Abstract
Many proteins of two-component signal transduction systems (TCS) have domain structures that do not comply with a phosphate flow as observed in linear TCS, phosphorelays, or simple branched pathways. An example is RodK, which is essential for fruiting body formation in Myxococcus xanthus and, in addition to a sensor domain, consists of a kinase domain and three receiver domains (RodK-R1, -R2, and -R3), all of which are functionally important. We identified the RokA response regulator as part of the RodK pathway. In vitro the isolated RodK kinase domain engages in phosphotransfer to RodK-R3 and RokA, with a kinetic preference for RokA. However, in the context of the full-length protein, the RodK kinase domain has a preference for phosphotransfer to RodK-R3 over RokA. We suggest that in full-length RodK, the spatial proximity of the RodK kinase domain and RodK-R3 compensate for the kinetic preference of the isolated kinase domain for RokA. Thus, the kinetic preference observed using an isolated kinase domain of a hybrid kinase does not necessarily reflect the phosphotransfer preference of the full-length protein. We speculate that the phosphorylation status of RodK-R1 and RodK-R2 determines whether RodK engages in phosphotransfer to RodK-R3 or RokA in vivo.
In bacteria two-component signal transduction systems (TCS) have important functions in the sensing of and responding to a wide variety of internal and external cues and in this way regulate diverse processes such as motility, stress responses, cell cycle, differentiation, and development (42). A classical TCS consists of two proteins, a sensor histidine kinase (HPK), and a cognate response regulator (RR). HPKs are multidomain proteins and typically contain a nonconserved sensor domain, which is responsible for detecting a particular stimulus, and a conserved kinase domain. RRs are either single-domain proteins consisting only of the conserved receiver domain or multidomain proteins consisting of the receiver domain and a variable output domain. The HPK is activated in response to a specific stimulus, resulting in autophosphorylation on a conserved histidine residue in the kinase domain. Subsequently, the phosphoryl group is transferred to a conserved aspartate residue in the receiver domain of the cognate RR. The change in the phosphorylation state of the receiver domain typically results in the activation of an attached output domain, leading to altered gene expression, altered protein-protein interactions, or the regulation of enzyme activity (42). Phosphorelay systems represent more complex TCS and consist of four signaling modules, i.e., the HPK, a receiver domain, a histidine phosphotransferase (Hpt) domain, and, finally, a second response regulator. Phosphorelays function as classical TCS except that phosphotransfer occurs in three sequential steps from the HPK over the first receiver domain to the Hpt domain and, finally, to the second RR (42). The domain organization of proteins in phosphorelays is highly modular. Thus, all four domains may reside on separate proteins (5): the HPK and first receiver, referred to as a hybrid HPK, may be present in the same protein (28) or the HPK, and the first receiver and the Hpt domain, also referred to as a hybrid kinase, may be present in the same protein (45). The simple TCS and phosphorelays are typically organized as linear 1:1 and 1:1:1:1 pathways, respectively. In addition to the linear pathways, there are examples of branched TCS and phosphorelay systems, in which one HPK engages in phosphotransfer to several RRs (42) or several HPKs engage in phosphotransfer to the same Hpt protein or the same RR (13, 17, 21).
In bacteria, the number of TCS proteins correlates with the genome size (9, 10), and species such as Myxococcus xanthus contain more than 250 TCS proteins (38, 47). Given the modular structure of TCS proteins, mechanisms must exist to avoid unwanted cross talk between noncognate TCS proteins (4, 23). Global in vitro phosphotransfer profiling of TCS proteins in Caulobacter crescentus and Escherichia coli demonstrated that the specificity of phosphotransfer reactions is maintained in vitro and that cognate pairs of TCS proteins display a clear kinetic preference for each other (3, 40, 48). Therefore, it has been argued that the molecular mechanism that ensures specificity in TCS signaling and precludes unwanted cross talk depends on the kinetic preference of cognate TCS proteins for each other (23, 40).
Several TCS proteins have domain structures that do not easily comply with a phosphate flow as described for simple TCS, phosphorelays, or simple branched pathways. For example, RetS of Pseudomonas aeruginosa (12) and LtnB of Synechococcus elongatus (27) are HPKs containing two C-terminal receiver domains, and RodK of M. xanthus is an HPK with three C-terminal receiver domains (Fig. 1A) (33). Moreover, analyses of TCS proteins in sequenced bacterial genomes have revealed other atypical domain combinations in TCS proteins (38). These observations suggest the existence of pathway architectures involving proteins of TCS yet to be resolved.
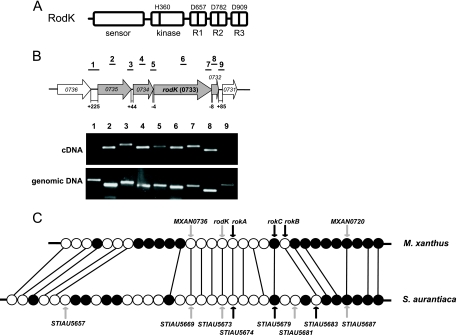
FIG. 1.
Analysis of the rodK locus. (A) RodK domain organization. The site of autophosphorylation (H360) is indicated. Potential phosphorylation sites (D657, D782, and D909) in the receiver domains are indicated. (B) Organization of the rodK transcriptional unit. Arrows indicate the direction of the transcription of rodK and flanking ORFs. Gray arrows indicate the rodK transcriptional unit. Lines above the arrows indicate intergenic and internal regions amplified by PCR with the indicated primer pairs. Numbers under the arrows indicate the sizes of the intergenic regions in base pairs. The PCR products amplified using cDNA and genomic DNA as templates were separated on a 2% agarose gel and shown as indicated. (C) Comparative analysis of the rodK locus in M. xanthus and S. aurantiaca. Open and closed circles indicate the ORFs transcribed in the same direction as and in the opposite direction of rodK, respectively. Orthologous ORFs are connected by lines. The ORFs encoding HPKs are indicated by gray arrows, and the ORFs encoding response regulators are indicated by black arrows. The HPKs encoded by STIAU5657 and STIAU5681 do not have orthologs in M. xanthus based on a pairwise reciprocal best BLASTP hit method (see Table S2 in the supplemental material for a complete list of the similarities and identities of proteins encoded by ORFs in rodK loci in M. xanthus and S. aurantiaca).
To further the understanding of how TCS proteins with atypical domain compositions function, we focused on the RodK protein of M. xanthus. M. xanthus responds to starvation with the formation of multicellular spore-filled fruiting bodies (7). This developmental program requires the temporal and spatial coordination of two morphogenetic processes, the aggregation of cells into nascent fruiting bodies, and the sporulation of cells located inside the fruiting bodies. Normally, cells that remain outside fruiting bodies do not sporulate (29). A ΔrodK mutant makes abnormally structured fruiting bodies and sporulates at reduced levels and with many spores being formed outside the fruiting bodies (33). Kinase activity is essential for RodK activity in vivo (35). Moreover, single substitutions of the conserved aspartate residues in the three receiver domains cause different defects in fruiting body formation, suggesting that each receiver domain has a distinct function in fruiting body formation, which depends on phosphorylation (35). In addition, mutant strains carrying pairwise substitutions of the conserved aspartate residues in the receiver domains have additive phenotypes, suggesting that the three receiver domains act in parallel pathways to regulate fruiting body formation (35). In in vitro phosphorylation experiments, full-length RodK autophosphorylates on the conserved histidine residue (H360) in the kinase domain, and then the phosphoryl group is transferred to the aspartate (D909) in the third receiver domain (RodK-R3), followed by the rapid dephosphorylation of D909 (35). Similar experiments did not provide evidence for the phosphotransfer from H360 to the aspartates (D657 and D782, respectively) in the two remaining receiver domains (RodK-R1 and RodK-R2) in full-length RodK (35). Based on these observations, it was suggested that D657 and D782 in RodK-R1 and RodK-R2 engage in phosphotransfer with other HPKs in vivo (35).
To further the understanding of the signaling mechanism of RodK, we analyzed the phosphotransfer properties of RodK. We identified RokA, a single receiver domain protein, as a novel component in the RodK pathway. Moreover, we show that the isolated RodK kinase domain engages in phosphotransfer to RodK-R3 and RokA with a clear kinetic preference for RokA. Despite the kinetic preference of the isolated kinase domain for RokA, full-length RodK preferentially engages in phosphotransfer to RodK-R3. Based on in vitro phosphorylation experiments with mutant RodK proteins, we suggest that in full-length RodK, the spatial proximity of RodK-R3 to the RodK kinase domain compensates for the kinetic preference of the kinase domain for RokA, thus allowing the preferential phosphotransfer to RodK-R3. These observations have implications for the interpretation of global phosphotransfer profiling experiments and suggest a novel mechanism for the regulation of phosphate flow within TCS.
MATERIALS AND METHODS
Growth conditions and development.
E. coli strains were grown in LB broth in the presence of relevant antibiotics (37). M. xanthus was grown in Casitone-Tris (CTT) medium in liquid cultures or on CTT agar plates (15). Aggregation was monitored on clone-fruiting (CF) agar (39) or in MC7 submerged culture as described previously (41). Briefly, cells were grown to a density of 5 × 108 cells/ml in CTT, harvested, and resuspended in MC7 buffer (10 mM MOPS [morpholinepropanesulfonic acid; pH 7.0], 1 mM CaCl2) at a calculated density of 5 × 109 cells/ml. A total of 20 μl of concentrated cell suspensions were spotted on CF agar, or 50-μl aliquots of concentrated cells were resuspended in a total volume of 400 μl MC7 in a 15-mm microtiter well and incubated at 32°C. Aggregation was followed by visual inspection using a Leica MZ8 stereomicroscope or a Leica IMB/E inverted microscope. The cells were photographed using a Leica DFC 350FX camera. The levels of sporulation were determined after development for 120 h as the number of sonication and heat-resistant CFU (41).
Bacterial strains and plasmids.
M. xanthus strains and plasmids are listed in Table 1. The plasmids were constructed by PCR using the primers listed in Table S1 in the supplemental material and cloning, followed by propagation in E. coli strain TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL endA1 nupG] unless otherwise stated. After cloning, the correct sequences of the DNA fragments were verified by sequencing. DK1622 was used as the wild type throughout. All strains constructed were confirmed by PCR. SA2800, SA2801, and SA2809 carry markerless in-frame deletions in rokA, rokB, and rokC, respectively. In-frame deletions were generated as described after cloning of the appropriate constructs in pBJ114 (38). SA2803 was generated by making a rokB in-frame deletion in SA2800. SA2808 was generated by making a rodK in-frame deletion in SA2800. SA2813 was generated by replacing the in-frame deletion of rokA in SA2800 with the rokA allele encoding RokAD56N.
TABLE 1.
M. xanthus strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| DK1622 | Wild type | 19 |
| SA1708 | ΔrodK (in-frame deletion from positions 307 to 2728)b | 33 |
| SA2800 | ΔrokA (in-frame deletion from positions 31 to 360)b | This study |
| SA2801 | ΔrokB (in-frame deletion from positions 31 to 360)b | This study |
| SA2803 | ΔrokA (in-frame deletion from positions 31 to 360),b ΔrokB (in-frame deletion from positions 31 to 360)b | This study |
| SA2808 | ΔrodK (in-frame deletion from positions 307 to 2728),b ΔrokA (in-frame deletion from positions 31 to 360)b | This study |
| SA2809 | ΔrokC (in-frame deletion from positions 28 to 627)b | This study |
| SA2813 | rokA with mutation in position 166 (exchange of G to A)b | This study |
| Plasmids | ||
| pBJ114 | Vector for constructing in-frame deletions, Kmr and galK+ | 18 |
| pUHE24-2 | Expression vector, Apr | 26 |
| pGEX-2T | Expression vector, Apr | GE Healthcare |
| pSW01 | 1,100 bp (positions −520 to 910 of rokA [positions 31 to 360 deleted])b in pBJ114 | This study |
| pMA16 | 1,100 bp (positions −520 to 910 of rokB [positions 31 to 360 deleted])b in pBJ114 | This study |
| pMA25 | 1,106 bp (positions −244 to 3607 of rodK [positions 307 to 2728 as well as positions 2969 to 3298 of rokA deleted])b in pBJ114 | This study |
| pMA29 | 1,098 bp (positions −522 to 1177 of rokC [positions 28 to 627 deleted])b in pBJ114 | This study |
| pSW33 | 1,430 bp (positions −520 to 910 of rokA [position 166 exchange of G to A])b in pBJ114 | This study |
| pMA134 | pGEX-2T derivative for the expression of GST-RodK-R1 (aa 606 to 717 of RodK) | This study |
| pMA234 | pGEX-2T derivative for the expression of GST-RodK-R1D657N (aa 606 to 717 of RodK) | This study |
| pMA156 | pGEX-2T derivative for the expression of GST-RodK-R2 (aa 731 to 841 of RodK) | This study |
| pMA256 | pGEX-2T derivative for the expression of GST-RodK-R2D782N (aa 731 to 841 of RodK) | This study |
| pMA178 | pGEX-2T derivative for the expression of GST-RodK-R3 (aa 858 to 972 of RodK) | This study |
| pMA278 | pGEX-2T derivative for the expression of GST-RodK-R3D909N (aa 858 to 972 of RodK) | This study |
| pMA10 | pGEX-2T derivative for the expression of GST-RokA (aa 1 to 136) | This study |
| pMA13 | pGEX-2T derivative for the expression of GST-RokAD56N (aa 1 to 136) | This study |
| pMA112 | pUHE24-2 derivative for the expression of His6-RodKKinase (aa 329 to 603) | This study |
| pMA1-4 | pUHE24-2 derivative for the expression of His6-RodKΔsensor (aa 329 to 981 of RodK) | This study |
| pAAR327 | pUHE24-2 derivative for the expression of His6-RodKHDDD (aa 1 to 981) | 33 |
| pAAR350 | pUHE24-2 derivative for the expression of His6-RodKHNDN (aa 1 to 981) | 35 |
| pAAR351 | pUHE24-2 derivative for the expression of His6-RodKHDNN (aa 1 to 981) | 35 |
| pAAR352 | pUHE24-2 derivative for the expression of His6-RodKHNND (aa 1 to 981) | 35 |
| pAAR353 | pUHE24-2 derivative for the expression of His6-RodKHNNN (aa 1 to 981) | 33 |
Kmr, kanamycin resistance; Apr, ampicillin resistance; aa, amino acids.
All coordinates are relative to the start codon of the relevant gene.
Reverse transcription-PCR.
Total RNA was extracted from DK1622 wild-type cells grown in liquid 1% CTT medium at 32°C to a density of 5 × 108 cells/ml as described previously (31). cDNA was synthesized using the cDNA archive kit (ABI) from 1.0 μg of DNA-free total RNA. PCR was carried out using standard reactions and cDNA, RNA, or genomic DNA from DK1622 as templates. The primers used are listed in Table S1 in the supplemental material.
Protein purification.
His6-RodKDDD, His6-RodKNDN, His6-RodKDNN, His6-RodKNND, His6-RodKNNN, His6-RodKΔsensor, and His6-RodKKinase were expressed from pAAR327, pAAR350, pAAR351, pAAR352 pAAR353, pAAR355, pMA1-4, and pMA112, respectively, in E. coli JM109 [F′ traD36 proA+B+ lacIq Δ(lacZ)M15 Δ(lac-proAB) glnV44 e14− gyrA96 recA1 relA1 endA1 thi hsdR17]. These plasmids are derivatives of the expression vector pUHE24-2 and are identical except for the specific rodK allele they contain. Glutathione S-transferase (GST)-RodK-R1, GST-RodK-R1D657N, GST-RodK-R2, GST-RodK-R2D782N, GST-RodK-R3, GST-RodK-R3D909N, GST-RokA, and GST-RokAD56N were expressed from pMA134, pMA234, pMA156, pMA256, pMA178, pMA278, pMA10, and pMA13, respectively, in BL21 [E. coli B F− ompT hsdS (rB− mB−) gal dcm]. These plasmids are derivatives of the expression vector pGEX-2T and are identical except for the specific gene they contain. His-tagged proteins were purified as described previously (33). GST-tagged proteins were purified using the GST-glutathione affinity system according to the manufacturer's protocol (GE Healthcare).
In vitro phosphorylation.
In vitro phosphorylation experiments were all performed in TGMNKD buffer (50 mM Tris-HCl [pH 8], 10% [vol/vol] glycerol, 5 mM MgCl2, 150 mM NaCl, 50 mM KCl, 1 mM dithiothreitol) as described previously (35). The in vitro phosphorylation experiments were carried out using two different setups. Either (i) RodKKinase (5 μM final concentration) or RodKNNN (5 μM final concentration) were incubated with 0.5 mM [γ-32P]ATP (14.8 GBq mmol−1; Amersham) as a substrate for 120 min at 25°C, followed by the removal of [γ-32P]ATP by ultrafiltration using Microcon-Y10 columns (Millipore) and then incubation with the receiver proteins (5 μM final concentrations) for various periods of time at 25°C, or (ii) various full-length His6-RodK proteins (5 μM final concentration) were incubated together with the individual receiver proteins (5 μM final concentration) for 120 min at 25°C in the presence of 0.5 mM [γ-32P]ATP. In all experiments, aliquots of 10 μl were quenched in 5 μl of 3× sodium dodecyl sulfate (SDS)/EDTA loading buffer (7.5% [wt/vol] SDS, 90 mM EDTA, 37.5 mM Tris-HCl [pH 6.8], 37.5% glycerol [vol/vol], 0.3 M dithiothreitol) and separated by SDS-polyacrylamide gel electrophoresis (PAGE) at 4°C without prior heating (35). Subsequently, the gel was exposed on a phosphorimager screen (GE Healthcare), scanned on a Typhoon phosphorimager (GE Healthcare) and analyzed using ImageQuant (GE Healthcare).
In vitro measurements of ATP hydrolysis.
The determination of ATP hydrolysis by His6-RodK proteins was carried out by incubating individual RodK proteins (5 μM final concentration) in TGMNKD buffer for 120 min at 25°C in the presence of 0.5 mM [α-32P]ATP (110 TBq mmol−1; Amersham) in the presence and absence of RokA (5 μM final concentration). As a negative control to show that ATP hydrolysis depends on RodK proteins, we used a mock control consisting of proteins purified as described for RodK proteins from E. coli JM109 containing the vector pUHE24-2. The nucleotides were separated from the proteins using Microcon-Y10 ultrafiltration columns (Millipore). Labeled adenine nucleotides were separated by thin-layer chromatography (TLC) as described previously (35). Briefly, aliquots of 0.5 μl of the eluate were applied to a poly(ethyleneimine)-cellulose F TLC plate (Merck) with 2.4 M formic acid as the solvent system. Labeled nucleotides on the TLC plates were visualized and quantified by phosphorimaging and analyzed using ImageQuant software. [α-32P]ATP and [α-32P]ADP were located on the TLC plates using the products of [α-32P]ATP hydrolysis by the Apyrase ATPase (Sigma-Aldrich) as markers.
Immunoblot analyses.
To generate anti-RokA antibodies, GST-RokA was purified and used to immunize a rabbit using standard procedures (37). RodK and RokA immunoblot analyses were carried out as described previously (33).
RESULTS
Identification of TCS proteins potentially interacting with RodK.
To identify TCS proteins that could be part of the RodK signal transduction pathway, we took advantage of the observation that cognate partners of TCS and phosphorelays are often encoded by genes in the same operon or clustered in the genome (20, 38, 47). Therefore, we initially mapped the rodK transcriptional unit. Three and four genes upstream and downstream from rodK, respectively, are transcribed in the same direction as rodK (Fig. 1B and C). Based on the distances between the start and stop codons of these genes (Fig. 1B), we speculated that the rodK mRNA is polycistronic and that MXAN0735, MXAN0734, rodK, and MXAN0732 constitute an operon. To test this hypothesis, a reverse transcription-PCR approach was used. RodK protein accumulates in vegetative cells (33). Therefore, to map the rodK mRNA, total RNA was isolated from exponentially growing wild-type cells (DK1622). PCR primer pairs were designed to generate PCR products covering internal regions of the genes of interest as well as intergenic regions (Fig. 1B). The PCR products labeled 2 to 8, which cover the internal and intergenic regions from MXAN0735 to MXAN0732, were obtained using genomic DNA as well as cDNA as templates (Fig. 1B). The control experiments verified that no PCR products were generated using RNA as a template in the absence of reverse transcription (data not shown). With the primer pairs 1 and 9, which cover the intergenic regions MXAN0736-MXAN0735 and MXAN0732-MXAN0731, respectively, PCR products were generated using genomic DNA as a template, whereas no PCR products were generated using cDNA as a template (Fig. 1B). These data show that the four genes MXAN0735, MXAN0734, rodK, and MXAN0732 are organized in an operon, suggesting that the four proteins are involved in the same process. MXAN0735 and MXAN0734 encode conserved hypothetical proteins, and MXAN0732 encodes a single-domain receiver protein.
We reasoned that TCS proteins encoded by genes conserved in rodK loci in other species could also be part of the RodK signal transduction pathway. By searching nonredundant databases for RodK orthologs, RodK was found only to be conserved in Stigmatella aurantiaca, which like M. xanthus is a deltaproteobacterium belonging to the Myxococcales and able to form fruiting bodies. To identify orthologs encoded by genes conserved between the two rodK loci, a pairwise reciprocal best-BLASTP-hit method (1) was used in which the proteins encoded by 15 open reading frames (ORFs) upstream and 15 ORFs downstream from rodK in M. xanthus were searched against the S. aurantiaca genome and vice versa. An overall synteny between the two rodK loci was found (Fig. 1C). Among the genes conserved between the rodK loci in M. xanthus and S. aurantiaca, five showed significant homology to TCS proteins (see Table S2 in the supplemental material for a complete list of the similarities and identities of the proteins encoded by ORFs in rodK loci in M. xanthus and S. aurantiaca). These genes include two encoding HPKs (MXAN0736 and MXAN0720) and three encoding RRs (MXAN0732, MXAN0727, and MXAN0726). A mutant carrying an in-frame deletion of MXAN0736 aggregates and sporulates as a wild-type cell, but the mutant spores germinate at a threefold lower level than the wild-type spores (38). As mutations in rodK do not cause spore germination defects (33, 35), we did not further analyze a possible interaction between RodK and MXAN0736. An in-frame deletion of MXAN0720 does not cause any defects in fruiting body formation, sporulation, or spore germination (X. Shi and L. Søgaard-Andersen, unpublished data), suggesting that MXAN0720 is not part of the RodK signal transduction pathway. Therefore, we focused on the three receiver domain proteins located downstream of RodK. From here on, these genes are referred to as rokA (MXAN0732), rokB (MXAN0726), and rokC (MXAN0727) for receiver downstream of RodK.
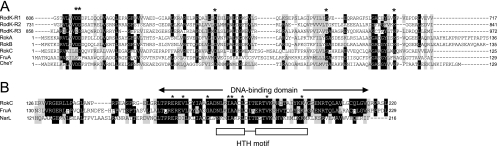
RokA and RokB are single receiver domain proteins lacking obvious output domains and share a high degree of similarity and identity (82%/72%) (Fig. 2A). RokC contains an N-terminal receiver domain and a C-terminal DNA binding output domain of the LuxR/FixJ family (Fig. 2C). RokC is most similar to the response regulator FruA (54% similarity/40% identity), which has a crucial role in fruiting body formation in M. xanthus (8, 30). Receiver domains are characterized by several conserved residues (36), which are all present in the receiver domains in RokA, RokB, and RokC, with the exception that RokC contains Glu instead of Asp at the position that corresponds to position 12 in E. coli CheY (Fig. 2A).
FIG. 2.
Comparison of TCS proteins encoded by ORFs in the rodK locus. (A) Alignment of receiver domains in RodK, RokA, RokB, and RokC with M. xanthus FruA and E. coli CheY. The alignment was generated using CLUSTAL W (44). Residues on black are identical in more than 50% of the proteins, and residues on gray indicate similar amino acids conserved in more than 50% of the proteins. Asterisks indicate conserved signature residues of receiver domains (36). (B) Alignment of DNA binding domain of RokC to those of E. coli NarL and M. xanthus FruA. The DNA binding domain in NarL is indicated. Asterisks indicate residues involved in maintaining the tertiary structure of the DNA binding domain in NarL (2). The α-helix-turn-α-helix (HTH) DNA binding motif in NarL is indicated below the sequences. Identical and similar residues are indicated as in panel A.
Genetic analysis of rokA, rokB, and rokC.
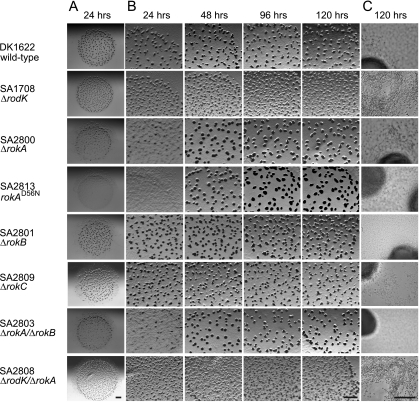
To analyze whether rokA, rokB, and rokC are important for fruiting body formation, we constructed in-frame deletion mutations in the three genes. The three mutant strains were exposed to starvation in parallel with the DK1622 wild type and SA1708 (ΔrodK). On CF agar, DK1622 wild-type cells formed fruiting bodies at 24 h of starvation, and spores were rarely observed outside fruiting bodies (Fig. 3). SA1708 (ΔrodK) aggregated to form many small, abnormally shaped fruiting bodies at 24 h. After 120 h of starvation, the ΔrodK strain sporulated at 1% of the level of the wild type, and many spores were formed outside fruiting bodies. The ΔrokA mutant (SA2800) displayed delayed aggregation and did not form fruiting bodies until 48 h of starvation. The strain sporulated at wild-type levels, and spores were rarely observed outside fruiting bodies. The ΔrokB (SA2801) and ΔrokC (SA2809) strains did not display defects in fruiting body formation or in spore germination. The high levels of similarity and identity between RokA and RokB suggested that these proteins could be functionally redundant. To test this hypothesis, we constructed a ΔrokA ΔrokB double mutant (SA2803). SA2803 displayed a phenotype similar to that of SA2800 (ΔrokA), suggesting that RokB is not important for fruiting body formation. To analyze whether rokA is in the same genetic pathway as rodK, we constructed a ΔrodK ΔrokA double mutant (SA2808). SA2808 had a phenotype indistinguishable from that of SA1708 (ΔrodK), suggesting that RodK and RokA act in the same pathway. Notably, the more severe developmental defects in the ΔrodK mutant compared to the ΔrokA mutant indicate that the output from RodK is not only generated through RokA.
FIG. 3.
Developmental phenotypes of mutants carrying mutations in genes encoding TCS proteins in the rodK locus. Strains were starved on CF agar for the indicated periods of time. Below the strain numbers, the genotype of a particular strain is indicated. Bars, 1.0 mm (A and B) and 50 μm (C).
To analyze the importance of the conserved aspartate residue (D56) in RokA for activity, the ΔrokA allele in SA2800 was replaced with the rokA(D56N) allele, which encodes a mutant RokA protein in which D56 was substituted with the nonphosphorylatable asparagine residue. The resulting strain SA2813 displayed defects in fruiting body formation similar to those of the ΔrokA strain (SA2800) (Fig. 3). These defects were not caused by lack of accumulation of RokAD56N because the mutant protein accumulated at levels and with a profile similar to that of RokA wild-type protein (Fig. 4, see below). Thus, the conserved aspartate residue is important for RokA activity in vivo. Taken together, these analyses suggest that RokA is part of the RodK signal transduction pathway whereas RokB and RokC are not part of this pathway and are not important for fruiting body formation.
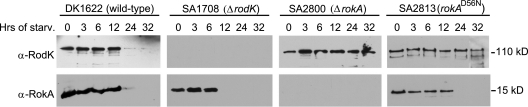
FIG. 4.
Immunoblot analyses of RodK and RokA accumulation. Total cell lysates were prepared from the indicated strains after starvation in submerged cultures for the indicated periods of time. Protein from an equal number of input cells (6 × 107 cells) was loaded per lane.
RodK and RokA have similar accumulation profiles.
To determine whether RodK and RokA display similar accumulation profiles, immunoblot analyses were carried out using antibodies against RodK (33) and RokA (Fig. 4). As previously shown (33), RodK accumulated in exponentially growing wild-type cells. During starvation, the accumulation level remained unchanged until 12 h, and at 24 h, the protein was no longer detectable. In the wild type, the antibodies against RokA recognized a protein with a size of 15 kDa corresponding well to the calculated molecular mass of RokA (14.9 kDa), and this protein was absent in the ΔrokA strain (SA2800). These observations suggest that the anti-RokA antibodies are specific to RokA. RokA was also detected in exponentially growing wild-type cells, the accumulation level remained unchanged until 12 h of starvation, and at 24 h, the protein was no longer detectable. In the ΔrodK strain (SA1708), RokA was only detectable until 6 h of starvation. In contrast, in the ΔrokA strain (SA2800) and the strain expressing mutant RokAD56N protein (SA2813), the level of RodK remained unchanged until 32 h of starvation. Thus, in wild-type cells, RodK and RokA display similar accumulation profiles, which would, in principle, allow the two proteins to interact in vivo. Moreover, lack of or alteration of one protein affects the accumulation of the other protein. These observations support the notion that RodK and RokA may interact in vivo.
Phosphotransfer in vitro from the RodK kinase domain.
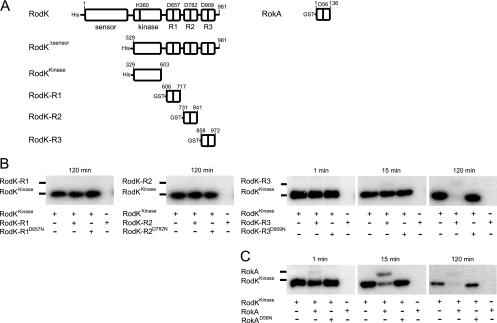
Previous experiments with full-length RodK proteins in vitro provided evidence for phosphotransfer from the RodK kinase domain (in the context of the full-length protein) to RodK-R3 and also suggested that full-length RodK does not engage in phosphotransfer to RodK-R1 and RodK-R2. To verify these observations and to examine whether the isolated RodK kinase domain engages in phosphotransfer to RokA, an N-terminal His6-tagged RodK protein (RodKKinase, residues 329 to 603) lacking the sensor domain and the three receiver domains, was expressed in E. coli and purified to near homogeneity by Ni2+-affinity chromatography under native conditions. Moreover, the three RodK receiver domains RodK-R1 (residues 606 to 717), RodK-R2 (residues 731 to 841), RodK-R3 (residues 858 to 972), and RokA (full-length) were expressed as GST-tagged proteins in E. coli and purified to near homogeneity by glutathione-affinity chromatography (Fig. 5A).
FIG. 5.
In vitro analysis of phosphotransfer from RodKKinase. (A) Schematic representation of proteins used in in vitro phosphotransfer analyses. Potential phosphorylation sites (D657, D782, and D909 in RodK and D56 in RokA) in the receiver domains are indicated. The amino acids of the respective proteins present in the different proteins are indicated. (B) Autoradiograms of in vitro phosphotransfer reactions from RodKKinase to the indicated receiver proteins. RodKKinase was autophosphorylated for 2 h in the presence of [γ-32P]ATP. After the removal of [γ-32P]ATP from the reaction, the sample was split and the indicated receiver proteins were added for the indicated periods of time. Samples were separated by SDS-PAGE without prior heating and detected by phosphorimaging. Note that the sizes of the receiver proteins are larger than of the RodKKinase protein due to the GST tag (calculated molecular mass, 24.2 kDa).
In in vitro phosphorylation experiments, RodKKinase (5 μM final concentration) was autophosphorylated by incubation for 2 h using [γ-32P]ATP as a phosphodonor. Subsequently, [γ-32P]ATP was removed, and the individual receiver proteins (5 μM final concentrations) added separately for 1, 15, and 120 min before the reactions were stopped. RodKKinase was efficiently autophosphorylated under these conditions as evidenced by the appearance of a radiolabeled band at the correct molecular mass (Fig. 5B). As expected, a RodKKinase protein, in which the conserved histidine residue corresponding to H360 in the full-length protein was substituted to alanine, was not autophosphorylated (data not shown). No phosphotransfer to RodK-R1 and RodK-R2 was detected even after 120 min. For RodK-R3, phosphotransfer was detected after 15 min and 120 min (Fig. 5B), as evidenced by the appearance of a faint radiolabeled band at the molecular mass corresponding to RodK-R3 and a decrease in the level of phosphorylated RodKKinase. The intensity of the radiolabeled band corresponding to RodK-R3 did not correlate with the decrease in the intensity of the radiolabeled band corresponding to RodKKinase, suggesting that RodK-R3 is rapidly dephosphorylated. When adding a mutant RodK-R3 (RodK-R3D909N), purified as described for the wild-type protein and in which the phosphorylatable D909 residue in the receiver domain was substituted to asparagine, no phosphotransfer was detected, demonstrating the specificity of the phosphotransfer reaction (Fig. 5B). Phosphotransfer to RokA from RodKKinase was also detected (Fig. 5C). Phosphotransfer to RokA was evident already after 1 min by the appearance of a radiolabeled band at the molecular mass corresponding to RokA and a decrease in the intensity of the radiolabeled band corresponding to phosphorylated RodKKinase. Also in the case of RokA, the intensity of the radiolabeled band corresponding to RokA did not correlate with the decrease in the intensity of the radiolabeled band corresponding to RodKKinase, suggesting that RokA is rapidly dephosphorylated. Phosphotransfer to RokA was dependent on the conserved D56 residue because no phosphotransfer was observed when using the mutant RokAD56N protein. For RodK-R1, RodK-R2, and RodK-R3, these results are in agreement with previous data and provide direct evidence for phosphotransfer from RodKKinase to RodK-R3 and support the notion that RodK-R1 and RodK-R2 are not substrates of the RodK kinase. The experiments also show that RokA is a direct substrate of RodKKinase. RodKKinase displays a clear kinetic preference for RokA compared to RodK-R3. From these data, we conclude that the phosphoryl group can be transferred from RodKKinase to RodK-R3 and RokA with a kinetic preference for RokA.
Phosphotransfer from the RodK full-length protein.
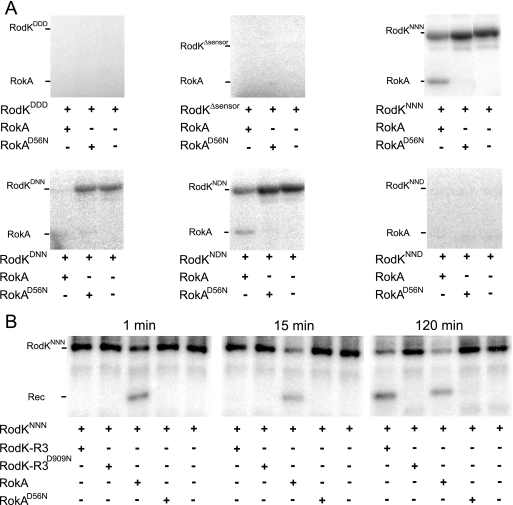
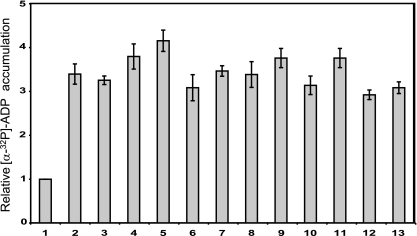
To determine whether phosphotransfer occurs from the full-length RodK protein to RokA, His6-tagged full-length RodK (RodKDDD) and various mutant full-length RodK proteins carrying substitutions of the potential phosphorylation sites in the three receiver domains (RodKD/ND/ND/N) were purified as described, and phosphotransfer to RokA was analyzed (Fig. 6). The autophosphorylation of RodKDDD cannot be detected directly due to the transfer of the phosphoryl group from H360 to D909 in RodK-R3, followed by rapid dephosphorylation (35). In contrast, autophosphorylation on H360 is detected in full-length RodK proteins in which the D909 is substituted to the nonphosphorylatable asparagine (35). Therefore, to be able to detect phosphotransfer from full-length RodK proteins to RokA, RodK proteins (5 μM final concentration) and RokA (5 μM final concentration) were incubated together for 2 h in the presence of [γ-32P]ATP. In agreement with previous results, phosphorylated wild-type RodK protein (RodKDDD) was not detected (Fig. 6A). Phosphotransfer from RodKDDD to RokA was also not detected (Fig. 6A). To verify that RodKDDD has kinase activity we used a previously described assay for kinase activity in which the accumulation of [α-32P]ADP in the in vitro phosphorylation assays was determined using [α-32P]ATP as a substrate and TLC to separate [α-32P]ATP and [α-32P]ADP (35). As shown in Fig. 7, similar amounts of [α-32P]ADP accumulated in reactions containing only RodKDDD and in reactions containing RodKDDD plus RokA. These data verify that RodKDDD has kinase activity and that the lack of detectable autophosphorylation of RodKDDD is caused by the rate of dephosphorylation being higher than the rate of autophosphorylation. Most importantly, despite the kinetic preference of RodKKinase for RokA, RokA cannot efficiently compete for the phosphoryl group on H360 in the context of the RodKDDD full-length protein. These data suggest that even in the presence of RokA, the phosphoryl group on H360 in RodKDDD is transferred to RodK-R3, followed by rapid dephosphorylation.
FIG. 6.
In vitro analysis of the phosphotransfer from RodK full-length proteins. (A) Autoradiograms of in vitro phosphotransfer reactions from the indicated RodK full-length proteins to RokA and RokAD56N. The indicated proteins were incubated together for 2 h in the presence of [γ-32P]ATP. Samples were separated by SDS-PAGE without prior heating and detected by phosphorimaging. (B) Autoradiograms of in vitro phosphotransfer reactions from RodKNNN to the indicated receiver proteins. RodKNNN was autophosphorylated for 2 h in the presence of [γ-32P]ATP. After the removal of [γ-32P]ATP from the reaction, the sample was split and the different receiver proteins were added for the indicated periods of time. Samples were separated by SDS-PAGE without prior heating and detected by phosphorimaging.
FIG. 7.
In vitro kinase activity of RodK proteins. Histogram of the relative level of [α-32P]ADP accumulation in reactions containing the following protein combinations: mock purified proteins from E. coli JM109 (1), RodKDDD (2), RodKDDD and RokA (3), RodKΔsensor (4), RodKΔsensor and RokA (5), RodKNNN (6), RodKNNN and RokA (7), RodKDNN (8), RodKDNN and RokA (9), RodKNDN (10), RodKNDN and RokA (11), RodKNND (12), and RodKNND and RokA (13). Proteins were incubated with [α-32P]ATP followed by the separation of the nucleotides by TLC and autoradiography. The relative level of [α-32P]ADP accumulation was determined by calculating the [α-32P]ADP/[α-32P]ATP ratio for a given reaction and then correcting this ratio with the [α-32P]ADP/[α-32P]ATP ratio of the reaction containing proteins from the mock purification. The relative levels of [α-32P]ADP accumulation are presented as the mean values of three technical replicates, and the error bars indicate standard deviations.
To analyze whether the difference in the kinetic preference of RodKKinase and RodKDDD was dependent on the presence or absence of the sensor domain, in vitro phosphorylation experiments were carried out using a His6-tagged RodK protein lacking the sensor domain, RodKΔsensor (residues 329 to 981) (Fig. 5A). Also in the case of RodKΔsensor, neither autophosphorylation nor phosphotransfer to RokA were detected (Fig. 6A). By assaying for kinase activity, we observed that [α-32P]ADP accumulated in the reactions containing RodKΔsensor and RodKΔsensor plus RokA at levels similar to that observed in the reactions containing RodKDDD and RodKDDD plus RokA (Fig. 7). These data suggest that also in the case of the RodKΔsensor protein, the phosphoryl group on H360 is preferentially transferred to RodK-R3 followed by rapid dephosphorylation. Thus, the presence or absence of the sensor domain does not influence the phosphate flow within full-length RodK.
Our data suggested that even in the presence of RokA, the phosphoryl group on H360 in RodKDDD is transferred to RodK-R3 followed by rapid dephosphorylation. To test this hypothesis and to determine whether full-length RodK proteins can engage in phosphotransfer to RokA, we analyzed phosphotransfer to RokA from various mutant full-length RodK proteins containing different combinations of asparagine substitutions of the potential phosphorylation sites in the three receiver domains. In in vitro phosphorylation experiments using RodKNNN, RodKDNN, and RodKNDN, autophosphorylation on H360 was detected as reported (33). Importantly, phosphotransfer to RokA was detected by the appearance of a radiolabeled band corresponding to the molecular mass of RokA and by a decrease in the intensity of the radiolabeled band corresponding to phosphorylated RodK proteins (Fig. 6A). Phosphotransfer from RodKNNN, RodKDNN, and RodKNDN was dependent on D56 in RokA because no phosphotransfer was detected to RokAD56N (Fig. 6A). In these experiments, the decrease in the level of phosphorylated RodK proteins in the presence of RokA indicates that the rate of phosphotransfer to RokA is higher than the rate of autophosphorylation of the RodK proteins. In contrast, neither autophosphorylation nor phosphotransfer to RokA was observed using RodKNND (Fig. 6A). By assaying for kinase activity as described, we observed that [α-32P]ADP accumulated in the reactions containing RodKNNN, RodKDNN, RodKNDN, and RodKNND with or without RokA at levels similar to those observed in the reactions containing RodKDDD and RodKDDD plus RokA (Fig. 7). Thus, all full length, RodK proteins display similar levels of kinase activity. Moreover, in full-length RodK proteins in which D909 has been substituted to the nonphosphorylatable asparagine, the phosphoryl group on H360 is transferred to RokA, whereas in full-length RodK proteins with an intact D909 residue, the phosphoryl group on H360 is transferred to D909. These data suggest that in the context of full-length RodK, the spatial proximity of RodK-R3 to the kinase domain compensates for the kinetic preference of the isolated kinase domain for RokA.
To rule out the possibility that full-length RodK has a different kinetic preference in trans for receiver domains than the isolated kinase domain, we determined the kinetic preference of RodKNNN for RodK-R3 and RokA in trans. To this end, RodKNNN was autophosphorylated for 2 h in the presence of [γ-32P]ATP, and subsequently [γ-32P]ATP was removed, the sample was split, and RodK-R3 or RokA was added for different periods of time (Fig. 6B). After 1 min, phosphotransfer to RokA was detected by the appearance of a radiolabeled band at the molecular mass of RokA and a decrease in the radiolabeled band corresponding to phosphorylated RodKNNN. Phosphotransfer in trans from RodKNNN to RodK-R3 was not detected until 120 min. From these analyses, we conclude that full-length RodKNNN has a kinetic preference in trans for RokA over RodK-R3.
DISCUSSION
To understand the signaling mechanism of the RodK kinase, we took advantage of the observation that in in vitro phosphotransfer profiling experiments, purified kinase domains exhibit a kinetic preference for their cognate in vivo substrates (3, 40, 48). In vitro phosphorylation experiments provided direct evidence for phosphotransfer from H360 in the isolated RodK kinase domain to D909 in RodK-R3, whereas phosphotransfer to D657 and D782 in RodK-R1 and RodK-R2 was not detected. These observations strongly support the notion that RodK-R3 does not inhibit phosphotransfer to D657 and D782 in RodK-R1 and RodK-R2 in the context of the full-length protein. Moreover, these observations strongly suggest that the lack of detectable phosphotransfer to D657 and D782 in RodK-R1 and RodK-R2 in full-length RodK is not due to lack of stimulation of the sensor domain. From these observations, we conclude that in full-length RodK, the RodK kinase domain only engages in phosphotransfer to RodK-R3, which is then rapidly dephosphorylated.
To further the understanding of how RodK regulates fruiting body formation we identified candidate RR proteins that could act in the RodK signal transduction pathway using two strategies. First, rodK was shown to be part of a four-gene operon, which in addition to rodK includes rokA, which encodes a RR consisting of a single receiver domain, and two genes (MXAN0735 and MXAN0734) encoding conserved hypothetical proteins. We are currently testing whether the MXAN0735 and MXAN0734 proteins are important for fruiting body formation. Second, we identified three genes (rokA, rokB, and rokC) encoding RRs that are conserved between the different rodK loci. In addition to genetic organization, three lines of evidence suggest that RodK and RokA act in the same signal transduction pathway. First, among the three RRs, only RokA is essential for fruiting body formation, and epistasis experiments suggest that RokA acts in the RodK pathway. Second, RodK and RokA display similar accumulation profiles, and a lack of RodK affects RokA accumulation and vice versa. Third, in vitro phosphorylation experiments showed that the isolated RodK kinase domain engages in phosphotransfer to D56 in RokA. From these observations, we conclude that RokA is part of the RodK signaling pathway. The ΔrodK mutant has more severe developmental defects than the ΔrokA mutant, suggesting that the output from RodK is not only generated through RokA consistent with the observation that RodK-R1, -R2, and -R3 have important functions in development (35).
Interestingly, the isolated RodK kinase domain has a strong kinetic preference for RokA over RodK-R3 in in vitro phosphotransfer experiments, whereas full-length RodK with an intact RodK-R3 domain exclusively engaged in phosphotransfer to RodK-R3. In contrast, full-length RodK proteins in which the D909 residue in RodK-R3 was substituted exclusively engaged in phosphotransfer to RokA. Moreover, full-length RodK (RodKNNN) that cannot transfer the phosphoryl group to RodK-R3 exhibits a kinetic preference for RokA over RodK-R3 in trans. Thus, the isolated RodK kinase domain and the RodK kinase domain in the context of the full-length protein both have a kinetic preference for RokA over RodK-R3 in trans. However, when the RodK kinase domain and the D909 residue in RodK-R3 are present in the same protein, i.e., in cis, the RodK kinase has a preference for RodK-R3 over RokA.
Several hybrid HPKs are part of phosphorelay systems in which the phosphoryl group is transferred from the histidine in the kinase domain to the aspartate in the receiver domain in the hybrid kinase, then to a histidine in an Hpt domain, and then to the aspartate in the receiver domain in the ultimate RR. To our knowledge, it has only been observed in the ArcBA TCS in E. coli that the isolated kinase domain of a hybrid kinase (ArcB) can engage directly in phosphotransfer to the receiver domain in the ultimate RR (ArcA) (11), suggesting that the transfer of the phosphoryl group to the receiver domain in the ultimate RR in a phosphorelay generally depends on an Hpt domain protein that acts between the two receiver domains (3, 16, 43). Therefore, we consider it unlikely that the phosphoryl group in full-length RodK in vivo is shuttled from RodK-R3 by an unknown Hpt protein to RokA. Based on these observations, we suggest that the recognition determinants in RokA involved in the interaction with the RodK kinase domain are more optimal than the corresponding determinants in RodK-R3. However, in the context of full-length RodK the spatial proximity of RodK-R3 results in a higher local concentration of RodK-R3 close to the kinase domain and thus compensates for the kinetic preference of the kinase domain for RokA. Hence, in full-length RodK, the spatial proximity of RodK3 supersedes the kinetic preference for RokA as observed with separate domains. Several phosphotransfer profiling experiments have revealed that HPKs in vitro have a strong kinetic preference for their in vivo cognate substrate (3, 23, 40, 48). The data presented here for the first time demonstrate that in the case of hybrid HPKs, the kinetic preference in vitro using isolated kinase domains do not necessarily reflect the phosphotransfer preference of the full-length proteins.
The conserved aspartate residues in RodK-R1 and RodK-R2 are important for RodK activity in vivo (35). The lack of phosphotransfer from the isolated RodK kinase domain to RodK-R1 and RodK-R2 as well as within full-length RodK suggests that RodK-R1 and RodK-R2 are phosphorylated by other HPKs. It has been suggested that the hybrid HPK RetS in P. aeruginosa, which contain two C-terminal receiver domains, could be phosphorylated by other HPK(s) (12, 22). However, to our knowledge, there are no documented examples in which HPK(s) engages in phosphotransfer to receiver domain(s) in another hybrid kinase. A pathway design in which HPK(s) phosphorylates receiver domains within a hybrid HPK would allow the integration of different input signals into a single protein. Several HPKs have been identified in M. xanthus that are essential for the spatial coupling of aggregation and sporulation, i.e., MXAN3290 (38), TodK (34), EspA (6), EspC (24), and RedC (14). We are currently testing whether any of these HPKs could be involved in RodK-R1 and RodK-R2 phosphorylation. Interestingly, a search of the SMART domain database (25) revealed that of more than 5,000 hybrid HPKs without an Hpt domain, 271 and 83 of these hybrid kinases contain two and three C-terminal receiver domains, respectively, as in RetS and RodK. These observations suggest that signal integration mechanisms operating at the level of hybrid HPKs and containing more than one receiver domain and no Hpt domain may not be uncommon.
RodK plays a crucial role in coordinating aggregation and sporulation during fruiting body formation. We speculate that RodK serves as an integration point for different signaling pathways. According to this model, one signal would modulate the RodK kinase activity, and other signals would modulate the activities of HPK(s) that phosphorylate RodK-R1 and RodK-R2. Moreover, we speculate that phosphotransfer from the RodK kinase domain to either RodK-R3 or RokA is determined by a novel regulatory mechanism that depends on the phosphorylation status of RodK-R1 and RodK-R2. RokA is a single receiver domain protein with no evident output domain. Therefore, it remains an open question how outputs are generated in response to changes in the phosphorylation status of RodK and RokA. One alternative would be that upon phosphorylation, the receiver domains engage in phosphotransfer to proteins containing Hpt domains, which could then engage in phosphotransfer to other RRs to generate the output responses. In addition to CheA kinases, bioinformatics analyses have only identified two proteins containing Hpt domains in M. xanthus, the HPK MXAN2317 and the RR MXAN7362 (38). Hpt domains are difficult to identify due to the low level of sequence conservation (3); thus, M. xanthus may encode more proteins containing Hpt domains. Alternatively, as has been described for the CheY single receiver domain protein and FliM (46) and for the Ssk1 single receiver domain protein and the Ssk2 MAP kinase (32), the receiver domains could engage in protein-protein interactions in a phosphorylation-dependent manner with proteins that are involved in the regulation of fruiting body formation.
Supplementary Material
Acknowledgments
We thank Marie Anders and Steffi Lindow for excellent technical assistance, Xingqi Shi for constructing the in-frame mutation in MXAN0720, Stuart Huntley for help with bioinformatics analyses, and Penelope Higgs for helpful discussions.
The Max Planck Society supported this work.
Footnotes
Published ahead of print on 9 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baikalov, I., I. Schröder, M. Kaczor-Grzeskowiak, K. Grzeskowiak, R. P. Gunsalus, and R. E. Dickerson. 1996. Structure of the Escherichia coli response regulator NarL. Biochemistry 3511053-11061. [DOI] [PubMed] [Google Scholar]
- 3.Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444899-904. [DOI] [PubMed] [Google Scholar]
- 4.Bourret, R. B. 2008. Signal transduction meets systems biology: deciphering specificity determinants for protein-protein interactions. Mol. Microbiol. 691336-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64545-552. [DOI] [PubMed] [Google Scholar]
- 6.Cho, K., and D. R. Zusman. 1999. Sporulation timing in Myxococcus xanthus is controlled by the espAB locus. Mol. Microbiol. 34714-725. [DOI] [PubMed] [Google Scholar]
- 7.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 6070-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellehauge, E., M. Nørregaard-Madsen, and L. Søgaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in M. xanthus development. Mol. Microbiol. 30807-817. [DOI] [PubMed] [Google Scholar]
- 9.Galperin, M. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 1884169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgellis, D., A. S. Lynch, and E. C. C. Lin. 1997. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 1795429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7745-754. [DOI] [PubMed] [Google Scholar]
- 13.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 1866902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgs, P. I., K. Cho, D. E. Whitworth, L. S. Evans, and D. R. Zusman. 2005. Four unusual two-component signal transduction homologs, RedC to RedF, are necessary for timely development in Myxococcus xanthus. J. Bacteriol. 1878191-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 742938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, J. L., H. C. Chen, H. L. Peng, and H. Y. Chang. 2008. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2839933-9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38535-542. [DOI] [PubMed] [Google Scholar]
- 18.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 979098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 765952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiil, K., J. B. Ferchaud, C. David, T. T. Binnewies, H. Wu, T. Sicheritz-Ponten, H. Willenbrock, and D. W. Ussery. 2005. Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiology 1513447-3452. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 10411568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski, M. A., and B. I. Kazmierczak. 2006. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 744462-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41121-145. [DOI] [PubMed] [Google Scholar]
- 24.Lee, B., P. I. Higgs, D. R. Zusman, and K. Cho. 2005. EspC is involved in controlling the timing of development in Myxococcus xanthus. J. Bacteriol. 1875029-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 251203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, S., C. Sugita, M. Sugita, and T. Omata. 2006. A new class of signal transducer in His-Asp phosphorelay systems. J. Biol. Chem. 28137868-37876. [DOI] [PubMed] [Google Scholar]
- 28.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 1733318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22757-767. [DOI] [PubMed] [Google Scholar]
- 31.Overgaard, M., S. Wegener-Feldbrügge, and L. Søgaard-Andersen. 2006. The orphan response regulator DigR is required for synthesis of extracellular matrix fibrils in Myxococcus xanthus. J. Bacteriol. 1884384-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 171385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen, A. A., S. L. Porter, J. P. Armitage, and L. Søgaard-Andersen. 2005. Coupling of multicellular morphogenesis and cellular differentiation by an unusual hybrid histidine protein kinase during fruiting body morphogenesis in Myxococcus xanthus. Mol. Microbiol. 561358-1372. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen, A. A., and L. Søgaard-Andersen. 2003. TodK, a putative histidine protein kinase, regulates timing of fruiting body morphogenesis in Myxococcus xanthus. J. Bacteriol. 1855452-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen, A. A., S. Wegener-Feldbrügge, S. L. Porter, J. P. Armitage, and L. Søgaard-Andersen. 2006. Four signalling domains in the hybrid histidine protein kinase RodK of Myxococcus xanthus are required for activity. Mol. Microbiol. 60525-534. [DOI] [PubMed] [Google Scholar]
- 36.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7626-633. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Shi, X., S. Wegener-Feldbrügge, S. Huntley, N. Hamann, R. Hedderich, and L. Søgaard-Andersen. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J. Bacteriol. 190613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimkets, L. J., and D. Kaiser. 1982. Induction of coordinated movement of Myxococcus xanthus cells. J. Bacteriol. 152451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3e334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Søgaard-Andersen, L., F. J. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10740-754. [DOI] [PubMed] [Google Scholar]
- 42.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC to YojN to RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40440-450. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhl, M. A., and J. F. Miller. 1996. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component regulatory phosphorelay. J. Biol. Chem. 27133176-33180. [DOI] [PubMed] [Google Scholar]
- 46.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 908787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitworth, D. E., and P. J. A. Cock. 2008. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology 154360-372. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2801448-1456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.