Abstract
The Escherichia coli HtrA protein is a periplasmic protease/chaperone that is upregulated under stress conditions. The protease and chaperone activities of HtrA eliminate or refold damaged and unfolded proteins in the bacterial periplasm that are generated upon stress conditions. In the absence of substrates, HtrA oligomerizes into a hexameric cage, but binding of misfolded proteins transforms the hexamers into bigger 12-mer and 24-mer cages that encapsulate the substrates for degradation or refolding. HtrA also undergoes partial degradation as a consequence of self-cleavage of the mature protein, producing short-HtrA protein (s-HtrA). The aim of this study was to examine the physiological role of this self-cleavage process. We found that the only requirement for self-cleavage of HtrA into s-HtrA in vitro was the hydrolysis of protein substrates. In fact, peptides resulting from the hydrolysis of the protein substrates were sufficient to induce autocleavage. However, the continuous presence of full-length substrate delayed the process. In addition, we observed that the hexameric cage structure is required for autocleavage and that s-HtrA accumulates only late in the degradation reaction. These results suggest that self-cleavage occurs when HtrA reassembles back into the resting hexameric structure and peptides resulting from substrate hydrolysis are allosterically stimulating the HtrA proteolytic activity. Our data support a model in which the physiological role of the self-cleavage process is to eliminate the excess of HtrA once the stress conditions cease.
The cell envelope of gram-negative bacteria mediates the communication of the cell with the environment, and it is responsible for many vital functions, including nutrient uptake and interaction with other bacteria and host cells. These activities are performed by a large collection of proteins that make the periplasm a cellular compartment with an even higher protein concentration than the cytoplasm (2). Bacteria are frequently exposed to multiple stresses such as heat shock, osmotic stress, and pH changes and are regularly challenged by the host immune system. Thus, the maintenance of periplasmic proteins in a fully functional state is a challenging task undertaken by the protein quality control system (5). It is generally accepted that under stress conditions misfolded proteins, protein fragments, and mislocalized membrane proteins appear, activating a stress response through three different signal transduction pathways (σE, Cpx, and Bae) (21, 22). Activation of this stress response in the periplasm triggers the upregulation of molecular chaperones, peptidases, proteases, and other enzymes with a role in eliminating or refolding damaged periplasmic proteins.
The Escherichia coli HtrA protein (also called DegP or protease Do) is a periplasmic protein (4) that is upregulated under stress conditions such as heat shock (14, 15). HtrA functions as a chaperone and a protease in a temperature-dependent fashion (24). Recent studies have also shown that HtrA substrates targeted for degradation or refolding are recognized differently, suggesting that the mechanisms through which HtrA recognizes the substrate may play a role in the protease-chaperone switch (8).
HtrA contains an N-terminal protease domain, followed by the PDZ1 and PDZ2 domains. In the absence of substrates, HtrA oligomerizes into a hexameric cage (12) that represents the resting state of the protein (10, 13). Upon binding to protein substrates, HtrA transforms into bigger cages formed by 12 or 24 monomers that encapsulate substrates for degradation or refolding (9, 13).
HtrA is a 474-residue protein whose first 26 amino acids are removed at the N terminus most likely by a signal peptidase rendering the mature 48-kDa protein (14, 15). This form of the protein will hereon be referred to as full-length HtrA. However, it has been described (11, 23) that mature HtrA undergoes partial degradation both in vivo and in vitro as a consequence of self-cleavage occurring after Cys69 and Gln82 of the mature protein. These forms of the protein have been named short-HtrA (s-HtrA) (23).
A similar phenomenon of autocleavage has been observed in other members of the HtrA family such as the human homologs HtrA1 (7) and HtrA2 proteins (6). The autocleavage process is not specific for proteases of the HtrA family. Several prokaryotic proteins involved in regulation of gene expression, such as the SOS response proteins LexA (16-18) and UmuD (3), are inactivated through a self-cleaving mechanism. Conversely, many mammalian proteases are produced as longer inactive precursors and depend on an intramolecular cleavage event to become active. This is the case for some gastric proteases such as pepsin and chymosin or the lysosome cathepsins D and E (1).
Although autocleavage as a mechanism of activation or inactivation of certain proteases is well documented, the physiological role and the events triggering the self-cleavage of HtrA are poorly understood. In this study, we observed that the hexameric cage structure is required to observe autocleavage of HtrA. In addition, we analyzed the conditions that led to self-cleavage of HtrA and we found that the only requirement to observe accumulation of the s-HtrA form in vitro was the hydrolysis of protein substrates. In fact, peptides resulting from the degradation of protein substrates were sufficient to induce autocleavage. Therefore, considering the current functional model for HtrA (9, 13), our data suggest that the physiological role of the HtrA autocleavage is to eliminate the excess of HtrA protein expressed under stress conditions when the enzymatic activities of the protein are no longer needed.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The pET21b-HtrA plasmid and the ΔPDZ2 HtrA mutant were obtained as described previously (10). The C57A+C69G HtrA mutant was obtained from the pET21b-HtrA plasmid using the QuikChange site-directed mutagenesis method (Stratagene).
Protein expression and purification.
Wild-type and HtrA mutants were expressed as C-terminal His-tagged proteins. The method to express and purify the proteins with a HiTrap metal chelating column (GE Healthcare Life Sciences) was performed as described previously (10).
When a protein preparation containing mostly full-length HtrA was required, E. coli BL21(DE3) cells with the expression vector containing wild-type or mutant HtrA were induced for 30 min at 37°C with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). However, to obtain a protein preparation with higher proportion of s-HtrA form, cells were induced for 3 h at 37°C with 1 mM IPTG in LB medium containing 5 mM dithiothreitol (DTT) freshly added at the moment of induction.
Size exclusion chromatography.
Gel filtration chromatography experiments were performed on a Superdex 200 10/300 GL column (GE Healthcare Life Sciences). The column was equilibrated in 50 mM HEPES (pH 7.3)-100 mM NaCl at 4°C. Protein samples (100 μl) at a concentration between 0.5 and 3.5 mg/ml were injected to the column. A gel filtration calibration kit (HMW; GE Healthcare Life Sciences) was used for column calibration.
Protease activity assays.
Protease activity assays in Fig. 1 were assembled as 250-μl reaction mixtures containing HtrA at 1.3 μM and substrate at 13 μM in 50 mM HEPES (pH 7.3). As substrates, we used malate dehydrogenase (MDH) from porcine heart (Roche and Sigma), bovine milk β-casein (Sigma), and egg white lysozyme (Bioshop). Reactions were incubated at 37°C for the β-casein and lysozyme assays and at 43°C for the MDH experiments. DTT was added to the lysozyme reactions up to a final concentration of 2 mM. At the indicated times, 10-μl samples were taken and mixed with 2× concentrated sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, resolved by SDS-PAGE, and stained with Coomassie brilliant blue (GE Healthcare Life Sciences). The remaining β-casein hydrolysis assays performed followed the same procedure as the one in Fig. 1. However, reaction mixtures contained HtrA at 4.1 μM and β-casein at 66 μM.
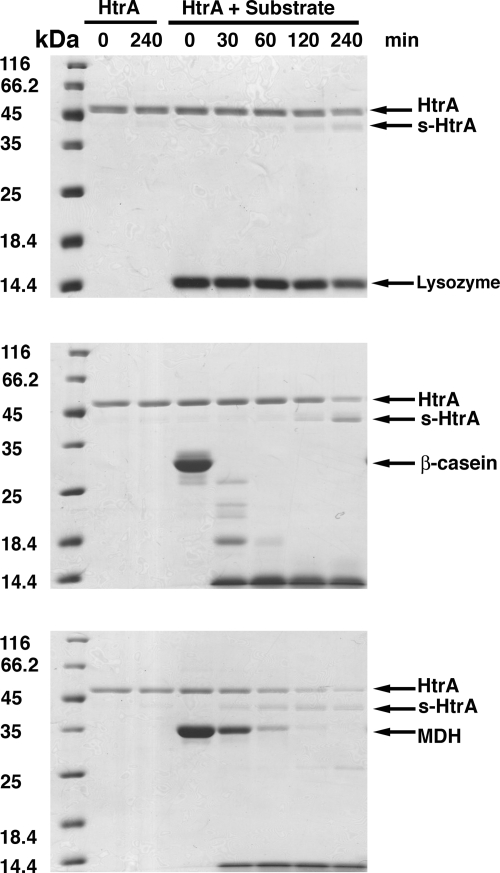
FIG. 1.
HtrA autocleavage in the presence of substrate. Proteolytic reactions were assembled in a total volume of 250 μl containing 1.3 μM HtrA and 13 μM concentrations of substrate. The substrates tested were lysozyme (top panel), β-casein (middle panel), and MDH (bottom panel) (lanes labeled “HtrA + Substrate”). A control reaction was performed for each degradation assay consisting of HtrA protein incubated in the absence of substrate (lanes labeled “HtrA”). Reactions were incubated and, at the indicated time points, samples were obtained, and the products of the reaction resolved by SDS-PAGE and stained by Coomassie brilliant blue. Arrows indicate the bands for the corresponding substrate, full-length HtrA and s-HtrA protein in each gel.
To obtain the accumulation profiles of s-HtrA protein for C57A+C69G HtrA mutant, the hydrolysis reactions were performed by mixing β-casein (66 μM) with HtrA or the C57A+C69G HtrA mutant (4.1 μM) in a reaction volume of 250 μl (50 mM HEPES [pH 7.3]). Where indicated, DTT was added to a final concentration of 2 mM. At the indicated times, 10-μl samples were resolved by SDS-PAGE and stained with Coomassie brilliant blue. The proteolysis reaction for the C57A+C69G HtrA mutant in each condition (i.e., the presence or absence of DTT) was run in the same gel together with the analogous reaction performed with the wild type to account for differences in staining between gels. Gels were scanned in a flatbed scanner, and the total intensity of the bands representing s-HtrA protein was measured and subtracted from the intensity of the band at time zero min by using ImageQuant TL software. These intensities were used to plot the accumulation profiles. Each proteolysis reaction was repeated three times, and an average and standard deviation was calculated.
To generate and purify β-casein peptides, 250-μl reaction mixtures were assembled in 50 mM HEPES (pH 7.3) containing a concentration of 4.1 μM HtrA and 66 μM β-casein. In the case where the peptides were generated by trypsin, the reaction contained 4.1 μM trypsin and 132 μM β-casein. The buffer conditions were the same, but MgCl2 was added up to a final concentration of 5 mM. Reaction mixtures were incubated at 37°C for 40 min and then boiled for 10 min and immediately chilled at 4°C to terminate the reaction and to precipitate HtrA and trypsin. Subsequently, reactions were spun down at 12,000 rpm in a benchtop centrifuge at 4°C, and the supernatant containing the β-casein peptides was topped up to 250 μl with 50 mM HEPES (pH 7.3) buffer containing HtrA. The final concentration of HtrA in the mixture was 4.1 μM. The reaction was then incubated at 37°C for 4 h. At the indicated times, 20-μl samples were taken, mixed with 2× concentrated SDS-PAGE loading buffer, and 12.5 μl was resolved by SDS-PAGE (12%). Gels were stained with Coomassie brilliant blue.
Western blotting.
Samples were resolved by SDS-12% PAGE and transferred to an Immobilon-P filter (Millipore) according to manufacturer's protocols. The membrane was then saturated with 5% skim milk in TBS-T buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) for 12 h at 4°C. The filters were incubated with a 1:12,500 dilution of the anti-HtrA serum for 1 h at 4°C. They were washed one time for 15 min and three times for 5 min with TBS-T buffer and then incubated with a 1:4,000 dilution of goat anti-rabbit immunoglobulin G conjugated to peroxidase. Finally, the filters were washed one time for 15 min and three times for 5 min as described above, plus an additional wash for 5 min in TBS buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). The filter was developed by using enhanced chemiluminescence.
The anti-HtrA serum was prepared by immunization of rabbits with C-terminal His-tagged HtrA protein that was purified as previously described (10).
RESULTS
Only substrate degradation is required for the autocleavage of HtrA and production of s-HtrA protein in vitro.
It was shown previously that both proteolytic activity of HtrA and reducing conditions were essential to observe autocleavage of HtrA and production of s-HtrA protein (23). To initiate our study aimed to unveil the physiological role of the s-HtrA protein, we investigated whether autocleavage of HtrA occurs during degradation of lysozyme, β-casein, and MDH. In the performed assays the concentrations of HtrA (1.3 μM) and substrate (13 μM) were similar to the ones used by Skorko-Glonek et al. (23). Consistent with that study, we observed autocleavage of HtrA in a lysozyme degradation assay under reducing conditions (Fig. 1, top panel). β-Casein and MDH did not require reducing conditions to be degraded by HtrA (11). However, s-HtrA protein was also obtained upon degradation of these substrates (Fig. 1, middle and bottom panels). The amount of s-HtrA generated in these reactions was very similar in the presence or absence of reducing agents (data not shown). Furthermore, no s-HtrA was produced in any of the conditions when HtrA was incubated in the absence of substrate (Fig. 1). The N-terminal sequences of the two degradation products observed in our proteolysis reactions (data not shown) were consistent with previously published data (23), establishing that the two s-HtrA protein forms arise as a consequence of cutting occurring after Cys69 or after Gln82.
Consistent with the study by Skorko-Glonek et al. (23), we observed that the amount of s-HtrA protein generated in these assays correlated with the amount of full-length substrate degraded and the speed of hydrolysis. The amount of s-HtrA protein accumulated over 4 h of incubation time was most prominent in the β-casein assay and followed by the experiment using MDH. In these experiments full-length substrate was eliminated in ∼30 (β-casein) and ∼120 (MDH) minutes (Fig. 1, middle and bottom panels), and the s-HtrA protein started to accumulate soon after the full-length substrate disappeared. Lysozyme hydrolysis progressed slowly, and there was still a significant amount of full-length substrate left after 4 h. As a result, only very little s-HtrA protein was generated even in the presence of reducing agents (Fig. 1, top panel).
In the previous experiment, the concentrations of HtrA and substrates were higher than those used in previous HtrA studies (19) and also those occurring in vivo. Therefore, these experiments were repeated at concentrations 10 times lower (HtrA at 0.13 μM and substrate at 1.3 μM). Similarly to the previous experiment, only substrate degradation was needed to observe autocleavege of HtrA (see Fig. S1 in the supplemental material) and reducing conditions were only essential in cases where they were required to unfold the substrate and make it susceptible for degradation by HtrA. The amount of s-HtrA protein generated in the assay also correlated with the amount of full-length substrate degraded and the speed of hydrolysis. Therefore, we concluded that autocleavage of HtrA occurs in a similar manner in a broad range of HtrA and substrate concentrations.
Cys57 and Cys69 residues in HtrA do not confer additional stability to the protein against autocleavage and are not essential to maintain the HtrA hexameric cage.
It has been previously suggested that reducing stress could induce autocleavage of HtrA because these conditions disrupt the disulfide bridge formed by the only two existing cysteine residues (Cys57 and Cys69) in the HtrA sequence (23). In our experiments, the presence of reducing agents was not a requirement in order to observe autocleavage of HtrA. However, we decided to test whether Cys57 and Cys69 residues are essential to maintain the HtrA hexameric cage or the disulfide bridge formed conferred additional stability to the protein against autocleavage.
Residues Cys57 and Cys69 in HtrA are located within the LA loop of the protease domain. The LA loop protrudes from the HtrA monomer and acts as a molecular spacer between the two trimers forming the HtrA hexameric cage. Two LA loops from opposite monomers in the hexamer wrap around each other and form the corner pillars of the cage. Our previous studies found that residues 39 to 78 in the LA loop (containing the Cys57 and Cys69) are essential to maintain the hexameric cage (10). To test specifically whether these two cysteine residues are essential to maintain the integrity of the hexamer, we constructed and purified the C57A+C69G HtrA mutant. The purified mutant behaved similarly to wild-type HtrA on a size exclusion chromatography column and produced an elution profile consistent with the hexameric form of the protein (Fig. 2A) ruling out any role of Cys57 and Cys69 on maintaining the HtrA hexamer.
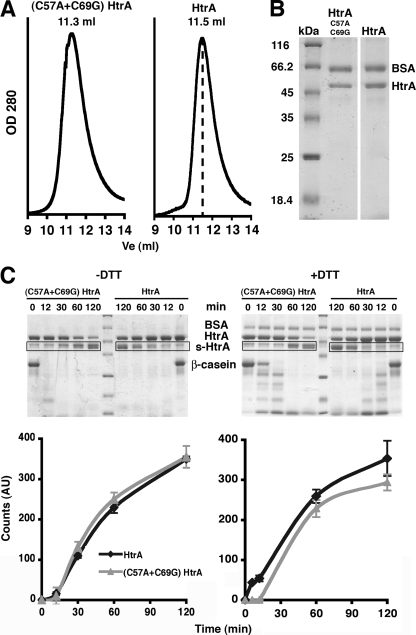
FIG. 2.
Characterization of the oligomeric state and stability of the C57A+C69G HtrA mutant. (A) Elution profile of the C57A+C69G HtrA (left) and wild-type HtrA (right) proteins from a Superdex-200 column. The dashed line in the plot on the right-hand side indicates the expected elution volume (Ve) for the hexameric form of the protein (11.5 ml). (B) C57A+C69G HtrA and wild-type HtrA proteins were eluted from a metal chelating column and resolved by SDS-PAGE and stained with Coomassie brilliant blue. Both full-length HtrA and s-HtrA proteins coeluted from this column. Bovine serum albumin (BSA) protein (2 μg) was added to each sample as a loading control. (C) Accumulation profiles of s-HtrA protein for the C57A+C69G HtrA mutant and wild-type HtrA in the presence or absence of DTT. Profiles were obtained by performing proteolysis reactions containing β-casein and HtrA or the C57A+C69G HtrA mutant. Reactions shown on the left were performed either in the absence (left panels) or in the presence (right panels) of DTT. Samples of the reactions at the specified time points were resolved by SDS-PAGE (upper panel). Each proteolysis reaction for the C57A+C69G HtrA mutant was run in the same gel, together with the analogous reaction performed with wild-type HtrA. BSA protein (2 μg) was added to samples as a loading control. Gels were stained by Coomassie brilliant blue and scanned, and the intensity of the bands representing s-HtrA protein (boxed areas, upper panel) was measured and plotted with respect to time (bottom panel). The plots represent the average and standard deviations obtained upon repeating each experiment three times.
Furthermore, two additional observations suggested that the disulfide bridge formed by these two cysteine residues does not confer additional stability to the protein. First, the autocleavage of wild-type HtrA also occurs in vivo during protein expression (23) and both full-length and s-HtrA coelute together from the metal chelating column used in our protein purification strategy. Thus, to minimize the amount of s-HtrA in our protein preparations, the expression time was limited to 30 min (10). Higher proportions of s-HtrA protein would be anticipated to coelute from the column with a preparation of an HtrA mutant with higher autocleavage rates. However, when the C57A+C69G HtrA was eluted under conditions identical to those used for the wild type, we did not observe increased amounts of s-HtrA protein coeluting with full-length HtrA (Fig. 2B). Second, the C57A+C69G HtrA mutant degraded β-casein as efficiently as wild-type HtrA both in the presence and in the absence of DTT (Fig. 2C, top panel). However, we did not observe any statistically significant differences between the accumulation profiles of s-HtrA protein from wild-type HtrA and the Cys mutant for any of the conditions tested (Fig. 2C, bottom panel). A similar experiment performed with MDH as a substrate did not show any statistically significant differences either (data not shown).
Altogether these experiments indicate that the cysteine residues present in the HtrA protein (Cys57 and Cys69) are not essential to maintain the integrity of the hexameric cage, and they do not stabilize HtrA in a manner that significantly prevents or delays autocleavage of the protein.
Continuous addition of full-length substrate delays autocleavage of HtrA.
To further investigate the conditions leading to the production of s-HtrA protein, we assembled a proteolysis reaction by mixing HtrA with β-casein. In the tested conditions, full-length β-casein was degraded after 30 min, and smaller peptides products started to accumulate (Fig. 3A). A significant amount of s-HtrA was already present after 2 h, and approximately 90% of the HtrA protein became autocleaved after 7 h of incubation (Fig. 3A). Incubation of the reaction for 24 h produced almost a complete self-degradation of the HtrA protein into the s-HtrA protein first and subsequent shorter HtrA forms, most of them not observed in the SDS-PAGE. When fresh β-casein was added at this point in the reaction mixture, we observed that substrate hydrolysis was significantly slowed down, suggesting that these fragments are proteolytically inactive (Fig. 3B, left panel). In a replica of this experiment, HtrA autocleavage progressed slightly faster during the overnight incubation, and we in fact observed a complete self-degradation of the enzyme, and no HtrA or s-HtrA form was left after 24 h of incubation. In this case, the reaction mixture showed no further degradation of newly provided β-casein (Fig. 3B; right panel). These reactions show that HtrA degrades the substrate first before the autocleavage becomes significant, and then it progresses until complete self-degradation and consequent inactivation of HtrA. A control experiment was carried out simultaneously by incubating HtrA protein alone for 24 h to ensure that the protein remains stable and active upon such a long incubation at 37°C. As expected, no s-HtrA protein was formed in the absence of substrate, and the enzyme remained proteolytically active, as demonstrated by fast degradation of the β-casein after 24 h of incubation of HtrA at 37°C (Fig. 3C).
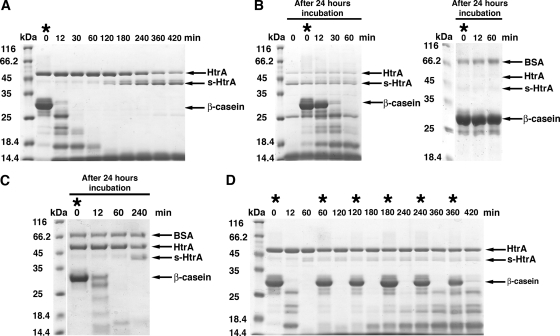
FIG. 3.
Full-length substrate delays autocleavage of HtrA. (A) A proteolysis reaction containing β-casein substrate (66 μM) and HtrA (4.1 μM) was incubated at 37°C for 7 h, and at the indicated times samples of the reaction were obtained and loaded on an SDS-PAGE gel and stained by Coomassie brilliant blue. The asterisk indicates when fresh β-casein was added in the reaction. (B) The previous reaction was kept up to 24 h at 37°C. Then, fresh β-casein was added (asterisk) and incubated for one additional hour. The left panel shows an SDS-PAGE gel resolving the indicated time points of this reaction. The SDS-PAGE results on the right replicate the experiment shown in the left panel, where autocleavage of HtrA progressed slightly faster during the overnight incubation and degraded all of the HtrA in the reaction. (C) HtrA protein (4.1 μM) was preincubated at 37°C for 24 h before the substrate β-casein (66 μM) was added (asterisk). Then, at the indicated times samples were extracted and resolved by SDS-PAGE. Gels were stained by Coomassie brilliant blue. (D) This proteolysis reaction was assembled as in panel A and then kept up to 24 h as in panel B. Asterisks indicate the time points when fresh β-casein was added during the reaction. The indicated time point samples were obtained from the reaction, resolved by SDS-PAGE, and stained with Coomassie brilliant blue. Where indicated an identical amount of BSA protein (2 μg) was added to each sample as a loading control.
To confirm that the presence of full-length substrate delays autocleavage of HtrA, we performed an additional proteolysis reaction. In this case, fresh β-casein was added at the beginning of the reaction and after every 60 min of incubation up to 6 h. Interestingly, very small amounts of s-HtrA protein accumulated in the SDS-PAGE even after 7 h of incubation, suggesting that constant presence of full-length β-casein or long degradation peptides inhibit autocleavage of the HtrA protein (Fig. 3D).
Peptides generated during proteolytic degradation of the substrate are sufficient to stimulate HtrA autocleavage.
The previous experiments showed that significant amounts of s-HtrA protein did not accumulate until all of the full-length β-casein and large degradation products were proteolyzed into small peptides. Thus, we wanted to test whether the small peptides generated during the proteolytic reaction are sufficient to stimulate HtrA autocleavage. A proteolysis reaction using β-casein as a substrate was assembled and incubated at 37°C. As previously shown, most of the substrate became proteolyzed after 30 min of reaction and most of the HtrA was self-cleaved after 4 h (Fig. 4A). The reaction was kept up to 24 h at the incubation conditions, and then fresh HtrA was added to the reaction. Under these conditions the newly added HtrA underwent autocleavage right from the beginning of the reaction and, similarly, most of the HtrA was autocleaved in 4 h (Fig. 4A).
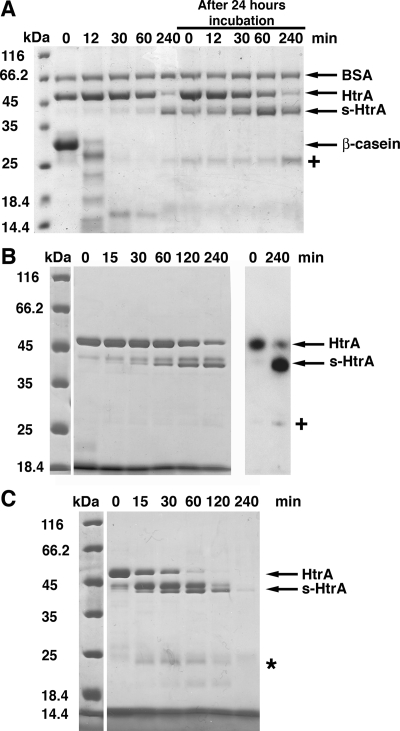
FIG. 4.
Degradation peptides products stimulate HtrA autocleavage. (A) A proteolytic reaction containing β-casein (66 μM) in the presence of HtrA (4.1 μM) was incubated for 4 h at 37°C. Samples were obtained at the indicated times, resolved by SDS-PAGE, and stained with Coomassie brilliant blue. After the reaction was kept at 37°C for 24 h, fresh HtrA (4.1 μM) was added, and the incubation was continued for four additional hours. Samples were taken at the indicated times to monitor the accumulation of s-HtrA protein. BSA protein (2 μg) was added to each sample as a loading control. (B) β-Casein peptides produced by HtrA hydrolysis were incubated with HtrA (4.1 μM) for 4 h at 37°C. At the indicated time points, samples were obtained and resolved by SDS-PAGE (left). The samples at time zero and 240 min were also processed for Western blotting with a rabbit anti-HtrA serum (right). The “+” sign indicates the 25-kDa self-cleavage form of HtrA recognized by the anti-HtrA serum. (C) β-Casein peptides produced by trypsin hydrolysis were mixed with HtrA (4.1 μM) in an autocleavage reaction for 4 h at 37°C. Time points of the reaction were obtained and resolved by SDS-PAGE. The asterisk indicates small peptides produced from HtrA and s-HtrA proteins by a residual amount of trypsin incorporated in the autocleavage reaction along with the β-casein peptides.
In order to confirm that the peptides generated during substrate hydrolysis are sufficient to stimulate HtrA autocleavage, we degraded β-casein substrate in the presence of either HtrA or trypsin for 40 min. Then, the reaction mixtures were boiled for 10 min to terminate the reaction and precipitate HtrA and trypsin. The precipitated proteins were spun down, and the supernatants containing the peptides were incubated with HtrA (4.1 μM) in two different autocleavage reactions for 4 h at 37°C. Consistent with the results presented in Fig. 4A, the freshly added HtrA underwent autocleavage and produced s-HtrA form in the presence of β-casein peptides starting at very early times of the reaction (Fig. 4B and C).
We noticed that an additional protein band of ∼25 kDa accumulated at later times of incubation in the proteolytic reaction (Fig. 4A, “+” sign, and 4B, left panel). To determine whether this protein was an additional self-cleavage product of HtrA, we analyzed the products of the reaction by Western blotting. Samples of the autocleavage reaction containing HtrA and β-casein peptides produced by HtrA were resolved by SDS-PAGE and processed for Western blot with a rabbit anti-HtrA serum (Fig. 4B, right panel). The 25-kDa band reacted with the anti-HtrA serum, suggesting that this reaction product is also a self-cleavage form of HtrA. This finding is consistent with our previous experiment (Fig. 3B), suggesting that the self-cleavage process of HtrA does not stop with the generation of the described ∼43-kDa s-HtrA forms. Rather, subsequent self-cleavage events hydrolyzed HtrA until complete degradation of the enzyme.
We also observed that a small amount of trypsin remained in the supernatant after the β-casein hydrolysis reaction was spun down. However, this amount of trypsin did not contribute to the amount of s-HtrA form generated in the autocleavage reaction. Trypsin is a highly specific enzyme that cleaves peptide bonds on the carboxyl side of residues Arg and Lys (20). HtrA contains more than 30 potential cleaving sites for trypsin, and it is quickly degraded by this protease into small peptides, none of them corresponding to the length of the s-HtrA forms. Some of these small peptides were observed on the SDS-PAGE (Fig. 4C, asterisk).
Similarly to the experiments where we looked at the production of s-HtrA form in the presence of several substrates at high (Fig. 1) and low (see Fig. S1 in the supplemental material) concentrations, we wanted to test whether peptides also stimulate HtrA autocleavage in a broad range of concentrations. Therefore, we repeated the autocleavage of HtrA by β-casein peptides but at significantly lower concentrations (HtrA at 0.13 μM). As expected, HtrA also underwent autocleavage and produced s-HtrA protein in the presence of β-casein peptides (see Fig. S2 in the supplemental material).
For the experiments performed at both high and low concentrations, we carried out control experiments similar to the ones shown in Fig. 1 and Fig. S1 in the supplemental material1, where HtrA alone was incubated at 37°C for 4 h. s-HtrA protein was not produced in any of the tested conditions.
Therefore, we concluded that peptides generated during the proteolytic reaction are sufficient to stimulate HtrA autocleavage in vitro, and this process occurs in a similar manner in a broad range of HtrA and peptide concentrations.
The hexameric cage structure is required for the autocleavage of HtrA.
We reported that the hexameric structure of HtrA is not essential for proteolytic activity since trimeric mutants of the protein are able to hydrolyze substrates similarly to wild-type HtrA (10). Therefore, the question then arose as to whether the HtrA hexameric cage is required for the autocleavage of HtrA.
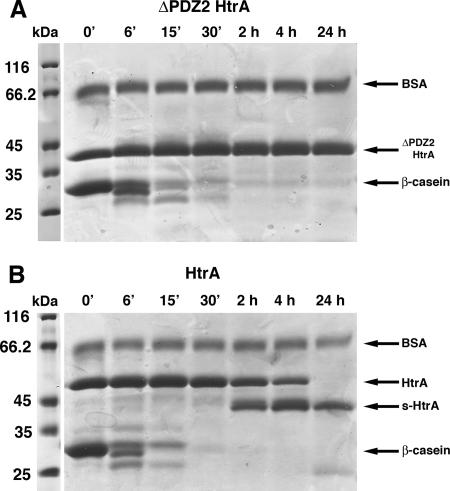
To answer this question, we tested the ability of a previously characterized ΔPDZ2 HtrA mutant (10) to produce s-HtrA protein. This HtrA construct is able to bind protein substrates and has proteolytic activity similar to wild-type HtrA; however, an enclosed cavity no longer exists in this mutant since the HtrA hexamer has been separated into two trimers (10). A proteolysis reaction was assembled with this mutant (Fig. 5A) by mixing the mutant protein (4.1 μM) with β-casein (66 μM) and incubating the reaction up to 24 h at 37°C. A proteolysis reaction was carried out in parallel containing wild-type HtrA and β-casein (Fig. 5B). We found that while the control reaction containing wild-type HtrA produced s-HtrA protein in large amounts (Fig. 5B), the reaction containing the ΔPDZ2 HtrA mutant did not produce any s-HtrA, even after 24 h of incubation (Fig. 5A), suggesting that the hexameric cage is required for the autocleavage of HtrA.
FIG. 5.
A trimeric HtrA mutant does not undergo autocleavage. (A) SDS-PAGE resolving samples obtained at the indicated time points from a β-casein proteolysis reaction performed with the ΔPDZ2 HtrA mutant. (B) Control reaction containing HtrA and β-casein showing production of s-HtrA protein at the same time points as for the results shown in panel A.
The s-HtrA form has a decreased proteolytic activity.
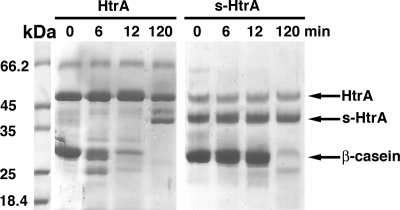
A previous study indicated that upon HtrA autocleavage, the hexameric cage undergoes irreversible dissociation into monomers (23). These results suggested that the autocleavage process compromises the oligomeric structure of the protein. In our study, we sought to determine whether the autocleavage of HtrA also compromises the HtrA proteolytic activity.
To do this, we obtained a protein preparation with a high proportion of s-HtrA, and the proteolytic activity was compared to a protein preparation containing mostly full-length HtrA. In order to maximize the amount of s-HtrA in our protein preparation, we extended the expression time to 3 h, and DTT was added to the LB medium at the moment of induction. These conditions stimulated the hydrolysis of substrates that require reducing conditions for unfolding and as a consequence HtrA autocleavage. This approach yielded a preparation of HtrA containing 60% of the protein as s-HtrA (Fig. 6). The obtained protein preparation (4.1 μM) was mixed with β-casein (66 μM) and incubated at 37°C for 2 h. This reaction was carried out in parallel with another β-casein proteolytic reaction containing mostly full-length HtrA. Samples obtained at 6 and 12 min showed that the proteolytic degradation of β-casein was significantly delayed in the reaction containing high proportions of s-HtrA protein, suggesting that the autocleavage process inhibits HtrA proteolytic activity. Nevertheless, most of the β-casein substrate was degraded after 2 h of incubation as a consequence of the remaining 40% full-length HtrA protein in this preparation (Fig. 6).
FIG. 6.
Proteolytic activity of the autocleaved HtrA protein. A preparation containing mostly full-length HtrA protein (lanes labeled “HtrA”) or 60% s-HtrA (lanes labeled “s-HtrA”) were incubated with β-casein for 2 h at 37°C. Samples were taken at the indicated time points and resolved by SDS-PAGE.
DISCUSSION
This study addresses a new facet of HtrA regulation: the mechanism to eliminate excess of HtrA from the cell once the stress conditions are overcome.
E. coli upregulates HtrA expression to quickly eliminate unfolded and damaged proteins associated with heat shock conditions and other form of stress. Based on our observations, particularly (i) that HtrA autocleavage mostly occurred after full-length substrate is degraded, (ii) that it is induced by the peptides produced during substrate hydrolysis, and (iii) that autocleavage affects the proteolytic activity of the enzyme, we hypothesized that the autocleavage of HtrA is in fact a self-regulatory mechanism aiming to eliminate the excess of HtrA produced under stress conditions once its enzymatic activities are no longer needed.
Our hypothesis integrates well with the current functional model for this protease: the hexameric HtrA cage is the resting state of the protein but upon substrate binding the protein reorganizes into large 12-mer and 24-mer oligomeric cages encapsulating the substrate for degradation. When substrate degradation has been completed HtrA reverts to its hexameric resting state (9, 13). Proteolytic activity of HtrA is also allosterically stimulated by oligopeptides, and this activation is mediated by the binding of the activating peptide to the PDZ1 domain (19).
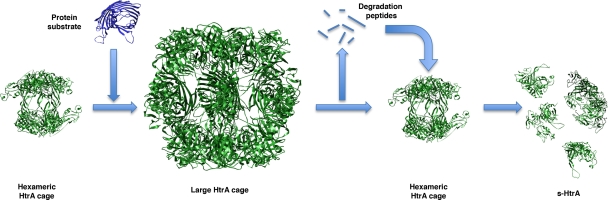
In the context of this functional model, our data can be interpreted as follows: while full-length substrate or long degradation peptides are present, HtrA remains mostly as 12-mer and 24-mer oligomeric cages, and limited amounts of s-HtrA form are produced. Our finding that the hexameric cage structure is required for the autocleavage of HtrA and that the s-HtrA form appeared late in the reaction suggests that the self-cleavage occurs when HtrA reassembles back into the resting hexameric cage once the degradation is completed and HtrA is in the presence of stimulating peptides resulting from substrate hydrolysis (Fig. 7). Existing structural data (9, 12, 13) support this hypothesis. The X-ray structure of the larger oligomers of HtrA showed that the LA loop has been extracted from the active site and the L1 and L2 loops are set up as a functional proteolytic site (13). However, in the hexameric form the LA loop protrudes into the active site of the subunit in the opposite trimer forcing the L1 and L2 loops into a twisted inactive conformation. Upon completion of the degradation, the protease returns to its hexameric form, but peptides resulting from substrate hydrolysis allosterically stimulate HtrA proteolytic activity (19). It is likely, although there is no experimental support yet, that peptides allosterically stimulating HtrA may keep the catalytic L1 and L2 loops in an active conformation. Consequently, the LA loop may be cleaved because it is placed back in close proximity to the catalytic site that may be now in an active conformation.
FIG. 7.
Schematic model of the autocleavage of HtrA. HtrA reorganizes into large cages that encapsulate the substrate for degradation. Once full-length substrate has been degraded, the HtrA reverts to its hexameric resting state, and peptides generated during substrate hydrolysis allosterically activate HtrA, inducing the self-cleavage process. PDB files 1ky9 and 2zle for the hexameric and 12-mer HtrA cages, respectively, were obtained from the Protein Data Bank and are displayed with the program UCSF Chimera (http://www.cgl.ucsf.edu/chimera).
Consistent with our hypothesis that the autocatalytic process is a regulatory suicide mechanism, we found that cleavage of the LA loop during autocleavage decreases HtrA proteolytic activity. A previous study reported (but did not show) (23) that the s-HtrA form remains fully active. Our results implicitly suggest that the s-HtrA form is inactive (Fig. 6). However, we were unable to overexpress and purify the s-HtrA forms (Δ1-69 HtrA and Δ1-82 HtrA) since the deletion regions compromise the proper folding of the protease domain and the trimer formation. Instead, our purification method yielded a preparation of HtrA containing 60% of the protein as s-HtrA, and therefore, at extended incubation times, substrate degradation was always observed due to the still existing full-length HtrA. Nevertheless, our results provide enough evidence to state that the s-HtrA form has a decreased proteolytic activity compared to full-length HtrA (Fig. 6).
There are several aspects of this regulatory model that are still not fully understood. For instance, the PDZ1 domain in HtrA mediates both substrate recognition and allosteric stimulation by binding to exposed C-terminal residues from the substrate and also short oligopeptides, respectively. Both full-length substrates and oligopeptides may bind to the same site in the PDZ1 domain; however, only long protein substrates seem to efficiently induce reorganization of HtrA into larger oligomeric cages. Further research is required to understand how these two substrates trigger a different response upon binding to HtrA.
In addition, proposing that HtrA has a clearance mechanism that eliminates the enzyme when it is no longer needed implicitly suggests that HtrA is harmful to the cell in the absence of adequate levels of substrate. Interestingly, during the course of our experiments we observed that expression of low levels of HtrA protein delayed bacterial growth, and expression of a noncleavable HtrA mutant (ΔPDZ2 HtrA), with a significantly longer clearance time, delayed bacterial growth even further (data not shown). These observations suggest that bacteria benefits from clearing HtrA when its enzymatic activities are no longer required and are an indication of the physiological relevance of these regulatory mechanism in vivo. However, testing our hypothesis in vivo is beyond the scope of the present study but will be the focus of our future work.
We noticed that some of the results presented here differ from a previous study on autocleavage of HtrA (23). Mainly, we found that the only requirement for autocleavage of HtrA is degradation of a substrate and, in fact, the peptides generated by HtrA during substrate degradation are sufficient to induce the self-cleavage of HtrA. The previous study indicates that reducing conditions must coexist along with substrate degradation to reduce the Cys57 and Cys69 disulfide bridge in HtrA and induce self-cleavage. Consistent with our own results, the C57A+C69G HtrA mutant did not show increased instability or a higher degree of self-cleavage compared to wild-type HtrA under any of the conditions tested. The reason for this discrepancy is unclear to us. There are some minor differences in the experimental conditions between both studies. In addition, the mutant used to test the influence of Cys57 and Cys69 in the stability of HtrA against autocleavage in the previous study (23) introduced different mutations on the cysteine residues (C57S+C69S). Potentially, in this mutant the oxygens that replace the sulfurs from the thiol groups could introduce an electrostatic repulsion between the two side chains that may destabilize the protein. Consistently, Skorko-Glonek et al. (23) found that the C57S+C69S mutant produced higher amounts of s-HtrA than did the wild type. Conversely, the C57A+C69G HtrA mutant in our study shortens or deletes the side chains of the two cysteine residues and probably has a milder effect on the three-dimensional structure of HtrA. In consequence, the C57A+C69G HtrA mutant is ideal for assessing the contribution of the disulfide bond to the stability of HtrA against autocleavage.
A previous publication (23) also reported that purified s-HtrA is stable. This result does not contradict our hypothesis that the autocleavage of HtrA is a mechanism aiming to eliminate the excess of HtrA. In fact, we also observed that in a purified HtrA preparation with a high proportion of the autocleaved form (used for experiment in Fig. 6), s-HtrA remained stable upon extended incubations at 37°C in the absence of substrate (data not shown). However, consistent with our hypothesis, if peptides resulting from substrate hydrolysis are present, s-HtrA is not stable and undergoes subsequent cleavages (Fig. 3B).
An alternative interpretation for our results is that the autocleavage of HtrA is just a side effect of the allosteric activation mechanism of HtrA. In this scenario, peptides from the hydrolysis reaction activate HtrA and, as a result, this hyperactive protease degrades itself. This simpler model sees the self-cleavage process as a “defect” of the mechanism of allosteric activation. According to this model, s-HtrA production should be produced in significant amounts as long as peptides resulting from the substrate hydrolysis are present in the reaction. However, our experiment illustrating that repetitive addition of full-length substrate drastically delays the autocleavage process contradicts this hypothesis. As described, we did not observe substantial autocleavage in spite of the presence of abundant peptides resulting from substrate hydrolysis. It was only after all of the recurrently added full-length substrate was hydrolyzed and HtrA reverted to its hexameric form that s-HtrA protein did accumulate significantly, indicating that bacteria effectively take advantage of the HtrA “defective” allosteric activation mechanism to eliminate the excess of HtrA in the cell.
Supplementary Material
Acknowledgments
We thank Daniela Damjanovic for constructing the C57A+C69G HtrA mutant. We are also grateful to Alba Guarne for insightful comments and critical reading of the manuscript.
This study was supported by a grant from the Canadian Institutes of Health Research (CIHR). J.O. is a recipient of a CIHR salary award and an Early Researcher Award from the Ministry of Research and Innovation.
Footnotes
Published ahead of print on 19 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Blair, W. S., and B. L. Semler. 1991. Self-cleaving proteases. Curr. Opin. Cell Biol. 31039-1045. [DOI] [PubMed] [Google Scholar]
- 2.Brass, J. M., C. F. Higgins, M. Foley, P. A. Rugman, J. Birmingham, and P. B. Garland. 1986. Lateral diffusion of proteins in the periplasm of Escherichia coli. J. Bacteriol. 165787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burckhardt, S. E., R. Woodgate, R. H. Scheuermann, and H. Echols. 1988. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc. Natl. Acad. Sci. USA 851811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10443-455. [DOI] [PubMed] [Google Scholar]
- 5.Ehrmann, M., and T. Clausen. 2004. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 38709-724. [DOI] [PubMed] [Google Scholar]
- 6.Gray, C. W., R. V. Ward, E. Karran, S. Turconi, A. Rowles, D. Viglienghi, C. Southan, A. Barton, K. G. Fantom, A. West, J. Savopoulos, N. J. Hassan, H. Clinkenbeard, C. Hanning, B. Amegadzie, J. B. Davis, C. Dingwall, G. P. Livi, and C. L. Creasy. 2000. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem. 2675699-5710. [DOI] [PubMed] [Google Scholar]
- 7.Hu, S. I., M. Carozza, M. Klein, P. Nantermet, D. Luk, and R. M. Crowl. 1998. Human HtrA, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem. 27334406-34412. [DOI] [PubMed] [Google Scholar]
- 8.Iwanczyk, J., D. Damjanovic, J. Kooistra, V. Leong, A. Jomaa, R. Ghirlando, and J. Ortega. 2007. The role of the PDZ domains in Escherichia coli DegP protein. J. Bacteriol. 1893176-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang, J., X. Zhang, Y. Chen, Y. Wu, Z. H. Zhou, Z. Chang, and S. F. Sui. 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. USA 10511939-11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jomaa, A., D. Damjanovic, V. Leong, R. Ghirlando, J. Iwanczyk, and J. Ortega. 2007. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 189706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, K. I., S. C. Park, S. H. Kang, G. W. Cheong, and C. H. Chung. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 2941363-1374. [DOI] [PubMed] [Google Scholar]
- 12.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416455-459. [DOI] [PubMed] [Google Scholar]
- 13.Krojer, T., J. Sawa, E. Schafer, H. R. Saibil, M. Ehrmann, and T. Clausen. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453885-890. [DOI] [PubMed] [Google Scholar]
- 14.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 1711574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipinska, B., S. Sharma, and C. Georgopoulos. 1988. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1610053-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 811375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little, J. W. 1983. The SOS regulatory system: control of its state by the level of RecA protease. J. Mol. Biol. 167791-808. [DOI] [PubMed] [Google Scholar]
- 18.Little, J. W., B. Kim, K. L. Roland, M. H. Smith, L. L. Lin, and S. N. Slilaty. 1994. Cleavage of LexA repressor. Methods Enzymol. 244266-284. [DOI] [PubMed] [Google Scholar]
- 19.Meltzer, M., S. Hasenbein, P. Hauske, N. Kucz, M. Merdanovic, S. Grau, A. Beil, D. Jones, T. Krojer, T. Clausen, M. Ehrmann, and M. Kaiser. 2008. Allosteric activation of HtrA protease DegP by stress signals during bacterial protein quality control. Angew Chem. Int. Ed. Engl. 471332-1334. [DOI] [PubMed] [Google Scholar]
- 20.Olsen, J. V., S. E. Ong, and M. Mann. 2004. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell Proteomics 3608-614. [DOI] [PubMed] [Google Scholar]
- 21.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 451599-1611. [DOI] [PubMed] [Google Scholar]
- 22.Raivio, T. L., and T. J. Silhavy. 1999. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2159-165. [DOI] [PubMed] [Google Scholar]
- 23.Skorko-Glonek, J., D. Zurawa, F. Tanfani, A. Scire, A. Wawrzynow, J. Narkiewicz, E. Bertoli, and B. Lipinska. 2003. The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim. Biophys. Acta 1649171-182. [DOI] [PubMed] [Google Scholar]
- 24.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97339-347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.