Abstract
Contact-dependent growth inhibition (CDI) is a mechanism identified in Escherichia coli by which bacteria expressing two-partner secretion proteins encoded by cdiA and cdiB bind to BamA in the outer membranes of target cells and inhibit their growth. A third gene in the cluster, cdiI, encodes a small protein that is necessary and sufficient to confer immunity to CDI, thereby preventing cells expressing the cdiBA genes from inhibiting their own growth. In this study, the cdiI gene was placed under araBAD promoter control to modulate levels of the immunity protein and thereby induce CDI by removal of arabinose. This CDI autoinhibition system was used for metabolic analyses of a single population of E. coli cells undergoing CDI. Contact-inhibited cells showed altered cell morphology, including the presence of filaments. Notably, CDI was reversible, as evidenced by resumption of cell growth and normal cellular morphology following induction of the CdiI immunity protein. Recovery of cells from CDI also required an energy source. Cells undergoing CDI showed a significant, reversible downregulation of metabolic parameters, including aerobic respiration, proton motive force (Δp), and steady-state ATP levels. It is unclear whether the decrease in respiration and/or Δp is directly involved in growth inhibition, but a role for ATP in the CDI mechanism was ruled out using an atp mutant. Consistent with the observed decrease in Δp, the phage shock response was induced in cells undergoing CDI but not in recovering cells, based on analysis of levels of pspA mRNA.
Intercellular communication mechanisms enable bacteria to coordinate biological phenomena such as DNA uptake, differentiation for fruiting body development, light production, and swarming (7, 8). These cell-to-cell interactions enable individual bacteria to form a multicellular community, such as in a biofilm on a solid surface, under specific environmental conditions (20, 52). Similarly to multicellular organisms, bacteria have signal transduction mechanisms to facilitate cellular cross talk, including two-component regulatory systems and other cell surface ligand-receptor interactions that control cellular processes.
We previously described a cross talk phenomenon designated as contact-dependent growth inhibition (CDI) in which one bacterial isolate (CDI+) blocks the growth of another bacterium when mixed together (4). CDI requires two contiguous genes, cdiB and cdiA, which encode proteins that are in the two-partner secretion (TPS) family. Overlapping the stop codon of cdiA is a downstream open reading frame designated cdiI, which encodes a 79-amino-acid protein that provides immunity to growth inhibition from cells expressing cdiBA (4). Evidence strongly indicates that cell-to-cell contact is required for growth inhibition. First, separation of CDI+ inhibitor cells and target cells by a 0.4-μm nitrocellulose membrane blocked CDI, distinguishing CDI from the class of soluble bacterial growth inhibitors known as bacteriocins. Second, to address the possibility that a very short-lived bacteriocin-like molecule might be released from CDI+ inhibitor cells, we separated inhibitor-target cell aggregates by fluorescence-activated cell sorting. Target cells within aggregates with CDI+ inhibitor cells lost viability, as measured by growth on LB medium, more rapidly than did unbound target cells, supporting the conclusion that cell-to-cell contact is required for CDI (4).
In our previous work, analysis of CDI was carried out with a bipartite system using Escherichia coli inhibitor cells containing cdiB, -A, and -I on a multicopy plasmid that constitutively expressed CDI activity, cocultured with E. coli K-12 target cells. Mixing inhibitor cells with E. coli K-12 target cells resulted in a 5- to 6-log decrease in target cell number after only 1 to 2 h (4). Because this bipartite CDI assay contains both inhibitor and target cells, monitoring of target cell growth in real time is not possible. Thus, we have not been able to determine if CDI is a reversible process or a nonreversible toxin-like system. This is important in assessing the role of CDI in the biology of E. coli as well as its potential role in many gram-negative bacteria, including uropathogenic E. coli, Burkholderia pseudomallei, and Yersinia pestis that contain genes with significant sequence identity to cdiB and cdiA (4).
Recently, in collaboration with J. Malinverni and T. Silhavy (Princeton University), we showed that the target cell receptor for CDI is BamA, an essential outer membrane protein (OMP). Homologues of bamA are conserved in genomes from bacteria to mitochondria (3). BamA is a key component of the β-barrel assembly machine required for biogenesis of many other OMPs (34, 43, 53). Our results indicated that the BamA receptor facilitates CdiB/CdiA-dependent cell-to-cell binding and growth inhibition since antibodies to BamA blocked formation of inhibitor-target cell aggregates and CDI (3). The ligand for BamA is not known, but it seems probable that it is CdiA, which is at the surface of inhibitor cells (4), and may form a short 40- to 50-nm fiber, such as filamentous hemagglutinin in Bordetella pertussis, based on sequence similarities (42). AcrB is also required for sensitivity of target cells to CDI, which acts downstream of the BamA receptor (3). AcrB is a protein that exports small molecules, including drugs/antibiotics, through the inner membrane, with further transport through the outer membrane in conjunction with AcrA and TolC (47, 55, 56). This export machine requires proton motive force (Δp) as an energy source (46). Markedly, only BamA and AcrB, independent from their respective export machines, are required for CDI (3). Based on these results, we developed a model in which CdiA of inhibitor cells bind to BamA on target cells, transmitting a signal into target cells such as a CdiA peptide (Fig. 1A). This signal could enter cells through an AcrB portal to interact with an unidentified cytosolic target. It is also possible that AcrB could play an indirect role in CDI-mediated growth inhibition.
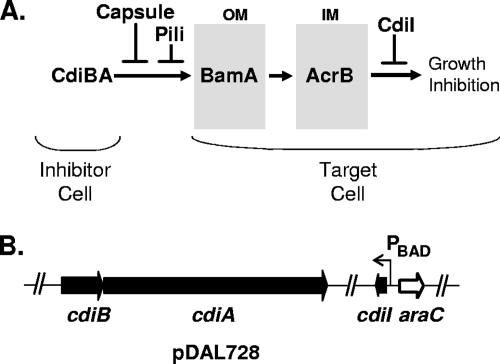
FIG. 1.
Construction of a contact-dependent autoinhibition system in Escherichia coli. (A) CDI model. CdiB and CdiA (CdiBA) expressed by one cell (inhibitor cell) binds to BamA in the outer membrane (OM) of an adjacent cell (target cell) (3). A signal, such as a CdiA peptide, is transferred through BamA to AcrB located in the inner membrane (IM), causing inhibition of cell growth by an unknown mechanism. CDI is modulated at the cell surface by certain pili, including pyelonephritis-associated pili (4), and by colanic acid capsule (3) and inside cells by CdiI immunity protein (4). (B) Map of the CDI autoinhibition plasmid (pDAL728).
Here, we describe the development of a CDI autoinhibition system in which growth inhibition is regulated by controlled expression of the CdiI immunity protein, enabling examination of a single population of cells undergoing CDI. Using this system, we show that cells undergoing CDI have significantly reduced cellular respiration, Δp, and steady-state ATP levels. Notably, this metabolic downregulation, as well as cellular growth inhibition, is reversible. To the best of our knowledge, this is the first report of a natural, reversible system controlling bacterial metabolism and growth.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
E. coli strains and plasmids used in this study are listed in Table 1. Cells were grown in modified Luria-Bertani broth (LBM) containing 50 mM KPO4 at pH 7.3. Antibiotics were used at the following concentrations (unless otherwise noted): ampicillin (Amp), 100 μg/ml; chloramphenicol (Cam), 34 μg/ml; kanamycin (Kan), 40 μg/ml; tetracycline (Tet), 12.5 μg/ml. When indicated, carbohydrates [l-(+)-arabinose and d-(+)-maltose monohydrate (Sigma)] were used at 0.2% to supplement the media. For autoinhibition studies, cultures were incubated at 37°C in an environmental shaker apparatus (New Brunswick series 25) at 225 rpm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| AN120 | atpA401 argE3 xylA5 thi-1 mtl-1 galK2 rpsL704 λ− | 16 |
| AN180 | argE3 xylA5 thi-1 mtl-1 galK2 rpsL704 λ− | 16 |
| DH5α | F−supE44 hsdR17(rK− mK+) recA1 φ80dlacZΔM15 ΔlacU169 gyrA96 endA1 thi-1 relA1 deoR λ− | 26 |
| DL5236 | DH5α pDAL728 | This study |
| DL5263 | LMG194 pDAL728 | This study |
| DL5280 | AN180 made Δara714 leu::Tn10 from LMG194 | This study |
| DL5281 | AN120 made Δara714 leu::Tn10 from LMG194 | This study |
| DL5283 | AN180 Δara714 leu::Tn10 pDAL728 | This study |
| DL5285 | AN120 Δara714 leu::Tn10 pDAL728 | This study |
| DL5361 | LMG194 pDAL728 pDAL724 | This study |
| DL5362 | LMG194 pDAL728 pBR322 | This study |
| DL5400 | LMG194 pBAD33 | This study |
| DL5414 | AN180 Δara714 leu::Tn10 pBAD33 | This study |
| DL5415 | AN120 Δara714 leu::Tn10 pBAD33 | This study |
| DL5417 | MC1061 pDAL728 pTSV-1 | This study |
| DL5418 | MC1061 pBAD33 pTSV-1 | This study |
| DL5711 | MC1061 acrB3 pDAL728 pTSV-1 | 3 |
| DL5735 | MC1061 ΔpspA740::kan pDAL728 pTSV-1 | This study |
| JW1297-1 | BW25113 ΔpspA740::kan (Keio collection) | 5 |
| LMG194 | F−ΔlacX74 galE galK thi rpsL ΔphoA (PvuII) Δara714 leu::Tn10 | 25 |
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL hsdR2(rK− mK+) mcrA mcrB1 | 17 |
| Plasmids | ||
| pBAD33 | Cloning vector containing an arabinose PBAD promoter; Camr | 25 |
| pBR322 | Cloning vector; Ampr, Tetr | 10 |
| pDAL230B | pBR325-pap21; Ampr, Camr | 4 |
| pDAL660Δ1-39 | pDAL660 deletion containing 16,734 bp DNA including cdiABI | 4 |
| pDAL724 | pap21 operon from pDAL230B ligated into pBR322 at the SalI and BamHI sites; Ampr | This study |
| pDAL726 | cdiI from pDAL660Δ1-39 ligated into pBAD33 at the SacI and SphI sites, resulting in arabinose-inducible cdiI; Camr | This study |
| pDAL728 | cdiAB from pDAL660Δ1-39 (BstZ17I-BstZ17I) blunt ligated into pDAL726 at the NsiI site; Camr | This study |
| pTSV-1 | Constitutive lacY plasmid; Tetr | 54 |
CDI autoinhibition system. (i) Plasmid construction.
cdiI and its ribosomal binding site were amplified from plasmid pDAL660Δ1-39 by PCR using oligonucleotides 5′-CAACAAGAGCTCAGGAAGGGGCAAAATGAAGAAGAAAC-3′ and 5′-CAACAAGCATGCCTATTTTCTGTCTAAGATACTAAGGCCCCAC-3′, containing SacI and SphI sites, respectively. The PCR product was digested with SacI and SphI and ligated into the corresponding restriction sites on pBAD33 (25) to construct plasmid pDAL726 with arabinose-inducible cdiI. pDAL726 was digested with NsiI, and the ends were blunted using an End-It DNA end repair kit (Epicentre). The cdiBA DNA region lacking most of the cdiI gene (162/237 bp) was excised from pDAL660Δ1-39 by using BstZ17I and ligated into the blunted NsiI site of pDAL726 to construct pDAL728 (Fig. 1B). This plasmid contains constitutively expressed cdiAB and arabinose-inducible cdiI.
(ii) Autoinhibition assay.
CDI+ autoinhibition strains and CDI− control strains were grown overnight in LBM on a tube roller apparatus (New Brunswick) with 0.2% arabinose and appropriate antibiotics. Overnight cultures were pelleted at 2,500 × g for 10 min and washed three times with arabinose-free medium. Washed pellets were resuspended in half the original volume of LBM with Cam, and the optical densities at 600 nm (OD600) of the cell suspensions were determined. Prepared cell suspensions were used to inoculate fresh LBM with appropriate antibiotics at a starting OD600 of 0.05. After 5 h of incubation with shaking at 225 rpm, cultures were supplemented with 0.2% arabinose to induce CdiI and maltose at 0.2% unless otherwise noted.
Measurement of ATP, membrane potential, and respiration.
The ATP measurements shown in Fig. 4 and 5 were obtained using a BacTiter-Glo microbial cell viability assay as described by the manufacturer (Promega). Briefly, at the times indicated, cells were pelleted by centrifugation and then resuspended in LBM to an OD600 of 1.0; then, 25 μl of each sample or known concentrations of ATP in LBM were mixed with 25 μl of BacTiter-Glo reagent in a luminometer tube. Reactions were incubated at room temperature for 5 min before relative luminescence units were determined using a luminometer set for a 1-second integration time (20/20n luminometer; Turner Biosystems).
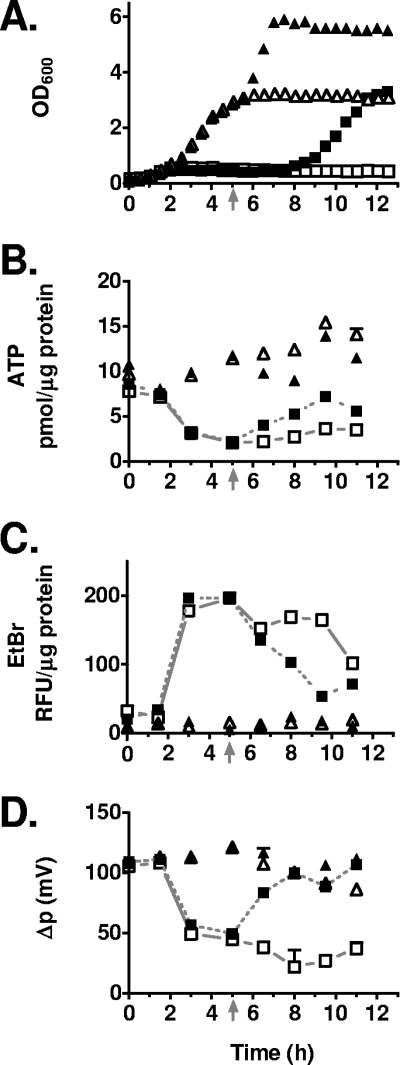
FIG. 4.
Measurement of membrane potential (Δp), EtBr export, and ATP level in cells entering and exiting CDI. CDI+ (DL5417; squares) and CDI− (DL5418; triangles) E. coli cells were prepared and incubated in LBM as described for Fig. 2. Arabinose and maltose (closed symbols) were added to two cultures at 5 h (as indicated by an arrow on the x axis). For clarity, data points for CDI+ cells receiving arabinose are connected by dotted lines and CDI+ cells without arabinose are connected by solid lines. Error bars show the standard deviation for duplicate measurements. (A) Growth curve. (B) Intracellular ATP level (pmol/μg protein). (C) EtBr fluorescence (relative fluorescence units [RFU per μg protein]). (D) Estimated Δp.
FIG. 5.
Analysis of CDI in E. coli lacking ATP synthase. Results for CDI+ E. coli DL5283 (with ATPase [squares]) and CDI+ DL5285 (without ATPase [inverted triangles]) cells are shown in the left panels, and results for E. coli CDI− control strains DL5414 (with ATPase [triangles]) and DL5415 (without ATPase [diamonds]) are shown in the right panels. Strains receiving arabinose and maltose at 5 h are indicated with closed symbols, and no-addition controls are indicated with open symbols. For clarity, data points for CDI+ atp+ cells with arabinose are connected by solid lines, and CDI+ atp401 cells with arabinose are connected by dotted lines. (A) OD600. (B) Intracellular ATP level. (C) EtBr fluorescence.
The Δp level was measured by quantitation of [14C]lactose transport (1, 24, 32), using [d-glucose-1-14C]lactose (50 to 62 mCi/mmol; GE Healthcare). Cells for this analysis constitutively expressed LacY permease from plasmid pTSV-1 (54) but contained a deletion of the lac operon including lacZ and, thus, were unable to catabolize lactose. CDI+ strain DL5417 (MC1061 pDAL728 pTSV-1) and CDI− strain DL5418 (MC1061 pBAD33 pTSV-1) were cultured in LBM as described above for the CDI autoinhibition system, pelleted by centrifugation, and then resuspended in 0.5 ml of LBM to an OD600 of 0.5. The cell suspension was mixed with 0.5 ml LBM containing 0.5 μCi/ml [14C]lactose and 28 μM total lactose for a final concentration of 0.25 μCi/ml [14C]lactose and 14 μM total lactose. Cells were incubated for 25 min at 37°C, at which time a steady-state internal level of [14C]lactose was reached and then 0.5-ml aliquots were vacuum filtered onto Whatman GF/F filters prewet with ice-cold 0.1 M LiCl. Each filter was washed twice with 15 ml of ice-cold 0.1 M LiCl before being dried at 160°C for 10 min and counted in a Beckman scintillation counter using 10 ml of Liquiscint scintillation fluid (National Diagnostics). Nonspecific binding was determined after addition of 50 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) to dissipate the membrane potential. The Δp level was calculated using the equation Δp ≈ ΔΨ − (2.3 RT/F)ΔpH (mV), where Ψ is membrane potential, R is the gas constant, T is absolute temperature, and F is the Taraday constant (27). At pH 7.5, ΔpH ≈ 0, and thus Δp ≈ ΔΨ = 61.5 log [(lactosein)/(lactoseout)]. The internal concentration of lactose (lactosein) was calculated using the previous estimate of 5.85 μl of intracellular fluid per mg of cell protein (21, 22). The cell volume-to-protein ratios were similar for cells undergoing CDI and control cells not undergoing CDI, based on analysis of packed cell volume using a capillary device (Bullseye cell counting kit; TPP, Switzerland).
The ethidium bromide (EtBr) efflux assay was adapted from protocols previously described (18, 45). Cells were cultured and prepared as described above for the CDI autoinhibition assay. Briefly, prepared cells were pelleted by centrifugation and resuspended in 2 ml of 50 mM KPO4 buffer (pH 7.5) for a final OD600 of 0.2. EtBr was added to the cell suspension at a final concentration of 25 μM. Samples were continuously monitored using a Victor3 fluorometer (Perkin Elmer) at an excitation and emission of 531 nm and 595 nm, respectively, until accumulation reached equilibrium.
Aerobic respiration was measured at 23°C in a sealed stirred cuvette with a Clarke oxygen electrode as recommended by the manufacturer (Qubit Systems). Calibration was carried out after air was bubbled into the oxygen electrode sample chamber (100% O2 saturation) and after addition of a few grains of sodium sulfite (0% saturation). E. coli samples from each time point in the autoinhibition experiment were collected by centrifugation and then resuspended in LBM to an OD600 of 0.5. Three milliliters of E. coli sample was added to the calibrated cuvette equipped with a stir bar; the top plunger was lowered, and air bubbles were expelled through the top port. Readings were taken every second for 1 to 2 min, and the slope was calculated by least-squares analysis. Measurements of Δp, ATP, EtBr, and O2 consumption were made in duplicate.
DAPI staining and cell imaging.
CDI+ autoinhibition strain DL5263 and CDI− control strain DL5400 were cultured and prepared as described above (28, 29, 33). Cells were immobilized onto coverslips coated with 0.1% poly-d-lysine (Sigma) and fixed with methanol for 5 min at 23°C (44). A 4,6-diamidino-2-phenylindole (DAPI; Sigma) solution was prepared by mixing a 5-μg/ml stock solution of DAPI with SlowFade gold antifade reagent (Molecular Probes) at a 3-to-2 ratio (vol/vol). Five microliters of the DAPI solution was placed onto the coverslips, which were sealed onto slides with nail polish. Cells were observed by combined fluorescence and phase-contrast microscopy using an Olympus fluorescent microscope and a 100× oil objective. Images were captured using a six-megapixel digital camera and MicroFire imaging software (Optronix, Goleta, CA).
qPCR analysis.
Total RNA was isolated from 1-ml samples taken at the time points indicated (see Fig. 7) and equilibrated to an OD600 of 1. Bacterial samples were treated with 1 ml of RNAprotect bacteria reagent (Qiagen), frozen at −80°C, and analyzed within 2 weeks. RNA was extracted by lysozyme/proteinase K treatment (RNAprotect bacteria reagent handbook) and further purified using a Qiagen RNeasy mini kit (Qiagen, Valencia, CA.). RNA was treated with Turbo DNase (Ambion, Austin, TX) to remove DNA contaminants, and 0.5 μg RNA was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). A control without reverse transcriptase was prepared to assess chromosomal DNA contamination. Quantitative PCR (qPCR) was carried out as follows. Primer pairs for pspA mRNA quantification, 5′-TGACGCTGGTGGACGATAC-3′ and 5′-CGAGCCATTGCTTCATCC-3′, were designed and analyzed using Vector NTI software (Invitrogen, Carlsbad, CA) and NetPrimer Applet (PREMIER Biosoft International). qPCR reactions were carried out on a Bio-Rad MyiQ single-color real-time PCR detection system. Twenty-five-microliter reaction mixtures contained 1 μl cDNA, 12.5 μl iQ SYBR green supermix (Bio-Rad), 0.1 μM primer, and water. A minimum of two reactions were performed for each sample. Thermal cycling conditions were as follows: 95°C for 3 min for polymerase activation and collection of experimental well factors and 40 cycles at 95°C for 10 s and 55.7°C for 30 s. A melting curve (55°C to 95°C) was added to analyze the end product. Data were analyzed using the iQ5 optical system software (Bio-Rad) and exported to Microsoft Excel and Prism 5.0 for further analysis. Threshold was automatically set by the software, and the baseline was user defined. An estimated copy number was calculated from the cycle threshold of each sample, using CT(1) = 40 (37). Copy numbers were divided by the corresponding sample at the 1-h time point to determine relative expression over time for each gene and condition. Products were run on 1.5% agarose to check for amplicon size and amplification specificity.
FIG. 7.
Induction of pspA transcription during CDI. CDI+ E. coli DL5417 (squares; panels A and B) and CDI+ E. coli DL5735 ΔpspA740::kan (circles; panel A) were grown in LBM with 0.2% maltose. Cultures receiving arabinose (0.2%) at 5 h are shown as closed symbols. (A) OD600. (B) pspA mRNA expression level relative to expression at 1 h, as determined using qPCR.
RESULTS
CDI is reversible.
Our previous analysis of CDI used a bipartite system consisting of CDI+ E. coli inhibitor cells mixed with E. coli K-12 target cells (4). Although this analysis was sufficient for characterization of the phenomenon of CDI, it did not allow differential biochemical analysis of the effects of CDI on target cells due to the presence of large numbers of CDI+ E. coli inhibitor cells. Therefore, we developed a CDI autoinhibition system in which a single E. coli isolate can be induced to undergo CDI, with subsequent analysis of the physiological changes that occur during CDI. We constructed a plasmid, pDAL728, in which cdiBA, expressed under its own constitutive promoter, is separated from the normally linked cdiI immunity gene, and expression of cdiI was placed under arabinose control by cloning downstream of the pBAD promoter (Fig. 1B). Initial studies with pDAL728 were performed using an E. coli host that can metabolize arabinose (ara+ DL5236). Transfer of DL5236 cells to an arabinose-free LBM medium at time zero, which prevents cdiI immunity gene expression, resulted in growth stasis. Cells receiving arabinose at 5 h to induce CdiI expression resumed growth within 3 h, whereas cells that did not receive arabinose remained growth inhibited (data not shown). These results showed that CDI is a reversible mechanism. Because DL5236 metabolizes arabinose, it was possible that arabinose could have acted not only as an inducer of CdiI but also as an energy source that might be required for recovery. To distinguish between these possibilities, we carried out the same experiment with pDAL728 in a ΔaraBAD E. coli host (DL5263) that transports but cannot catabolize arabinose. Under these conditions, addition of arabinose at 5 h failed to reverse growth inhibition unless an energy source, such as maltose (Fig. 2A) or another carbohydrate (lactose, ribose, or glycerol) (unpublished data), was added. These results indicate that reversal of CDI observed after addition of arabinose alone to the araBAD+ strain, described above, was due to dual functions of arabinose as an inducer of both CdiI immunity protein expression and utilization as an energy source. Addition of maltose alone did not reverse CDI, indicating that an energy source alone was not sufficient for growth recovery (Fig. 2A).
FIG. 2.
Characterization of the CDI autoinhibition system. Reversibility and carbon source requirement. (A) CDI+ E. coli LMG194 Δara714 with pDAL728 (DL5263) were prepared and incubated at 37°C with shaking at 225 rpm as described in Materials and Methods for the CDI autoinhibition assay. Cultures were incubated in LBM (open squares), LBM plus arabinose (closed circles), LBM plus maltose (open circles), or LBM plus maltose plus arabinose (closed squares). Maltose (0.2%) was present at time zero, and arabinose (0.2%) was added at 5 h, where indicated. (B) CDI+ DL5263 (squares) and CDI− control E. coli DL5400 cells (triangles) were prepared as described for panel A for the CDI autoinhibition assay. Maltose and arabinose (0.2% each) were added to the indicated cultures at 5 h (closed squares and triangles; shown by arrow on x axis). (C) Extended time course. CDI+ DL5263 was grown and treated as described for panel A in LBM. After 20 h of incubation, cultures were supplemented with 0.2% arabinose (closed circles) or with 0.2% arabinose and maltose (closed squares). A no-supplement control was included (open squares). (D) Inhibition of CDI by Pap pili. CDI+ E. coli cells expressing Pap21 pili (half-shaded squares) or not expressing Pap21 pili (open squares) were prepared as described for panel A in LBM without supplements.
As shown in Fig. 2B, CDI− vector control cells were not growth inhibited (compared to CDI+ cells) and grew faster and to a higher density after the addition of arabinose and maltose at 5 h, as expected. Total cellular protein levels closely paralleled OD values, indicating that during growth inhibition no net synthesis of protein occurred (unpublished data). We found that cells could remain in a growth-inhibited state for at least 20 h and still recover growth after the addition of arabinose and maltose (Fig. 2C), showing the reversibility of CDI after a long growth arrest period.
Previously, we showed that pyelonephritis-associated pili (Pap), expressed at the cell surface, block CDI (4). This Pap-mediated block of CDI is specific, since type 1 pili expression does not affect CDI (4). To address whether cell-to-cell contact in the CDI autoinhibition system (Fig. 2) is required for the growth-inhibitory phenotype observed, we examined the effect of Pap pili expression in CDI+ cells harboring plasmid pDAL728 (cdiB+A+I+). As shown in Fig. 2D, expression of Pap pili in CDI+ E. coli prevented growth arrest (Fig. 2D), suggesting that growth autoinhibition requires surface contact between cells, similar to the bipartite CDI system (4).
Cells undergoing CDI form multinucleoid filaments.
To explore the physiologic state of cells undergoing CDI, we initially examined cell morphology over the time course of growth inhibition and recovery shown in Fig. 2B, using DAPI to stain nucleoids (Fig. 3). CDI+ cells began to form filaments between 3 h and 5 h after removal of arabinose at time zero, with some cells containing multiple nucleoids without visible septation (Fig. 3, panel in first row, lane 5). At later times, cell “ghosts” lacking nucleoids were present in the CDI+ cell population (Fig. 3) but were generally absent in CDI+ cells receiving arabinose and maltose at 5 h. Cell morphology in the latter CDI+ cell population recovered to that of CDI− cells, which were short rods with one to two nucleoids (Fig. 3, lower panels). No change in cell morphology was observed after the addition of arabinose and maltose to CDI− control cells (Fig. 3, compare panels in bottom two rows). These results suggest that cells undergoing CDI are blocked at some stage(s) in the cell division process, consistent with results shown in Fig. 2 indicating that OD values do not increase in cells undergoing CDI and previous data showing that colony formation is blocked by CDI (3).
FIG. 3.
E. coli cell morphology during CDI-mediated autoinhibition and recovery. CDI+ E. coli (DL5263) and CDI− control E. coli (DL5400) cells were grown in arabinose-free LBM, as shown in Fig. 2B, performed in parallel with the morphological analysis shown here. After 5 h, the cultures were supplemented with 0.2% arabinose and 0.2% maltose, as indicated at the left of each row of panels (+ara +mal). At the times indicated, cells were stained with DAPI and imaged at the same magnification (×1,000), using phase optics and fluorescence overlay by using MicroFire imaging software (Optronix, Goleta, CA). Multiple nucleoids are indicated with arrows, and ghosts are indicated with blunt arrows.
CDI induces metabolic downregulation.
The observation that cells undergoing CDI show multiple nucleoids after a few hours of growth inhibition suggested that they may still be partially metabolically active. An important indicator of the metabolic state and health of cells is the steady-state ATP level, which is replenished in glycolysis by substrate-level phosphorylation and during oxidative phosphorylation through generation of Δp, which drives ATP formation via ATP synthase. Cellular ATP levels were measured and found to decrease concomitant with growth inhibition by the use of an in vivo test developed by Schneider and Gourse (50) in which cells expressing luciferase are permeabilized by polymyxin B to allow entry of luciferin substrate. However, cells that reached stationary phase appeared to become resistant to permeabilization by polymyxin B, complicating the comparison of ATP levels in CDI+ and CDI− cells over an extended time course (data not shown). Therefore, we employed an in vitro test in which ATP levels extracted from cells are measured using a thermostable luciferase (BacTiter-Glo, Promega), which showed a ≤5-fold reduction in ATP levels at the same time that growth inhibition was observed (compare Fig. 4A and B). Cells receiving arabinose/maltose at 5 h to induce CdiI showed partial recovery in ATP levels at 6.5 h (Fig. 4B), prior to growth recovery, which occurred at about 9.5 h (Fig. 4A). These results indicate that the steady-state ATP levels in cells undergoing CDI are significantly reduced compared to those of the CDI− control cells.
To address whether the reduction in ATP level observed for cells undergoing CDI is required for growth inhibition, we constructed a CDI+ autoinhibition system and a CDI− control in an E. coli atp401 strain, which contains a mutation that abolishes ATP synthase activity (12). As expected based on previous results (16), the atp401 mutant grew slower than its congenic atp+ counterpart in the CDI− background and reached a lower cell density in stationary phase (Fig. 5A, right panel). The CDI+ atp401 strain underwent CDI but recovered after the addition of arabinose and maltose at 5 h, similar to atp+ bacteria (Fig. 5A, left panel). Thus, the ATPase is not necessary for CDI or growth recovery. Notably, the ATP level of the atp401 CDI+ mutant undergoing growth inhibition (Fig. 5B, left panel) was as high as or higher than those of atp401 CDI− cells which did not undergo CDI (Fig. 5B, right panel). In contrast, the atp+ cells undergoing CDI had low ATP levels, as described above (compare the left panels of Fig. 4B and 5B). These results strongly indicate that CDI does not occur via reduction in ATP level, since E. coli atp401 mutants that are growth inhibited due to CDI have levels of ATP that are as high as or higher than those of the CDI− control cells that are growing normally. Together, these results show that there is no correlation between ATP level (Fig. 5B) and growth inhibition or recovery (Fig. 5A). Thus, although ATP level decreases during CDI, this does not appear to be causal in modulating growth inhibition.
Previous work indicated that the inner-membrane multidrug transport protein AcrB might play a role in CDI since acrB mutants are CDI resistant (4). AcrB uses Δp to transport small molecules including EtBr across the inner membrane (55). We incubated cells undergoing CDI (Fig. 4A) in EtBr and measured intracellular levels of EtBr by a fluorescence assay (see Materials and Methods). Under these conditions, AcrB is required for the majority of EtBr export, based on analysis of an acrB mutant (data not shown). The results indicated that EtBr transport in CDI+ cells (Fig. 4C) was repressed in cells concomitant with growth inhibition (compare Fig. 4A and C), whereas CDI− cells showed normal EtBr transport over the time course (Fig. 4C). Similar results were observed for E. coli cells lacking ATP synthase activity (Fig. 5C), indicating that the ATP synthase is not required for the observed EtBr export defect occurring during CDI. Notably, recovery of EtBr transport in CDI+ cells began by 6.5 h, prior to the resumption of growth which occurred after 8 h in cells receiving arabinose and maltose (Fig. 4A and C). These results indicated that AcrB function is blocked in cells undergoing CDI and recovers prior to the resumption of cell growth.
The CDI system could directly inhibit AcrB export function and/or affect AcrB function indirectly by a reduction in Δp, which supplies the energy for export. The latter possibility was tested by measuring [14C]lactose transport in lacIZY+ cells constitutively expressing the LacY permease but lacking β-galactosidase necessary to digest lactose (see Materials and Methods). A steady-state level of intracellular [14C]lactose was reached by 20 min, and this level was reduced to near background level (measured in the absence of bacterial cells) by the addition of 50 μM CCCP, which dissipates Δp. Experiments were carried out in buffered LBM, which maintained the medium pH at 7.25 ± 0.15 over an 8-h time course (Fig. 4A and unpublished data). Lactose transport was measured in cells suspended in a 50 mM potassium phosphate buffer at pH 7.5 as previously described to maintain internal and external pH values at approximately the same value (ΔpH ≈ 0). The results showed that calculated Δp was significantly reduced (≤5-fold) in cells undergoing CDI at the same time that growth inhibition and EtBr accumulation occurred (compare Fig. 4A, C, and D). Moreover, cells recovering from CDI showed an increase in Δp occurring within 1.5 h after the addition of arabinose and maltose and prior to growth recovery (Fig. 4D). These results indicate that Δp is significantly reduced in cells undergoing CDI. Notably, recovery of Δp began by 6.5 h, well ahead of growth recovery of arabinose-treated cells (Fig. 4A and D). Together, the EtBr and lactose transport assays indicate that growth inhibition induced by CDI is accompanied by a two- to fivefold drop in Δp (Fig. 4D), sufficient to block AcrB export activity (Fig. 4C).
The finding that Δp and ATP levels are reduced during CDI raised the question of whether respiration is also affected. Mitochondria respond to a reduction in Δp by increasing respiration, a process involving respiratory control in which the Δp regulates electron transport, maintaining a constant ATP/ADP ratio (13, 14). In contrast, E. coli shows respiratory control under conditions of starvation only (15, 40), possibly because under normal laboratory conditions, the rate of respiration is near maximal. The measurement of oxygen consumption indicated that respiration was significantly reduced in CDI+ cells cotemporally with growth inhibition, whereas respiration in CDI− control and CDI+ acrB mutant cells increased until a steady state was reached at about 3 h (Fig. 6B). These results indicate that downregulation of respiration is dependent upon CdiB and CdiA as well as AcrB. CDI+ cells receiving arabinose and maltose at 5 h recovered respiration to the levels observed in CDI− control cells, whereas CDI+ cells with no additions maintained a low respiration rate through 12 h (Fig. 6B). Taken together, these results indicate that ATP, Δp, and aerobic respiration are significantly reduced in E. coli undergoing CDI and that this metabolic downregulation is under reversible regulatory control.
FIG. 6.
Reversible downregulation of respiration in CDI. CDI+ E. coli DL5417 strains (left panels [squares]), CDI− E. coli DL5418 control strains (right panels [closed triangles]), and CDI+ acrB mutant E. coli DL5711 strains (open circles) were prepared and incubated as described for Fig. 2, with maltose (0.2%) added at time zero. Cells receiving arabinose at 5 h (arrows) are indicated by closed symbols. Error bars show the standard deviation of the mean calculated from two independent experiments.
Induction of the pspA (phage shock) transcription response in CDI.
The reduction in ATP, Δp, and respiration during CDI (Fig. 4 and 6) raised the question of how cells respond to these metabolic changes. Previous work has shown that reduction in membrane potential induces the phage shock response, which requires pspA expression (9, 19, 36). Therefore, we measured the levels of pspA mRNA by using qPCR over the time course of growth autoinhibition. We found a roughly 20-fold increase in pspA mRNA, peaking at about 8 h (Fig. 7A and B). Notably, this pspA response was absent in cells that recovered growth after addition of arabinose and maltose at 5 h (Fig. 7A and B). These results indicated that the phage shock pathway is activated during CDI. Previous results suggested that the phage shock pathway may contribute to recovery from certain environmental stresses, such as heat shock and hyperosmolarity (19). To determine if the phage shock response plays a direct role in CDI, we measured the effects of a pspA null mutation on CDI. Cells lacking PspA became growth inhibited after removal of CdiI and recovered from growth inhibition following the induction of CdiI, same as the wild-type cells (Fig. 7A). Thus, although the phage shock pathway is induced in cells undergoing CDI, it does not appear to play an essential role in CDI.
DISCUSSION
In this work, we developed a CDI autoinhibition system to study a single population of E. coli cells undergoing CDI. Our results showed that CDI is a reversible mechanism in which cell-to-cell contact induces a downregulation of the steady-state ATP level and Δp. Notably, respiration was repressed cotemporally with Δp and ATP (Fig. 4), the opposite of what has been observed to occur in mitochondria and starved bacteria exposed to uncoupling agents that dissipate Δp, such as CCCP (15, 31). In the case of starved bacteria, respiratory control is lost, and cellular respiration is constitutively increased. To the best of our knowledge, this is the first report of a bacterial response pathway in which multiple metabolic parameters are reversibly downregulated.
Previously, we studied CDI by in vitro competition assay using two populations of cells—CDI+ inhibitory cells expressing the cdiBAI genes and E. coli K-12 target cells (4). The mixed cell population of this bipartite CDI system precluded determination of whether growth-inhibited cells were dead or in a growth-repressed but viable state (4). Cell membranes of growth-inhibited cells appeared to be intact based on exclusion of propidium iodide (11), but it was difficult to assess whether they were capable of growth recovery due to the mixed cell population. Here, we examined a single population of cells undergoing growth inhibition by using the CDI autoinhibition system (Fig. 1B and 2A and B). Our results showed that cells (ΔaraBAD; unable to use arabinose as a carbon/energy source) undergoing CDI resumed growth within 2.5 h of induction of expression of the cdiI immunity gene with arabinose, but only if a utilizable carbohydrate was present (maltose, ribose, lactose, or glycerol). Although LB contains maltose and maltodextrins, these appear to be mostly degraded in cultures that have reached an OD600 of approximately <0.5 (6), which is less than the density reached by the growth-inhibited cells in our experiments (Fig. 2B). We note that although an energy source is required for recovery from CDI, growth inhibition does not appear to be caused by a limitation in energy substrate, since inclusion of 0.2% maltose in cell cultures from the beginning of a CDI experiment (Fig. 2A) does not affect the time at which growth arrest occurs. Since growth recovery occurred normally after the addition of arabinose, this indicates that at the time that cells were growth arrested (∼3 h), there was sufficient maltose present to allow recovery (Fig. 2A). In the case of E. coli ara+ cells, the addition of arabinose alone was sufficient for growth recovery (see Results), since arabinose acted as both an inducer of cdiI and an energy source.
Our results showing that growth recovery requires an energy source and occurs about 2.5 h after induction of the CdiI immunity protein (Fig. 2A and B) raise two important questions. First, what role does an energy source play in growth recovery? If the reduction in aerobic respiration or Δp observed (Fig. 4D and 6) were causal in growth inhibition, then an energy source could function to supply NADH for electron transport and proton export, respectively. It is also possible that CDI operates through a mechanism independent of Δp and respiration and that the downregulation observed in these metabolic parameters results indirectly from an effect on an unidentified CDI target. A second question that arises is why does growth arrest occur after about 3 h and growth recovery take a similar amount of time following induction of CdiI? Since growth inhibition and recovery occurred at about the same time whether maltose (0.2%) was present from the beginning of the experiment or added after 5 h, this delay does not appear to be due to the timing of addition of the energy source. Since induction of CDI was affected by washing arabinose out of the medium, the timing of when cells become growth inhibited should depend upon how quickly arabinose is cleared from cells after extracellular arabinose has been removed (note that cells could not catabolize arabinose) and also on the half-life of the CdiI immunity protein. It is likely that these two factors account for the temporal delay observed in cell growth inhibition. In contrast, the delay in growth recovery after induction of CdiI should depend, in part, on the time it takes for cells to attain a level of CdiI sufficient to confer immunity to CDI. Since growth-inhibited cells appear to have a low metabolic state (Fig. 4 and 6), protein expression may be slowed. Another factor that may contribute to the delay in growth recovery observed involves the fate of growth-inhibited cells after CdiI induction. Does CdiI expression reverse growth inhibition immediately, or does it take time for CdiI to counteract the effects of CDI? For example, a CdiA peptide, imported into cells via BamA, might form a complex with a cellular target with a slow complex dissociation rate. In this case, there might be a significant time delay for CdiI to displace CdiA from the complex and restore cellular growth.
We do not know the mechanism by which cellular respiration, Δp, and ATP are reduced in cells undergoing CDI. As discussed above, these cellular parameters could be reduced as an indirect consequence of interaction of components of the CDI system, such as CdiA, with an unidentified target(s). Alternatively, it is possible that Δp might be reduced by interaction with AcrB. AcrB is a potential downstream target since acrB mutants are CDI resistant (3). AcrB is a proton antiporter (41, 56), interacting with the periplasmic AcrA protein and OMP TolC to form a machine that exports small molecules outside of cells (55). Notably, the export machine is not necessary for CDI since null mutations in acrA and tolC do not affect growth inhibition (3). Since acrB is not an essential gene, growth inhibition could not occur by blocking AcrB activity but might occur by a gain of function effect that opened the proton channel of AcrB. This could result in the reduction in Δp and EtBr export observed (Fig. 4C and D) (38). The reduction in ATP occurring during CDI could result from ATP synthase-catalyzed breakdown of ATP in response to low Δp. This conclusion is consistent with the finding that atp401 CDI+ cells had a much higher ATP level (about 15-fold at 3 h) than did their atp+ counterparts in growth-inhibited cells (Fig. 5B, left panel). Consistent with the hypothesis that Δp may control cell growth in CDI, uncouplers of oxidative phosphorylation, such as CCCP and 2,3-dinitrophenol, reduce Δp and inhibit cell growth by a mechanism that is reversible at low levels of uncoupler (22, 23, 35). In contrast to CDI, cells recovering growth in the presence of CCCP do not appear to recover Δp, leading Kinoshita and coworkers to conclude that E. coli can grow in the absence of Δp after a lag period (35) during which the bacteria presumably alter gene expression to adapt to uncoupler (23). Addition of CCCP causes an increase in respiration in starved E. coli, the opposite response to that we observe in CDI (15, 40). Thus, it seems unlikely that the mechanism of CDI involves solely a reduction in Δp, although this may contribute to growth inhibition.
Morphological analysis of cells undergoing CDI indicated that, with increasing time of growth inhibition, filaments were observed (Fig. 3). In some cases, the filaments contained multiple nucleoids, and in others, nucleoids were absent (Fig. 3, last two panels in first row). These results raise the possibility that one or more steps in cell division/cell septation might be inhibited, either through the reduction in Δp, respiration, or another mechanism. Notably, within a few hours of induction of CdiI with maltose present, filaments were absent, and cell morphology appeared normal (Fig. 3, compare top two rows). These results suggest that during growth recovery, filament septation occurred to produce normally sized daughter cells.
Infection of cells by bacteriophages and other environmental stresses, such as exposure to proton ionophores, can reduce Δp by proton translocation across the membrane, inducing the phage shock response initiated by transcriptional activation of pspA (19). Therefore, we examined pspA mRNA levels over an 11-h time course of growth inhibition and recovery to determine if the phage shock response was activated in CDI. The results show that pspA transcription was induced by ∼20-fold at its peak, which occurred about 5 h after cells stopped growing (Fig. 7). Cells receiving arabinose and maltose at 5 h to induce recovery (Fig. 7B) did not show significant induction of pspA, consistent with the finding that Δp recovery initiates by 6.5 h (Fig. 4D). These results indicate that cells respond to the low Δp induced during CDI by activating the phage shock response. Since the phage shock response may function to maintain Δp and/or membrane stability (9, 30), we tested to see if CDI occurred in a pspA null mutant. The results showed that cells underwent CDI and could recover from CDI in the absence of pspA (Fig. 7A), indicating that the phage shock response does not play an essential role in CDI.
The finding that CDI is reversible and caused a multifactorial metabolic downregulation raised the question of what physiologic role this pathway plays in the biology of E. coli. This growth inhibitory system was discovered using an intestinal isolate of E. coli that constitutively expressed CdiB and CdiA and was protected from autoinhibition by expression of CdiI. CDI might have a toxin-like function to inhibit growth of competing intestinal bacteria. However, the reversible nature of CDI and our findings that colanic acid capsule (3) and certain types of pili (4) protect E. coli from growth inhibition suggest that CDI may function as an intraspecies communication mechanism (8, 51). The role of CDI could be to allow survival under conditions of oxidative stress by reduction in respiration. Another possibility is that the lowered membrane potential of cells in CDI might provide resistance to host antimicrobial agents, such as defensins (39). The reversible metabolic quiescence occurring in CDI shares some features with a phenomenon known as the “viable but not culturable” (VBNC) state (48). Recent work by Alam and colleagues indicates that Vibrio cholerae in the VBNC state within biofilms can be infectious for at least a year (2). It is not known if there are multiple mechanisms responsible for the VBNC state, but it is possible that they share aspects with CDI.
Functional homologs of CdiA and CdiB are present in uropathogenic E. coli, and potential homologs are present in a wide variety of pathogenic bacteria, such as Yersinia pestis and Burkholderia pseudomallei (4). It will be important to determine if these pathogens undergo CDI-dependent metabolic downregulation within their hosts and to determine if this contributes to their virulence and/or persistence in the host. In this regard, it is notable that Mycobacterium tuberculosis, which causes a chronic disease, has recently been shown to have a five- to sixfold-lower ATP level in its nonreplicating quiescent state (49). The mechanism by which this metabolic downregulation occurs in M. tuberculosis is not known, but it could involve features of the CDI system.
Acknowledgments
We thank B. Matsumoto for microscopy advice and S. Casjens and R. Gourse for plasmids.
We are very grateful to the National Science Foundation (NSF grant 0642052 to D.A.L.) and the Department of Homeland Security (DOE contract number DE-AC05-00OR22750 to J.W.) for their generous support of this project.
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Ahmed, S., and I. R. Booth. 1981. Quantitative measurements of the proton-motive force and its relation to steady state lactose accumulation in Escherichia coli. Biochem. J. 200573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M., M. Sultana, G. B. Nair, A. K. Siddique, N. A. Hasan, R. B. Sack, D. A. Sack, K. U. Ahmed, A. Sadique, H. Watanabe, C. J. Grim, A. Huq, and R. R. Colwell. 2007. Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc. Natl. Acad. Sci. USA 10417801-17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, S., J. Malinverni, K. Jacoby, B. Thomas, R. Pamma, B. Trinh, S. Remers, J. Webb, B. Braaten, T. Silhavy, and D. Low. 2008. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) and the inner membrane transport protein AcrB. Mol. Microbiol. 70323-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki, S. K., R. Pamma, A. D. Hernday, J. E. Bickham, B. A. Braaten, and D. A. Low. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 3091245-1248. [DOI] [PubMed] [Google Scholar]
- 5.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baev, M. V., D. Baev, A. J. Radek, and J. W. Campbell. 2006. Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of sugars, alcohols, and organic acids with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71310-316. [DOI] [PubMed] [Google Scholar]
- 7.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2582-587. [DOI] [PubMed] [Google Scholar]
- 8.Bassler, B. L., and R. Losick. 2006. Bacterially speaking. Cell 125237-246. [DOI] [PubMed] [Google Scholar]
- 9.Becker, L. A., I. S. Bang, M. L. Crouch, and F. C. Fang. 2005. Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol. Microbiol. 561004-1016. [DOI] [PubMed] [Google Scholar]
- 10.Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4121-136. [DOI] [PubMed] [Google Scholar]
- 11.Boulos, L., M. Prevost, B. Barbeau, J. Coallier, and R. Desjardins. 1999. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 3777-86. [DOI] [PubMed] [Google Scholar]
- 12.Bragg, P. D., and C. Hou. 1977. Purification and characterization of the inactive Ca2+, Mg2+-activated adenosine triphosphatase of the unc A− mutant Escherichia coli AN120. Arch. Biochem. Biophys. 178486-494. [DOI] [PubMed] [Google Scholar]
- 13.Brown, G. C. 1992. Control of respiration and ATP synthesis in mammalian mitochondria and cells. Biochem. J. 2841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, G. C., P. L. Lakin-Thomas, and M. D. Brand. 1990. Control of respiration and oxidative phosphorylation in isolated rat liver cells. Eur. J. Biochem. 192355-362. [DOI] [PubMed] [Google Scholar]
- 15.Burstein, C., L. Tiankova, and A. Kepes. 1979. Respiratory control in Escherichia coli K 12. Eur. J. Biochem. 94387-392. [DOI] [PubMed] [Google Scholar]
- 16.Butlin, J. D., G. B. Cox, and F. Gibson. 1971. Oxidative phosphorylation in Escherichia coli K12. Mutations affecting magnesium ion- or calcium ion-stimulated adenosine triphosphatase. Biochem. J. 12475-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138179-207. [DOI] [PubMed] [Google Scholar]
- 18.Chen, J., T. Kuroda, M. N. Huda, T. Mizushima, and T. Tsuchiya. 2003. An RND-type multidrug efflux pump SdeXY from Serratia marcescens. J. Antimicrob. Chemother. 52176-179. [DOI] [PubMed] [Google Scholar]
- 19.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57621-628. [DOI] [PubMed] [Google Scholar]
- 20.Dunny, G. M., T. J. Brickman, and M. Dworkin. 2008. Multicellular behavior in bacteria: communication, cooperation, competition and cheating. Bioessays 30296-298. [DOI] [PubMed] [Google Scholar]
- 21.Felle, H., J. S. Porter, C. L. Slayman, and H. R. Kaback. 1980. Quantitative measurements of membrane potential in Escherichia coli. Biochemistry 193585-3590. [DOI] [PubMed] [Google Scholar]
- 22.Fraimow, H. S., J. B. Greenman, I. M. Leviton, T. J. Dougherty, and M. H. Miller. 1991. Tobramycin uptake in Escherichia coli is driven by either electrical potential or ATP. J. Bacteriol. 1732800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage, D. J., and F. C. Neidhardt. 1993. Adaptation of Escherichia coli to the uncoupler of oxidative phosphorylation 2,4-dinitrophenol. J. Bacteriol. 1757105-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gober, J. W., and E. R. Kashket. 1985. Measurement of the proton motive force in Rhizobium meliloti with the Escherichia coli lacY gene product. J. Bacteriol. 164929-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 27.Harold, F. M., and P. C. Maloney. 2008. Energy transduction and ion currents. In R. Curtiss III (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC.
- 28.Hill, T. M., B. Sharma, M. Valjavec-Gratian, and J. Smith. 1997. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J. Bacteriol. 1791931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraga, S., H. Niki, T. Ogura, C. Ichinose, H. Mori, B. Ezaki, and A. Jaffe. 1989. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J. Bacteriol. 1711496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jovanovic, G., L. J. Lloyd, M. P. Stumpf, A. J. Mayhew, and M. Buck. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 28121147-21161. [DOI] [PubMed] [Google Scholar]
- 31.Kadenbach, B. 2003. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim. Biophys. Acta 160477-94. [DOI] [PubMed] [Google Scholar]
- 32.Kashket, E. R. 1982. Stoichiometry of the H+-ATPase of growing and resting, aerobic Escherichia coli. Biochemistry 215534-5538. [DOI] [PubMed] [Google Scholar]
- 33.Kato, J., Y. Nishimura, M. Yamada, H. Suzuki, and Y. Hirota. 1988. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J. Bacteriol. 1703967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S., J. C. Malinverni, P. Sliz, T. J. Silhavy, S. C. Harrison, and D. Kahne. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317961-964. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita, N., T. Unemoto, and H. Kobayashi. 1984. Proton motive force is not obligatory for growth of Escherichia coli. J. Bacteriol. 1601074-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15162-171. [PMC free article] [PubMed] [Google Scholar]
- 37.Kubista, M., J. M. Andrade, M. Bengtsson, A. Forootan, J. Jonak, K. Lind, R. Sindelka, R. Sjoback, B. Sjogreen, L. Strombom, A. Stahlberg, and N. Zoric. 2006. The real-time polymerase chain reaction. Mol. Aspects Med. 2795-125. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, B., and J. B. Le Pecq. 1984. Effect of mutation, electric membrane potential, and metabolic inhibitors on the accessibility of nucleic acids to ethidium bromide in Escherichia coli cells. Biochemistry 23166-176. [DOI] [PubMed] [Google Scholar]
- 39.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letellier, L., and B. Labedan. 1985. Release of respiratory control in Escherichia coli after bacteriophage adsorption: process independent of DNA injection. J. Bacteriol. 161179-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 1645-55. [DOI] [PubMed] [Google Scholar]
- 42.Makhov, A. M., J. H. Hannah, M. J. Brennan, B. L. Trus, E. Kocsis, J. F. Conway, P. T. Wingfield, M. N. Simon, and A. C. Steven. 1994. Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J. Mol. Biol. 241110-124. [DOI] [PubMed] [Google Scholar]
- 43.Malinverni, J. C., J. Werner, S. Kim, J. G. Sklar, D. Kahne, R. Misra, and T. J. Silhavy. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61151-164. [DOI] [PubMed] [Google Scholar]
- 44.Mazia, D., G. Schatten, and W. Sale. 1975. Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. J. Cell Biol. 66198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullin, S., N. Mani, and T. H. Grossman. 2004. Inhibition of antibiotic efflux in bacteria by the novel multidrug resistance inhibitors biricodar (VX-710) and timcodar (VX-853). Antimicrob. Agents Chemother. 484171-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami, S. 2008. Multidrug efflux transporter, AcrB—the pumping mechanism. Curr. Opin. Struct. Biol. 18459-465. [DOI] [PubMed] [Google Scholar]
- 47.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3215-218. [PubMed] [Google Scholar]
- 48.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 4393-100. [PubMed] [Google Scholar]
- 49.Rao, S. P., S. Alonso, L. Rand, T. Dick, and K. Pethe. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 10511945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider, D. A., and R. L. Gourse. 2004. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J. Biol. Chem. 2798262-8268. [DOI] [PubMed] [Google Scholar]
- 51.Slechta, E. S., and M. A. Mulvey. 2006. Contact-dependent inhibition: bacterial brakes and secret handshakes. Trends Microbiol. 1458-60. [DOI] [PubMed] [Google Scholar]
- 52.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9133-137. [DOI] [PubMed] [Google Scholar]
- 53.Werner, J., and R. Misra. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 571450-1459. [DOI] [PubMed] [Google Scholar]
- 54.Wyckoff, E., L. Sampson, M. Hayden, R. Parr, W. M. Huang, and S. Casjens. 1986. Plasmid vectors useful in the study of translation initiation signals. Gene 43281-286. [DOI] [PubMed] [Google Scholar]
- 55.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300976-980. [DOI] [PubMed] [Google Scholar]
- 56.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 967190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]