimbriae in K. pneumoniae. The diversity of fimH was compared to those of three other genes from the same isolates—mdh, fumC, and tonB. Both mdh and fumC are housekeeping loci, commonly used for MLST (4, 42, 47) because their variation is considered to be of a neutral nature due to high functional and structural conservation of the encoded proteins. The third locus, tonB, has been used for MLST (4, 42), although there is evidence in E. coli that tonB is under positive selection for structural variability (2). The variability of fimH is two- to threefold lower than that of the housekeeping and tonB loci, primarily due to a lower level of silent changes. This trend might indicate the relatively late evolutionary acquisition of the type 1 fimbrial operon by K. pneumoniae subspecies or homogenization of alleles due to selective sweeps, i.e., a periodic expansion of specific lineages across many habitats of the species. However, the fimH diversity pattern is generally similar to that of other K. pneumoniae genes, with a predominance of silent changes over replacement mutations. The K. pneumoniae fimH nucleotide diversity of 1.8% ± 0.06% is similar to that reported previously for E. coli, where nucleotide diversity is 1.64% ± 0.07%, and the dN/dS ratio is also significantly less than 1 (0.004/0.052 = 0.077) (34).
The phylogeny of fimH is less diverse and exhibits limited congruence with other loci, suggesting that fimH is prone to relatively frequent horizontal transfer within or between clonal groups of K. pneumoniae. Based on the level of between-group diversity, these clonal groups appear to represent distinct subspecies of K. pneumoniae. Frequent horizontal transfer of fimH has been reported for E. coli and is thought to be driven by strong diversifying selection on fimA (47), which encodes the major subunit of type 1 fimbriae, a major surface antigen. In contrast, the housekeeping loci mdh and fumC were, in general, phylogenetically congruent and thus appeared less prone to horizontal transfer. However, tonB exhibited signs of horizontal transfer, likely due to surface expression that subjects it to immune pressure for structural diversification.
The overall clonal diversity of K. pneumoniae obtained here, based on four loci, is comparable to the diversity obtained previously with MLST based on seven loci that included six housekeeping genes (rpoB, gapA, mdh, pgi, phoE, and infB), as well as tonB (4). Under our scheme, 34 clones with unique MLST types were identified among 54 isolates, while the seven-locus scheme identified 40 unique MLST clones among 67 isolates. The comparable clonal diversity identified in our study, which used only four loci, shows the advantage of adding fimH to the MLST scheme, the frequent horizontal transfer of which leads to the genetic diversification of K. pneumoniae populations. The role of fimH in clonal diversification is also evident in the analysis of clonal diversity of K. pneumoniae isolates from different sources, where fimH was a key locus in the diversification of several otherwise identical clones.
Among K. pneumoniae isolates from different sources, environmental isolates were the most diverse, with all isolates exhibiting unique MLST profiles and with multiple loci contributing to diversity. This is likely due to the fact that the environment (water, soil, and plants) is the main natural reservoir of K. pneumoniae, where clonal population diversity has been accumulating evolutionarily for a long time. The diversity of sepsis isolates tested was also high, in agreement with a previous study that grouped 43 sepsis isolates into 29 district sequence types by seven-gene MLST, suggesting that the ability to cause sepsis is a relatively common property of K. pneumoniae isolates. Further, it has been demonstrated that isolates of environmental origin are equal in virulence to isolates of clinical origin (39). This is supported by our observation that isolate Kp342, isolated from a plant and known to play a major role in nitrogen fixation (7), exhibited 100% similarity in all four loci to that of sp3, a sepsis isolate (Fig. 2).
In contrast, isolates from patients with liver abscesses are closely related. The earliest reports of a distinctive syndrome of community-acquired K. pneumoniae septicemia with liver abscess came from Taiwan in the 1980s (45). In 2007, it was suggested that isolates causing liver abscesses were members of a common clonal population, even though these strains were isolated on three different continents (42). Furthermore, similar to our findings, several reports have demonstrated that isolates causing liver abscesses in the majority of cases are serotypes K1 and K2 (8), representing only a small fraction of the 77 different serotypes known in K. pneumoniae (20). Still, the fimH gene from one of the isolates tested here (3859) was sufficiently different from those of the other liver abscess isolates to indicate its horizontal transfer. Thus, liver abscess isolates of K. pneumoniae have undergone some population diversification. It is possible either that the clone has been in circulation for a relatively long time or that pressure to exchange the type 1 fimbrial cluster is very high. Interestingly, isolates sp29 from a patient with sepsis and cas126 from the environment both displayed K1 serotypes. Both isolates belong to group A, according to all four genes, but are not phylogenetically linked to the eight liver isolates or to each other. Of the isolates in which all four genes were obtained, isolate 3861 was the only liver isolate of serotype K2, consistent with the absence of the magA locus by PCR amplification.
Urinary isolates of K. pneumoniae also exhibited low diversity, i.e., they were highly clonal in nature. This may be due in part to the similar geographical origins of these isolates, with 20 of 21 collected from hospitals and foster care homes in Denmark. However, except for the tonB locus, isolate C3091, collected in Washington, DC (18), is grouped with UTI isolates from Denmark, suggesting that even geographically unlinked urinary isolates of K. pneumoniae may be closely related. Also, the existence of the 13-isolate group, with identical MLST types isolated from patients in different locations, strongly suggests that this clone is involved in stable endemic circulation as a uropathogen among susceptible individuals in Denmark. Obtaining additional uropathogenic strains from more diverse geographic origins should provide a more definitive conclusion about the clonal diversity of uropathogenic K. pneumoniae and the extent of endemic circulation of uropathogenic strains.
Considering the high clonal diversity and prevalence of K. pneumoniae isolates of environmental and sepsis origin, the clonal nature of the liver and uropathogenic isolates demonstrates the importance of certain clone-specific traits in K. pneumoniae pathogenicity. This allows future investigations to focus on comparative analysis of clinically distinctive clones in elucidating the molecular basis of K. pneumoniae pathogenesis.
Another interesting phenomenon observed in the endemic uropathogenic clone is fimH diversification, which occurred in isolate cas665 through point mutation (S62A), rather than horizontal transfer. In E. coli, the same mutation is proposed to be acquired under positive selection in uropathogenic isolates (34), where A62 FimH variants demonstrate increased mannose-binding capability under static or low-shear conditions, increased urinary epithelium tropism, and enhanced urovirulence (32). Indeed, when the mannose-binding capabilities of K. pneumoniae FimH with and without mutation A62 were evaluated, the mutant variant exhibited dramatically increased binding, suggesting that S62A has been acquired in K. pneumoniae under positive selection (S. G. Stahlhut et al., submitted for publication). To our knowledge, this is the first evidence of pathoadaptive diversification of uropathogenic bacteria that has been detected in the course of endemic circulation.
Aside from A62, no other mutations in the mature FimH peptide have been found in K. pneumoniae that parallel the pathoadaptive mutations in FimH of E. coli. In E. coli FimH, about 50 different mutations have been characterized thus far, with over half occurring in hot-spot positions, i.e., at specific amino acid residues (34). Thus, K. pneumoniae FimH appears to be under less positive selection for the acquisition of pathoadaptive changes. This is possibly due to the fact that K. pneumoniae is primarily an environmental species that does not circulate continuously among the human population, in contrast to E. coli. Thus, pathoadaptive mutations do not accumulate in significant numbers in K. pneumoniae populations, especially considering the long-term instability of the pathoadaptive mutations due to their fitness trade-off in nonurinary habitats, where the gain of a functional advantage under novel conditions is accompanied by functionally detrimental effects in the original habitat. For FimH mutations, one such trade-off is increased sensitivity of the adhesin to inhibition by soluble mannosylated compounds (32). Nevertheless, the occurrence of the S62A mutation in the course of the endemic circulation of a nosocomial uropathogenic clone of K. pneumoniae strongly supports the importance of type 1 fimbriae in UTI caused by K. pneumoniae, as has been shown for E. coli.
Finally, it appears that the L(−12) deletion in the signal peptide of K. pneumoniae FimH has been acquired on multiple occasions, suggesting an adaptive significance of the mutation. Although the mutation does not affect the structure or, implicitly, the function of FimH per se, it may affect the translocation of nascent FimH across the inner membrane. It has recently been demonstrated that signal peptide mutations are acquired under positive selection in E. coli FimH and that they decrease the rate of protein translocation into the periplasm (28). Because FimH initiates the biogenesis of type 1 fimbriae, decreased periplasmic FimH leads to the expression of fewer but longer fimbriae. It is thus conceivable that the L(−12) mutation may result in such morphological changes in fimbrial structure. If so, longer fimbriae could be adaptive for K. pneumoniae strains by allowing the fimbrial tip (with FimH) to protrude through the thick capsule commonly produced by K. pneumoniae isolates. It has been shown that capsular material can in fact interfere with fimbrial function (29), making plausible the adaptive value of longer fimbriae. However, further studies are required to clarify the adaptive significance of the signal peptide mutation in K. pneumoniae FimH. Also, though other within-species changes in K. pneumoniae FimH do not overlap with the pathoadaptive changes observed in E. coli FimH, their adaptive significance cannot be excluded.
Analysis of the genetic and structural diversity of the FimH adhesin of K. pneumoniae in the context of the population structure of the species allows certain insights into the physiological significance of the type 1 fimbriae. In particular, the high level of horizontal transfer of FimH that likely indicates the advantage of high structural variability of the major fimbrial subunit, FimA, shows that the type 1 fimbria of K. pneumoniae plays an important role in the colonization of habitats where such variability is essential. Whether the pressure comes from the adaptive or innate immunity of yet-unidentified host organisms or from phage escape remains to be determined. Also, the population analysis provided a framework in which to determine the rapid within-clone evolution of FimH in the endemic uropathogenic strain, indicating its functional significance in the latter subpopulation of K. pneumoniae. Thus, evolutionary tools can be successfully applied by microbiologists studying molecular mechanisms that underlie the ecology and pathogenesis of bacterial pathogens.
Acknowledgments
S. G. Stahlhut and C. Struve were partially supported by Danish Research Agency grants 2101-06-0009 and 2052-03-0013, respectively.
Footnotes
Published ahead of print on 16 January 2009.
REFERENCES
- 1.Bagley, S. T. 1985. Habitat association of Klebsiella species. Infect. Control 652-58. [DOI] [PubMed] [Google Scholar]
- 2.Carlton, T. M., J. T. Sullivan, G. S. Stuart, K. Hutt, I. L. Lamont, and C. W. Ronson. 2007. Ferrichrome utilization in a mesorhizobial population: microevolution of a three-locus system. Environ. Microbiol. 92923-2932. [DOI] [PubMed] [Google Scholar]
- 3.Connell, I., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 939827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diancourt, L., V. Passet, J. Verhoef, P. A. Grimont, and S. Brisse. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 434178-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiMartino, P., D. Sirot, B. Joly, C. Rich, and A. Darfeuille-Michaud. 1997. Relationship between adhesion to intestinal Caco-2 cells and multidrug resistance in Klebsiella pneumoniae clinical isolates. J. Clin. Microbiol. 351499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang, F. C., N. Sandler, and S. J. Libby. 2005. Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43991-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouts, D. E., H. L. Tyler, R. T. DeBoy, S. Daugherty, Q. Ren, J. H. Badger, A. S. Durkin, H. Huot, S. Shrivastava, S. Kothari, R. J. Dodson, Y. Mohamoud, H. Khouri, L. F. Roesch, K. A. Krogfelt, C. Struve, E. W. Triplett, and B. A. Methé. 2008. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 254e1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fung, C. P., F. Y. Chang, S. C. Lee, B. S. Hu, B. I. Kuo, C. Y. Liu, M. Ho, and L. K. Siu. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlach, G. F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 1711262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, D. S., A. Gottschau, and H. J. Kolmos. 1998. Epidemiology of Klebsiella bacteraemia: a case control study using Escherichia coli bacteraemia as control. J. Hosp. Infect. 38119-132. [DOI] [PubMed] [Google Scholar]
- 11.Hommais, F., S. Gouriou, C. Amorin, H. Bui, M. C. Rahimy, B. Picard, and E. Denamur. 2003. The FimH A27V mutation is pathoadaptive for urovirulence in Escherichia coli B2 phylogenetic group isolates. Infect. Immun. 713619-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis, W. R., V. P. Munn, A. K. Highsmith, D. H. Culver, and J. M. Hughes. 1985. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect. Control 668-74. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, J. R., S. J. Weissman, A. L. Stell, E. Trintchina, D. E. Dykhuizen, and E. V. Sokurenko. 2001. Clonal and pathotypic analysis of archetypal Escherichia coli cystitis isolate NU14. J. Infect. Dis. 1841556-1565. [DOI] [PubMed] [Google Scholar]
- 14.Korhonen, T. K., R. Virkola, K. Lahteenmaki, Y. Bjorkman, M. Kukkonen, T. Raunio, A. M. Tarkkanen, and B. Westerlund. 1992. Penetration of fimbriate enteric bacteria through basement membranes: a hypothesis. FEMS Microbiol. Lett. 79307-312. [DOI] [PubMed] [Google Scholar]
- 15.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276607-611. [DOI] [PubMed] [Google Scholar]
- 16.Matsen, J. M., J. A. Spindler, and R. O. Blosser. 1974. Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl. Microbiol. 28672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 2821494-1497. [DOI] [PubMed] [Google Scholar]
- 18.Oelschlaeger, T. A., and B. D. Tall. 1997. Invasion of cultured human epithelial cells by Klebsiella pneumoniae isolated from the urinary tract. Infect. Immun. 652950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okano, H., K. Shiraki, H. Inoue, T. Kawakita, N. Yamamoto, M. Deguchi, K. Sugimoto, T. Sakai, S. Ohmori, K. Murata, and T. Nakano. 2002. Clinicopathological analysis of liver abscess in Japan. Int. J. Mol. Med. 10627-630. [PubMed] [Google Scholar]
- 20.Ørskov, I., and F. Ørskov. 1984. Serotyping of Klebsiella. Methods Microbiol. 14143-164. [Google Scholar]
- 21.Ottow, J. C. 1975. Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 2979-108. [DOI] [PubMed] [Google Scholar]
- 22.Podschun, R. 1990. Phenotypic properties of Klebsiella pneumoniae and K. oxytoca isolated from different sources. Zentralbl. Hyg. Umweltmed. 189527-535. [PubMed] [Google Scholar]
- 23.Podschun, R., A. Fischer, and U. Ullmann. 1992. Siderophore production of Klebsiella species isolated from different sources. Zentralbl. Bakteriol. 276481-486. [DOI] [PubMed] [Google Scholar]
- 24.Podschun, R., S. Pietsch, C. Holler, and U. Ullmann. 2001. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 673325-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podschun, R., and U. Ullmann. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11589-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouttu, R., T. Puustinen, R. Virkola, J. Hacker, P. Klemm, and T. K. Korhonen. 1999. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol. Microbiol. 311747-1757. [DOI] [PubMed] [Google Scholar]
- 27.Rahimian, J., T. Wilson, V. Oram, and R. S. Holzman. 2004. Pyogenic liver abscess: recent trends in etiology and mortality. Clin. Infect. Dis. 391654-1659. [DOI] [PubMed] [Google Scholar]
- 28.Ronald, L. S., O. Yakovenko, N. Yazvenko, S. Chattopadhyay, P. Aprikian, W. E. Thomas, and E. V. Sokurenko. 2008. Adaptive mutations in the signal peptide of the type 1 fimbrial adhesin of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 10510937-10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schembri, M. A., J. Blom, K. A. Krogfelt, and P. Klemm. 2005. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect. Immun. 734626-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidler, R. J., M. D. Knittel, and C. Brown. 1975. Potential pathogens in the environment: cultural reactions and nucleic acid studies on Klebsiella pneumoniae from clinical and environmental sources. Appl. Microbiol. 29819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokurenko, E. V., V. Chesnokova, R. J. Doyle, and D. L. Hasty. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J. Biol. Chem. 27217880-17886. [DOI] [PubMed] [Google Scholar]
- 32.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 958922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokurenko, E. V., H. S. Courtney, J. Maslow, A. Siitonen, and D. L. Hasty. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 1773680-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokurenko, E. V., M. Feldgarden, E. Trintchina, S. J. Weissman, S. Avagyan, S. Chattopadhyay, J. R. Johnson, and D. E. Dykhuizen. 2004. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 211373-1383. [DOI] [PubMed] [Google Scholar]
- 35.Spencer, R. C. 1996. Predominant pathogens found in the European Prevalence of Infection in Intensive Care Study. Eur. J. Clin. Microbiol. Infect. Dis. 15281-285. [DOI] [PubMed] [Google Scholar]
- 36.Struve, C., M. Bojer, E. M. Nielsen, D. S. Hansen, and K. A. Krogfelt. 2005. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J. Med. Microbiol. 541111-1113. [DOI] [PubMed] [Google Scholar]
- 37.Struve, C., M. Bojer, and K. A. Krogfelt. 2008. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect. Immun. 764055-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struve, C., and K. A. Krogfelt. 2003. Role of capsule in Klebsiella pneumoniae virulence: lack of correlation between in vitro and in vivo studies. FEMS Microbiol. Lett. 218149-154. [DOI] [PubMed] [Google Scholar]
- 39.Struve, C., and K. A. Krogfelt. 2004. Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 6584-590. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109913-923. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turton, J. F., H. Englender, S. N. Gabriel, S. E. Turton, M. E. Kaufmann, and T. L. Pitt. 2007. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 56593-597. [DOI] [PubMed] [Google Scholar]
- 43.Vakharia-Rao, H., K. A. Kastead, M. I. Savenkova, C. M. Bulathsinghala, and K. Postle. 2007. Deletion and substitution analysis of the Escherichia coli TonB Q160 region. J. Bacteriol. 1894662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, J. H., Y. C. Liu, S. S. Lee, M. Y. Yen, Y. S. Chen, J. H. Wang, S. R. Wann, and H. H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 261434-1438. [DOI] [PubMed] [Google Scholar]
- 45.Wang, K., H. J. Lin, C. L. Perng, H. Chiang, C. T. Lee, F. Y. Chang, and S. D. Lee. 1995. Pseudomelanosis duodeni: report of eight cases. J. Formos. Med. Assoc. 94632-634. [PubMed] [Google Scholar]
- 46.Weissman, S. J., V. Beskhlebnaya, V. Chesnokova, S. Chattopadhyay, W. E. Stamm, T. M. Hooton, and E. V. Sokurenko. 2007. Differential stability and trade-off effects of pathoadaptive mutations in the Escherichia coli FimH adhesin. Infect. Immun. 753548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissman, S. J., S. Chattopadhyay, P. Aprikian, M. Obata-Yasuoka, Y. Yarova-Yarovaya, A. Stapleton, W. Ba-Thein, D. Dykhuizen, J. R. Johnson, and E. V. Sokurenko. 2006. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]