Abstract
The antigenically related coaggregation receptor polysaccharides (RPS) of Streptococcus oralis strains C104 and SK144 mediate recognition of these bacteria by other members of the dental plaque biofilm community. In the present study, the structure of strain SK144 RPS was established by high resolution NMR spectroscopy as [6Galfβ1-6GalNAcβ1-3Galα1-2ribitol-5-PO4−-6Galfβ1-3Galβ1]n, thereby indicating that this polysaccharide and the previously characterized RPS of strain C104 are identical, except for the linkage between Gal and ribitol-5-phosphate, which is α1-2 in strain SK144 versus α1-1 in strain C104. Studies to define the molecular basis of RPS structure revealed comparable genes for six putative transferases and a polymerase in the rps loci of these streptococci. Cell surface RPS production was abolished by disrupting the gene for the first transferase of strain C104 with a nonpolar erm cassette. It was restored in the resulting mutant by plasmid-based expression of either wcjG, the corresponding gene of S. pneumoniae for serotype 10A capsular polysaccharide (CPS) biosynthesis or wbaP for the transferase of Salmonella enterica that initiates O-polysaccharide biosynthesis. Thus, WcjG, like WbaP, appears to initiate polysaccharide biosynthesis by transferring galactose-1-phosphate to a lipid carrier. In further studies, the structure of strain C104 RPS was converted to that of strain SK144 by replacing the gene (wefM) for the fourth transferase in the rps locus of strain C104 with the corresponding gene (wcrC) of strain SK144 or Streptococcus pneumoniae serotype 10A. These findings identify genetic markers for the different ribitol-5-phosphate-containing types of RPS present in S. oralis and establish a close relationship between these polysaccharides and serogroup 10 CPSs of S. pneumoniae.
The coaggregations observed between different viridans group streptococci and Actinomyces naeslundii (6) provided early evidence for the role of interbacterial adhesion in dental plaque biofilm formation. Interactions between these bacteria were subsequently attributed to binding of A. naeslundii type 2 fimbriae to specific Gal and GalNAc-containing cell wall polysaccharides, referred to as receptor polysaccharides (RPS), on strains of Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii (7, 9, 14). These streptococci inhabit the tooth surface (23), where they grow in close association with type 2 fimbriated A. naeslundii (26) and other members of the dental plaque biofilm community. Growth and biofilm formation were not observed in flow cells when coaggregating strains of S. oralis and A. naeslundii were cultured separately in dilute saliva (27). However, when cultured together, the two strains grew as a mixed-species community, thereby supporting a recognition role for cell surface RPS in biofilm development.
Six structural types of RPS have been identified by high resolution nuclear magnetic resonance (NMR) of the cell wall polysaccharides isolated from over 20 coaggregating strains of S. sanguinis, S. gordonii, and S. oralis (8). These polysaccharides are composed of structurally distinct repeating units that contain conserved Galf linked β1-6 to a host-like recognition motif, which is GalNAcβ1-3Gal (Gn) in certain types of RPS and Galβ1-3GalNAc (G) in others. The flexible β1-6 linkage from Galf (34) is thought to function as a hinge, exposing the adjacent host-like motif for adhesin-mediated recognition (21). Whereas both Gn and G types of RPS are recognized by type 2 fimbriated A. naeslundii, only Gn types are recognized by the GalNAc-binding adhesins present on non-RPS-bearing strains of S. sanguinis and S. gordonii (8). Conversely, only G types are coaggregation receptors of certain Veillonella spp. (25). The host-like features of these polysaccharides, although critical for interbacterial adhesion, contribute little to RPS serotype specificity, which instead reflects the immunogenic features of these molecules (21). As a result, the identification of RPS-bearing streptococci requires both serotyping (i.e., serotypes 1, 2, 3, 4, or 5) and receptor typing (i.e., types Gn or G) of these bacteria.
A possible molecular approach for the identification of these bacteria is evident from comparative studies of the chromosomal loci (rps) for RPS biosynthesis in different strains (33, 35-37). In this regard, the genes wchA and wchF, which were first identified in Streptococcus pneumoniae (5, 15), encode the first two transferases for synthesis of RPS serotypes 1, 2, and 3. WchA transfers Glc-1-phosphate from UDP-Glc to a carrier lipid, and WchF adds Rha β1-4 to Glc. Subsequent synthesis of both the antigenic and receptor regions in these polysaccharides depends on other encoded transferases (35-37), many of which are distinguishable from those identified in S. pneumoniae. In addition to Glc- and Rha-containing types of RPS, other types have been described that lack these sugars but contain ribitol-5-phosphate (3), in addition to GalNAc, Galp, and Galf, which are common constituents of all types. The ribitol-5-phosphate-containing group, represented by type 4Gn RPS of S. oralis C104 and type 5Gn RPS of S. oralis SK144, is the subject of the present study. The results define the structural and genetic basis of the antigenic difference noted between these polysaccharides. They also reveal a close molecular relationship between these types of RPS and certain capsular polysaccharides (CPS) of S. pneumoniae, most notably those in CPS serogroup 10.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table 1 lists the wild-type and mutant streptococcal strains used in this study. The identification of isolates as strains of S. oralis was based on sequencing of sodA and in some cases, additional housekeeping genes, as previously described (13). Streptococci were grown at 37°C in Todd-Hewitt broth (Difco Laboratories), brain heart infusion (BHI) broth (Oxoid), or a previously described (6) complex medium containing 0.05% Tween 80. Erythromycin at 10 μg/ml or kanamycin at 750 μg/ml was added to these media as needed for the cultivation of antibiotic-resistant strains.
TABLE 1.
Streptococci and plasmids used in this study
| Strain or plasmid | Descriptionc | Source or reference |
|---|---|---|
| Wild-type strains | ||
| S. oralis SK144 | Type 5Gn RPS | 14 |
| S. oralis C104 | Type 4Gn RPS | 6 |
| S. gordonii 38 | Type 2Gn RPS | 6 |
| S. oralis 34 | Type 1Gn RPS | 22 |
| S. pneumonia 8334 | Serotype 10A CPS | ATCC |
| S. oralis SK143 | Type 4Gn RPS | 14 |
| S. oralis 4080 | Infant oral isolate, type 4Gn RPSa | M. Cole |
| S. oralis R4 | Adult supragingival plaque, type 4Gn RPSb | This study |
| S. oralis EK2 | Adult supragingival plaque, type 4Gn RPSb | This study |
| S. oralis E1 | Adult supragingival plaque, type 4Gn RPSb | This study |
| S. oralis A3 | Infant supragingival plaque, type 4Gn RPSb | This study |
| S. oralis K2 | Infant supragingival plaque, type 5Gn RPSb | This study |
| S. oralis A6 | Infant supragingival plaque, type 5Gn RPSb | This study |
| Mutant strains | ||
| S. oralis YC1 | S. oralis C104 with erm in place of wefM, RPS− | This study |
| S. oralis YC2 | S. oralis YC1 with wcrC of strain SK144 in place of erm, RPS+ | This study |
| S. oralis YC3 | S. oralis C104 with erm in place of wcjG, RPS− | This study |
| S. oralis YC4 | S. oralis C104 with erm in place of wciB, RPS− | This study |
| S. oralis YC5 | S. oralis C104 with erm in place of wefD, RPS− | This study |
| S. oralis YC6 | S. oralis C104 with erm in place of wefE, RPS− | This study |
| S. gordonii GC14 | S. gordonii 38 with erm in place of wefD, RPS− | 35 |
| S. gordonii XC3 | S. gordonii 38 with erm in place of wefE, RPS− | 33 |
| Plasmids | ||
| pCM18 | Streptococcus-E. coli shuttle vector, Emr | 11 |
| pCM18-1 | pCM18 digested with EcoRI to delete CP25-gfp-CAT fragment, Emr | This study |
| pCM18-2 | pCM18-1 with kan in place of erm, Kmr | This study |
| pJY | pCM18-2 with Kpn2I site used for insertion of synthetic promoter (CP25) and multiple cloning site, Kmr | This study |
| pJY-1 | pJY expressing wcjG of S. pneumonia 8334 | This study |
| pJY-2 | pJY expressing wbaP of Salmonella enterica serovar Typhi Ty2 (ATCC 700931) | This study |
| pJY-3 | pJY expressing coding region for C-terminal 214 amino acids of WbaP of S. enterica serovar Typhi Ty2 | This study |
| pJY-4 | pJY expressing wciB of S. pneumonia 8834 | This study |
| pJY-5 | pJY expressing wefD of S. gordonii 38 | This study |
| pJY-6 | pJY expressing wefD of S. oralis C104 | This study |
| pJY-7 | pJY expressing wefE of S. gordonii 38 | This study |
| pJY-8 | pJY expressing wefE of S. oralis C104 | This study |
| pJY-9 | pJY expressing wcrC of S. pneumonia 8834 | This study |
Type 4Gn RPS identified by NMR of the isolated polysaccharide.
RPS type identified by dot immunoblotting of bacteria with anti-4Gn RPS-specific IgG and anti-4Gn/5Gn RPS-specific IgG; only the latter antibody reacted with type 5Gn RPS-bearing strains.
RPS+ and RPS− denote the presence or absence of cell surface RPS as detected by coaggregation with A. naeslundii 12104.
Isolation and structural characterization of RPS.
RPS were solubilized by mutanolysin digestion of protease-treated streptococcal cell walls and purified by DEAE Sephacel (GE Healthcare) column chromatography as previously described (8). Purified polysaccharides were eluted from the column and recovered in fractions containing 110 to 140 mM NaCl in 0.01 M Tris-HCl buffer (pH 8.0).
The RPS (250 μg) of strain SK144 was hydrolyzed with 4 N HCl (250 μl, 100°C, 3 h). After evaporation of the HCl with nitrogen gas, the sample was dissolved in 250 μl of deionized water and passed through a 0.22-μm filter. The hydrolysate was analyzed for neutral and phosphorylated sugars by high-performance anionic exchange (HPAE) chromatography using a Carbopac PA1 column (4 by 250 mm; Dionex) and a pulsed amperometric detector (Dionex). Neutral monosaccharides were eluted with 16 mM NaOH at a flow rate of 1 ml/min. Phosphorylated monosaccharides were eluted with 100 mM NaOH and 200 mM sodium acetate for 10 min and then with a constant concentration of 100 mM NaOH and a linear gradient of sodium acetate from 200 mM to 600 mM over 20 min. Peaks were identified by comparing their retention times with those of standard monosaccharides (Sigma). Ribitol-5-phosphate was prepared by NaBH4 reduction of ribose-5-phosphate (3).
HPAE chromatography with a Carbopac MA1 column (4 by 250 mm; Dionex) was used to analyze the polysaccharide for alditols. SK144 RPS (250 μg) was dissolved in hydrofluoric acid (0.2 ml, 48%, cold) and incubated at 5°C for 3 days to cleave phosphate. After evaporating the hydrofluoric acid under a stream of nitrogen, the sample was hydrolyzed with 4 N HCl (300 μl, 2 h, 100°C). The hydrolysate was evaporated with nitrogen to remove HCl, dissolved in 250 μl deionized water, and passed through a 0.22-μm filter. Sample volumes of 20 μl were applied to the column and eluted with 600 mM NaOH (flow rate of 0.4 ml/min).
NMR spectra were recorded on a Bruker DRX 500 and a General Electric GN500 spectrometer (500 MHz for 1H) at 25°C. Polysaccharides (5 mg) were exchanged in D2O (99.8 atom% D) by three cycles of lyophilization from this liquid. Dried samples were then dissolved in 0.5 ml of 99.996% D2O for NMR. All carbon and proton chemical shifts are reported relative to the internal standard of acetone (1H δ = 2.225 ppm and 13C δ = 31.07 ppm). Phosphorus chemical shifts are reported relative to an external reference signal for 85% H3PO4 (δ = 0.00 ppm).
The following two-dimensional NMR data were recorded with standard pulse sequences: nuclear Overhauser effect spectroscopy (NOESY), correlation spectroscopy, total correlation spectroscopy (TOCSY), 1H-detected 13C heteronuclear single quantum correlation, combination heteronuclear single quantum correlation-TOCSY, multiple quantum coherence spectroscopy, and heteronuclear multiple bond coherence spectroscopy. All NMR data were processed using the FELIX (Biosym Corp., San Diego, CA) or NMRPipe (NMR Science) programs.
NOESY and TOCSY spectra were recorded with mixing times of 300 ms and 70 ms, respectively. 1H-detected 31P spin echo experiments were performed using delay times of 0, 10, 20, 40, 60, 80, 100, and 120 ms.
PCR amplification and sequencing of genes for RPS production.
The 21,137-bp DNA sequence (GenBank accession number EF587720) containing the rps locus of S. oralis SK144 was assembled from the sequences of overlapping PCR products. These products, which included the 10-kb fragment from wzg to wzx, were PCR amplified from genomic DNA of strain SK144 using primers designed from sequences in the rps loci of S. oralis strain 34 (36) or S. gordonii strain 38 (33). Inverse PCR (24) was performed to extend certain sequences. The 21,008-bp DNA sequence (GenBank accession number EF587719) containing the rps locus of S. oralis C104 was PCR amplified from genomic DNA of this strain using primers designed from the rps locus of strain SK144. Selected genes were also PCR amplified from genomic DNA of other type 4Gn or 5Gn RPS-producing strains (Table 1). Primers for the amplification of wefK from these strains were designed from the sequence of this gene or those of the flanking genes (i.e., wzx and glf) in strain C104. Primers for the amplification of wefM or wcrC were designed from common sequences in the flanking genes (i.e., wefL or wefD) of strains SK144 and C104. Sequencing of PCR products was performed at Sequetech Corp., Mountain View, CA. The resulting sequences were assembled and annotated using Vector NT1 software (Invitrogen) and the National Center for Biotechnology Information BLAST program (4).
Antibodies and immunological methods.
Type 4Gn/5Gn RPS-specific immunoglobulin G (IgG) from rabbit antiserum R100 against S. oralis C104 (8) was affinity purified by 4 M MgCl2 elution from a small column of immunoadsorbent, prepared by coupling partially oxidized type 4Gn RPS to Affi-Gel Hz (Bio-Rad) as previously described (33). This antibody was rendered type 4Gn RPS-specific by absorption with type 5Gn RPS-producing S. oralis SK144. Absorption was performed by incubating 0.5 μg type 4Gn/5Gn RPS-specific IgG with about 109 washed S. oralis SK144 cells for 1 h at 4°C in 1 ml phosphate-buffered saline (pH 7.4) containing 4 mg/ml bovine serum albumin (Sigma). Following centrifugation of the mixture to remove added bacteria, the supernatant containing type 4Gn RPS-specific IgG was recovered and filter sterilized. The other primary antibody used in this study was type 2Gn RPS-specific IgG, which has been described previously (35).
Dot immunoblotting was performed as previously described (35) to detect binding of RPS-specific IgG to decreasing numbers of streptococci spotted on nitrocellulose membranes. The membranes were blocked and incubated with either 50 ng/ml RPS-specific IgG or a 1/10 dilution of the absorbed antibody described above, which was followed by peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad) prior to development with a metal-enhanced DAB substrate kit (Pierce Biotechnology) to detect bound antibody.
Genetic transformation.
Transformation of S. gordonii 38 and mutant strain GC14 was performed as previously described (35). The competence-stimulating peptide (CSP) of strains C104 and SK144 was identified by PCR amplification and sequencing of comC from these strains using the previously described primers (37). The predicted amino acid sequence of ComC (i.e., MKNTVKLEQFKEVTEAELQEIRGGDWRISETIRNLIFPRRK) was the same for both strains. The position of the putative Gly-Gly cleavage site in this sequence was used to identify the CSP, which is italicized above. This peptide was synthesized using automated 9-fluorenylmethoxy carbonyl chemistry and purified by high-performance liquid chromatography (CBER Research Central, NIH). Early-log-phase cultures of S. oralis C104 or mutants of this strain in Todd-Hewitt broth or BHI containing 5% heated inactivated fetal horse serum (Sigma) were incubated 30 min at 37°C with CSP at a final concentration of 100 μg/ml to induce competence in these bacteria. Reaction mixtures were then incubated 3 h at 37°C with transforming DNA (i.e., purified PCR products or plasmids) prior to plating on BHI agar containing 5% fetal horse serum (Sigma) and appropriate antibiotics to select for transformants.
Construction of mutant strains.
S. oralis YC1, YC3, YC4, YC5, and YC6 (Table 1) were prepared from S. oralis C104 by transformation of this strain with DNA constructs containing a nonpolar erm cassette (i.e., ermAM) (19) flanked by 0.5- to 1.0-kb gene-targeting sequences for homologous recombination with identical sequences in the rps locus of strain C104. Transforming DNA was prepared by overlap extension PCR, performed as previously described (12, 35) with appropriately designed primers (18) and KOD Hot Start DNA polymerase (Novagen). The location of the erm cassette in each mutant strain was verified by PCR.
The RPS+ transformant S. oralis YC2 was obtained by transformation of RPS− S. oralis YC1 with PCR-amplified wcrC of S. oralis SK144 flanked by gene-targeting sequences for the insertion of this gene in place of the erm cassette in strain YC1. S. oralis YC2 was identified by colony immunoblotting (35) performed with type 4Gn/5Gn RPS-specific IgG. The presence of wcrC at the expected location in strain YC2 was verified by DNA sequencing.
Construction of plasmids.
Plasmid pJY (Fig. 1) was constructed from previously described pCM18 (11) to facilitate genetic complementation studies in erm-containing mutants of S. oralis C104. Initially, we converted pCM18 to pCM18-1 (Table 1) by digesting the former plasmid with EcoRI to delete the constitutive lactococcal promoter (CP25), the gene (gfp) for green fluorescent protein and the downstream multiple cloning site. Next, we replaced erm in pCM18-1 with kan from pSF151 (29) by selection of a Kanr transformant following cotransformation of S. gordonii DL1 with pCM18-1 and an overlap extension PCR product containing the kan gene. Finally, we inserted a specifically synthesized 139-bp DNA sequence (Blue Heron Biotechnology, Bothell, WA) containing a constitutive promoter (CP25) and multiple cloning site into the Kpn2I site of pCM18-2 to obtain pJY.
FIG. 1.
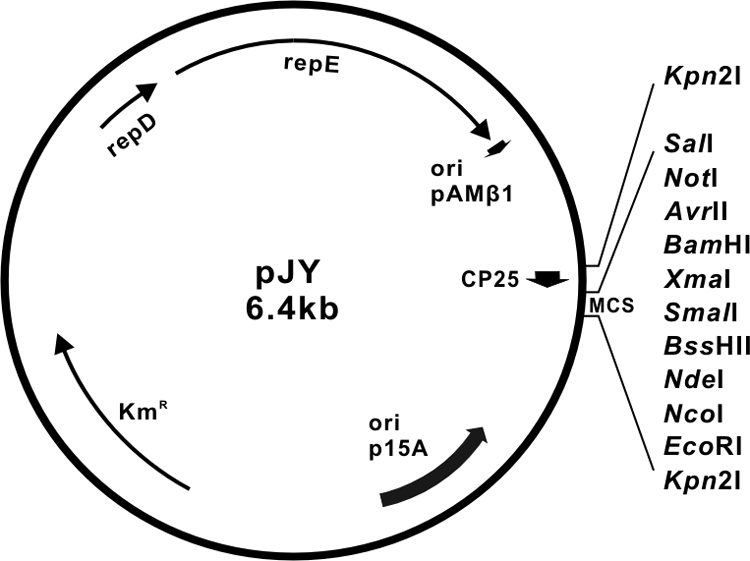
Physical map of the Escherichia coli-streptococcus shuttle vector pJY used for genetic complementation of RPS production in erm-containing mutants of S. oralis C104. Key features of the plasmid include a synthetic promoter (CP25), a multicloning site (MCS) and a selectable kan marker.
The derivatives of pJY listed in Table 1 were prepared by cloning PCR-amplified genes for various transferases of S. pneumoniae serotype 10A, Salmonella enterica serovar Typhi TY2, S. gordonii 38, or S. oralis C104 into the multiple cloning site of this plasmid. The primers used for PCR were designed to amplify not only the gene of interest but also the upstream Shine-Dalgarno sequence. The 708-bp insert present in pJY-3 was PCR amplified from S. enterica serovar Typhi Ty2 genomic DNA using forward and reverse primers (AGTCCTAGGCCCTCGTTTAGAGGATTGCCATT and CTATCCATGGCAATTATTATTCAGTACTTCTCGGTAAGC, respectively) that contained AvrII or NcoI restriction sites (underlined) for cloning into pJY. The integrity of the cloned sequence, which included the coding region for the C-terminal amino acids of WbaP and the Shine-Dalgarno sequence located immediately upstream (32), was verified by DNA sequencing.
RESULTS
The structural difference between types 4Gn and 5Gn RPS.
The structure of type 5Gn RPS from S. oralis SK144 was determined from the composition and NMR spectra of this polysaccharide. HPAE chromatography (16 mM NaOH, Carbopac PA1 column) of a 4 N HCl hydrolysate revealed peaks of galactose (14.67 min) and galactosamine (11.00 min) and a peak that eluted early (2.90 min) in the chromatogram, indicating an alditol. The last peak could not be identified due to the similar retention times of different alditols (i.e., ribitol, xylitol, arabitol, and lyxitol) under these conditions. The selectivity of the Carbopac MA1 column was improved by elution with 600 mM NaOH. Under these conditions, the retention times of the unknown alditol and the ribitol standard were indistinguishable (17.95 min) compared to 12.51 min for xylitol and 14.85 min for arabitol and lyxitol.
Strain SK144 RPS appeared to consist of Gal, GalNAc, and ribitol in a ratio of 2:1:1, based on quantitative analysis of the data from HPAC chromatography. However, the NMR spectra of this polysaccharide, which are presented and discussed as supplemental material, indicated four galactose residues, two furanosides, and two pyranosides. The observance of a total of only two Gal residues by HPLC was readily explained by the phosphorylation of one galactose residue and by variable and partial degradation of galactose under acidic hydrolysis conditions strong enough to cleave the amino sugar linkage.
The 1H and 13C chemical shifts of the five sugar residues and of the ribitol were assigned using standard methods of coherence transfer and NOE (2), as described in the supplemental material. The complete assignment of chemical shifts is presented in Table 2, and the NMR evidence (see Fig. S1 to S4 in the supplemental material) for the linkages between the residues is summarized in Fig. 2A. The results indicated that the structure of type 5Gn RPS (Fig. 2A) was identical to that of previously characterized type 4Gn RPS (3), except for the linkage between α-Gal (residue E) and ribitol (residue F), which is α1-2 in the former polysaccharide versus α1-1 in the latter.
TABLE 2.
Residue-by-residue comparison of HSQC 1H and 13C chemical shifts for the RPS of S. oralis strains SK144, C104, and YC5
| Strain | Chemical shift (ppm)a
|
||||||
|---|---|---|---|---|---|---|---|
| Residue | H-1 C-1 | H-2 C-2 | H-3 C-3 | H-4 C-4 | H-5 C-5 | H-6 C-6 | |
| SK144 | β-Galf (A) | 5.218 | 4.205 | 4.102 | 4.101 | 3.985 | 3.956, 3.981 |
| 110.09 | 82.29 | 77.47 | 83.29 | 70.17 | 67.18 | ||
| C104 | 5.218 | 4.208 | 4.101 | 4.108 | 3.99 | 3.97 | |
| 110.05 | 82.26 | 77.46 | 83.36 | 70.1 | 67.18 | ||
| YC5 | 5.219 | 4.213 | 4.102 | 4.112 | 3.996 | 3.98 | |
| 110.10 | 82.26 | 77.53 | 83.39 | 70.13 | 67.16 | ||
| SK144 | β-Galp (B) | 4.507 | 3.677 | 3.732 | 4.105 | 3.727 | 3.776 |
| 103.91 | 70.87 | 81.13 | 69.43 | 75.98 | 61.86 | ||
| C104 | 4.507 | 3.675 | 3.742 | 4.107 | 3.73 | 3.78 | |
| 103.91 | 70.84 | 81.09 | 69.38 | 75.96 | 61.85 | ||
| YC5 | 4.507 | 3.676 | 3.745 | 4.110 | 3.734 | 3.78 | |
| 103.88 | 70.84 | 81.14 | 69.34 | 75.96 | 61.84 | ||
| SK144 | β-Galf (C) | 5.071 | 4.075 | 4.102 | 4.006 | 4.027 | 3.767, 4.080 |
| 108.74 | 81.85 | 77.56 | 83.95 | 70.34 | 71.91 | ||
| C104 | 5.071 | 4.068 | 4.1 | 4.006 | 4.027 | 3.767, 4.077 | |
| 108.62 | 81.81 | 77.51 | 83.96 | 70.78 | 71.91 | ||
| YC5 | 5.056 | 4.096 | 4.100 | 4.003 | 4.029 | 3.768, 4.084 | |
| 108.36 | 81.97 | 77.57 | 83.94 | 70.40 | 72.02 | ||
| SK144 | β-GalNAc (D) | 4.651 | 3.952 | 3.757 | 3.946 | 3.827 | 3.776, 3.909 |
| 103.97 | 53.39 | 71.57 | 68.52 | 74.58 | 68.18 | ||
| C104 | 4.635 | 3.944 | 3.752 | 3.945 | 3.829 | 3.775, 3.911 | |
| 103.93 | 53.36 | 71.56 | 68.59 | 74.54 | 68.08 | ||
| SK144 | α-Galp (E) | 5.201 | 3.898 | 3.976 | 4.228 | 4.063 | 3.740 |
| 100.01 | 68.40 | 80.05 | 70.04 | 71.79 | 61.92 | ||
| C104 | 4.959 | 3.89 | 3.963 | 4.204 | 3.98 | 3.74 | |
| 99.92 | 68.26 | 80.22 | 70.08 | 71.43 | 62.06 | ||
| YC5 | 4.988 | 3.831 | 3.914 | 4.010 | 4.125 | 3.686, 3.900 | |
| 99.84 | 69.27 | 70.18 | 70.10 | 70.62 | 67.90 | ||
| SK144 | Ribitol (F) | 3.831, 3.921 | 4.035 | 4.023 | 3.858 | 3.988, 4.081 | |
| 60.68 | 80.27 | 72.31 | 71.17 | 67.67 | |||
| C104 | 3.599, 3.965 | 4.054 | 3.818 | 3.933 | 3.99, 4.085 | ||
| 69.48 | 71.63 | 72.29 | 71.66 | 67.4 | |||
| YC5 | 3.609, 3.964 | 4.064 | 3.817 | 3.923 | 4.003, 4.079 | ||
| 69.72 | 71.58 | 72.29 | 71.67 | 67.46 | |||
FIG. 2.
Interresidue connectivities used to establish the structures of (A) type 5Gn RPS of S. oralis SK144 and (B) modified type 4Gn RPS of S. oralis YC5, the wefD knockout mutant of S. oralis C104.
Identification and comparison of rps loci.
The rps loci of S. oralis C104 and SK144, like those of other S. oralis strains (36, 37), were identified downstream of two aliB-like open reading frames (ORFs) and upstream of aliA (Fig. 3). Each locus contained 13 comparable genes, including four (i.e., wzg, wzh, wzd, and wze) for proteins with regulatory or processing roles in polysaccharide biosynthesis, six for different transferases, wzy for a polymerase, wzx for a flippase, and glf for galactofuranose mutase, the enzyme that converts UDP-Galp to UDP-Galf (Table 2). The gene wefK for a putative O-acetyltransferase (Table 3) was also identified, but only in the rps locus of strain C104 (Fig. 3). This was unexpected, as type 4Gn RPS of this or other strains (3, 8) does not appear to be O-acetylated. In addition to strains C104 and SK144, we examined other type 4Gn or 5Gn RPS-producing strains (Table 1) for the presence of wefK by PCR and DNA sequencing. The results revealed copies of wefK (GenBank accession numbers FJ555240, FJ560891, FJ560892, FJ560893, and FJ560894) that were 99% identical from five additional type 4Gn RPS-producing isolates examined (i.e., strains SK143, 4080, R4, E1, and A3, respectively), always between wzx and glf, as in strain C104 (Fig. 3). The presence of wefK, however, was not detected by PCR of genomic DNA prepared from type 4Gn RPS-producing strain EK2 or from the two additional type 5Gn RPS-producing isolates examined. Thus, wefK was identified only from type 4Gn RPS-producing strains, not from all strains examined.
FIG. 3.
Molecular comparison of S. oralis C104 type 4Gn RPS and S. oralis SK144 type 5Gn RPS. ORF diagrams show comparable genes for four regulatory and processing proteins ( ), six glycosyl or ribitol-5-phosphate transferases (
), six glycosyl or ribitol-5-phosphate transferases ( ), a polymerase (
), a polymerase ( ), a flippase (
), a flippase ( ) and the enzyme galactofuranose mutase (
) and the enzyme galactofuranose mutase ( ), for synthesis of UDP-Galf, an essential RPS precursor. The gene for an O-acetyltransferase (
), for synthesis of UDP-Galf, an essential RPS precursor. The gene for an O-acetyltransferase ( ) was also found in the rps locus of strain C104. Comparable genes in strains C104 and SK144 are the same, except for wefM and wcrC, which account for the different linkages between Gal and ribitol-5-phosphate in types 4Gn and 5Gn RPS. The region downstream of wefE in strain C104 contains the 5′ and 3′ ends of aliA.
) was also found in the rps locus of strain C104. Comparable genes in strains C104 and SK144 are the same, except for wefM and wcrC, which account for the different linkages between Gal and ribitol-5-phosphate in types 4Gn and 5Gn RPS. The region downstream of wefE in strain C104 contains the 5′ and 3′ ends of aliA.
TABLE 3.
Selected homologues of genes in the rps locus of S. oralis C104
| Gene | Protein size (aa) | Selected homologue | Accession no. | % Identity (aa) | Proposed function |
|---|---|---|---|---|---|
| wcjG | 211 | WcjG of S. pneumoniae 10A | CAI34731 | 83 (211) | Gal-1-phosphate transferase |
| EpsE of S. thermophilus Sfi6 | AAC44012 | 50 (206) | |||
| WbaP of S. enterica LT2 | AAC44096 | 41 (203) | |||
| wciB | 264 | WciB of S. pneumoniae 10A | CAI33047 | 76 (264) | Galf transferase |
| wzy | 405 | Wzy of S. pneumoniae 47F | CAI34657 | 73 (405) | Polymerase |
| Wzy of S. pneumoniae 10A | CAI33048 | <10 (405) | |||
| wefL | 275 | WhaI of S. pneumoniae 47F | CAI34658 | 85 (275) | Ribitol-5-phosphate transferase |
| WcrB of S. pneumoniae 10A | CAI33049 | 29 (271) | |||
| wefM | 360 | WcrC of S. pneumoniae 10A | CAI33050 | 68 (360) | Gal transferase |
| wefD | 317 | WciF of S. pneumoniae 10F | CAI33109 | 95 (317) | GalNAc transferase |
| WciF of S. pneumoniae 10A | CAI33052 | 81 (317) | |||
| WefD of S. gordonii 38 | AAN64567 | 46 (315) | |||
| WefD of S. oralis 34 | BAD22623 | 45 (311) | |||
| wzx | 471 | Wzx of S. oralis 10557 | BAF44340 | 97 (471) | Flippase |
| Wzx of S. pneumoniae 10F | CAI33110 | 95 (470) | |||
| Wzx of S. pneumoniae 10A | CAI33054 | 88 (470) | |||
| wefK | 332 | WciG of S. pneumoniae 10F | CAI33111 | 96 (332) | Acetyltransferase |
| WefK of S. oralis 10557 | BAF44341 | 94 (332) | |||
| glf | 367 | Glf of S. pneumoniae 10A | CAI33055 | 96 (367) | UDP-Galf mutase |
| Glf of S. gordonii 38 | AAN64570 | 87 (367) | |||
| wefE | 350 | WcrH of S. pneumoniae 10F | CAI33113 | 95 (350) | Galf transferase |
| WefE of S. oralis 34 | BAD22627 | 83 (350) | |||
| WefE of S. gordonii 38 | AAN64571 | 81 (349) |
Initial steps in the synthesis of type 4Gn and 5Gn RPS.
Most of the genes for transferases in the rps locus of strain C104 (and SK144) were identified as homologues of genes in the cps loci of S. pneumoniae serotypes 10A and/or 10F (Table 3). These included wcjG, which was predicted to encode the membrane-associated transferase that initiates CPS biosynthesis (1, 5). Replacement of the wcjG homologue in strain C104 with a nonpolar erm cassette abolished cell surface RPS production, as shown by the failure of mutant strain YC3 to react in dot immunoblotting (Fig. 4). Cell surface RPS production was restored by transformation of this mutant with pJY-1, harboring wcjG for the initial transferase of S. pneumoniae serotype 10A. We also tested wbaP, the initial transferase for O-antigen synthesis by S. enterica (Table 3), for its ability to complement the deletion of wcjG in S. oralis YC3 (Fig. 4). To our surprise, the production of RPS was restored by the expression of intact wbaP from pJY-2 or the 3′ region of wbaP for the Gal-1-phosphate transferase domain (32) from pJY-3. These results strongly suggested that the first step in synthesis of type 4Gn RPS was the WcjG-mediated transfer of Galp-1-phosphate to a carrier lipid.
FIG. 4.
Identification of wcjG and wciB as the first two genes for transferases in the rps locus of S. oralis C104 by plasmid-based genetic complementation. Partial ORF diagrams depict chromosomal genes of S. oralis C104 (white) or erm (red) and plasmids expressing genes from S. pneumoniae serotype 10A (blue) or Salmonella enterica Typhi Ty2 (yellow). Plasmid pJY-3 harbored the coding sequence for the galactosyl-1-phosphate transferase domain (GT) of WbaP. Dot immunoblotting of streptococci was performed with anti-type 4Gn/5Gn RPS IgG as the primary antibody.
The gene for the second transferase in the rps locus of strains C104 (and SK144) was identified as a homologue of S. pneumoniae wciB (Table 3). The occurrence of this gene in S. pneumoniae was recently associated with the presence of β1-3-linked Galf as the second sugar in a number of CPS repeating units (1), and indeed, β1-3-linked Galf also occurs as the second sugar in the proposed biosynthetic repeating unit of type 4Gn RPS (Fig. 3). Results from studies of genetic complementation provided further support for the identification of wciB in S. oralis C104. Thus, cell surface RPS production was abolished by replacing the wciB homologue in S. oralis C104 with the erm cassette and restored in the resulting mutant (strain YC-4) by plasmid-based expression of wciB from S. pneumoniae serotype 10A CPS (Fig. 4).
The gene wefL for the third transferase in the rps locus of strain C104 (and SK144) was found to encode a LicD protein that we propose transfers ribitol-5-phosphate to Galf, forming ribitol-5-PO4-6Galf in types 4Gn and 5Gn RPS (Fig. 3). The sequence of WefL is 85% identical to that of WhaI of S. pneumoniae serotype 47F, the structure of which is unknown, and 29% identical to that of WcrB of S. pneumoniae serotype 10A (Table 3). The proposed role of WcrB in synthesis of CPS 10A involves the transfer of ribitol-5-phosphate to Galf, forming ribitol-5-PO4-5Galf (1). Thus, WcrB of S. pneumoniae and WefL of S. oralis are predicted to link ribitol-5-phosphate to different hydroxyl groups of Galf.
Molecular basis of RPS serotype.
The relatively low homology noted between wefM in strain C104 and its counterpart in strain SK144 (Fig. 3) suggested that these ORFs might represent different genes. Although 75% identical, the proteins encoded by these genes were only 50% identical over their N-terminal regions compared to 90% over their C-terminal regions. In contrast, the encoded protein of strain SK144 exhibited uniformly high homology (i.e., 88% identity) with WcrC of S. pneumoniae, which in a recent study (1) was proposed to catalyze the α1-2 transfer of Gal to ribitol-5-phosphate in synthesis of serotype 10A CPS. To establish the relationship between these genes in different species, we compared the wefM homologue of S. oralis SK144 (i.e., wcrC) and wcrC of S. pneumoniae 8334 (serotype 10A CPS) for their abilities to complement RPS production in a wefM mutant of S. oralis C104 (i.e., S. oralis YC1). The expression of these genes, either from the chromosome of S. oralis YC2 or from plasmid pJY-9 in strain YC1, restored cell surface RPS production, as shown by results of dot immunolabeling (Fig. 5). In these determinations, strains YC2 and YC1(pJY-9) were immunolabeled following incubation with anti-type 4Gn/5Gn RPS-specific IgG but not following incubation with anti-type 4Gn RPS-specific IgG (Fig. 5), thereby suggesting the production of type 5Gn RPS by these strains. This was confirmed by comparing the 1H and 13C NMR spectra of type 5Gn RPS from strain SK144 (Table 2) with those of polysaccharides isolated from strains YC2 and YC1(pJY-9) (results not shown). The spectra of these three polysaccharides were indistinguishable from each other within the estimated experimental error of ±0.02 ppm in the 13C dimension and ±0.005 in the 1H dimension.
FIG. 5.
Conversion of type 4Gn RPS to 5Gn RPS by replacement of wefM in S. oralis C104 with wcrC from S. oralis SK144 or CPS 10A S. pneumoniae. Partial ORF diagrams of each strain indicate the presence of genes from S. oralis C104 (white), S. oralis SK144 (black), S. pneumoniae CPS 10A (blue), or erm (red). Dot immunoblotting of streptococci was performed with different primary RPS-specific IgGs, one that reacted with both 4Gn and 5Gn RPS and the other that was type 4Gn RPS specific. The reactions of S. oralis YC2 and S. oralis YC1(pJY-9) with the former but not the latter antibody suggested the production of type 5Gn RPS by these strains, which was confirmed by NMR.
To extend these findings to other strains, we sequenced the region between wefL and wefD in six additional type 4Gn RPS-producing strains and two additional type 5Gn RPS-producing strains (Table 1). Six distinct nucleotide sequences (GenBank accession numbers EU559293 to EU559298) were identified from the strains that produced type 4Gn RPS, one that was identical to wefM of strain C104 and five that were at least 95% identical. The two additional type 5Gn RPS-producing strains that were examined contained identical copies of wcrC (GenBank accession numbers EU559299 and EU559300) which differed from wcrC of strain SK144 by a single base pair that had no effect on the amino acid sequence. The perfect correlation noted between RPS serotype and the presence of wefM or wcrC in these bacteria further established these genes as markers of type 4Gn and 5Gn RPS production.
Molecular basis of RPS receptor type.
The gene for the fifth transferase in the rps locus of S. oralis C104 (i.e., wefD), was found to share greater homology with wciF of S. pneumoniae serogroup 10 than with wefD of S. gordonii 38 or S. oralis 34 (Table 3). Previously, we showed that S. gordonii GC14, the mutant obtained by replacing wefD of S. gordonii 38 with the erm cassette, produced a type 2Gn-like polysaccharide that lacked β-GalNAc (35). In the present study, we found that erm replacement of the wefD/wciF homologue in S. oralis C104 had a similar effect on the structure of type 4Gn RPS. Thus, high resolution NMR of the type 4Gn-like polysaccharide isolated from mutant strain YC5 revealed four anomeric resonances rather than five, as in type 4Gn RPS (Table 2). The complete assignment of the 1H and 13C chemical shifts of the polysaccharide from strain YC5 (Table 2), determined using coherence transfer and NOESY data, is described in the supplemental material. Analysis of these data (see Fig. S5 to S9 in the supplemental material), including the sugar linkage data, which is summarized in Fig. 2B, led to the conclusion that the structural difference between the modified RPS of strain YC5 (Fig. 2B) and type 4Gn RPS (3) involved the absence of β- GalNAc (residue D) in the former polysaccharide and the presence of a β1-6 linkage between β-Galf (residue C) and α-Galp (residue E).
The cell surface phenotypes of wefD-deficient strain GC14 and wefD/wciF-deficient strain YC5 were also comparable. Each exhibited reduced RPS-specific immunoreactivity compared with that of the wild type in dot immunoblotting (Fig. 5), and each failed to coaggregate with A. naeslundii 12104. However, despite these similarities, it was unclear whether the encoded GalNAc transferases of these strains had the same acceptor specificity, as WefD of S. gordonii 38 acted on terminal Galα-1-PO4−, whereas the enzyme of S. oralis C104 acted on terminal Galα1-1ribitol-5- PO4−. Consequently, we tested the corresponding genes for their abilities to complement heterologous RPS production (Fig. 6). Plasmid-based expression of the wefD/wciF homologue from strain C104 in S. gordonii GC14 restored RPS production, as did the expression of wefD from strain 38 in S. oralis YC5 (Fig. 6). Thus, these genes, although not closely related (Table 3), appeared to be functionally equivalent. In view of this, we designated the wefD/wciF homologue of S. oralis C104 (and SK144) as wefD rather than wciF to maintain continuity in our studies and because the former gene was described first.
FIG. 6.
Identification of wefD and wefE in the rps locus of S. oralis C104 by plasmid-based genetic complementation. Partial ORF diagrams depict chromosomal genes of S. oralis C104 (white), S. gordonii 38 (green), or erm (red) and plasmids expressing either wefD or wefE from these strains. Dot immunoblotting of streptococci was performed with anti-type 2Gn or anti-type 4Gn/5Gn RPS IgG as the primary antibody.
The replacement of wefE in S. gordonii 38 and the wefE/wcrH homologue (Table 3) in S. oralis C104 with erm abolished cell surface RPS production in these strains, as shown by dot immunoblotting of the resulting mutants, S. gordonii XC3 or S. oralis YC6 (Fig. 6). RPS-specific immunoreactivity was restored in these mutants by plasmid-based expression of the wefE/wcrH homologue of S. oralis C104 in S. gordonii XC3 or wefE of S. gordonii 38 in S. oralis YC6, thereby indicating that these genes are complementary.
DISCUSSION
The present findings extend comparative molecular studies of RPS structure to the ribitol-5-phosphate-containing polysaccharides found on certain strains of S. oralis that coaggregate with A. naeslundii and other members of the oral biofilm community. These polysaccharides are of interest not only as receptors for biofilm formation but also because their structures resemble those of the CPS of S. pneumoniae serotypes 10A (16) and 10F (30). The similarities noted between these types of RPS and CPS are even more striking at the molecular level. Thus, the corresponding rps and cps loci of these streptococci share synteny and high homology between a number of individual genes (Table 3). Of the 15 genes identified in the rps loci of S. oralis C104 and SK144 (Fig. 3), 12 (i.e., wzg, wzh, wzd, wze, wcjG, wciB, wcrC, wefD/wciF, wzx, wefK/wciG, glf, and wefE/wcrH) occur in the cps loci of serogroup 10 of S. pneumoniae; the remaining three (i.e., wzy, wefL, and wefM) are alleles of genes found in these or other CPS serotypes. In addition, remnants of two other genes (i.e., wcrD and wcrG) identified in S. pneumoniae serotype 10A (1, 5) appear to be present in S. oralis C104 and SK144, where they occur in the intragenic regions that flank wefD (Fig. 3). These findings clearly point to a common evolutionary history for these polysaccharides.
We expected from earlier studies (37) that the last step in synthesis of the type 4Gn or 5Gn RPS repeating unit was the transfer of Galf to GalNAcβ1-3Gal. Consequently, we anticipated that the first step would involve the transfer of Gal-1-phosphate to a carrier lipid, as implied by the model shown in Fig. 3. Our identification of WcjG as the initial transferase is consistent with this proposal. Thus, the predicted activity of WcjG in S. pneumoniae involves the transfer of Galp rather than Galf in synthesis of 10A and 10F CPS (1). Homology also was noted between WcjG and two other Gal-1-phosphate transferases, EpsE of S. thermophilus Sfi6 (28) and WbaP of S. enterica (Table 3). The finding that intact wbaP or the 3′ region for the Gal-1-phosphate transferase domain of WbaP (32) complemented RPS production in a wcjG mutant of S. oralis provided experimental evidence for the similar WcjG-mediated transfer of Galp-1-phosphate as the first step in RPS biosynthesis.
Type 4Gn RPS of S. oralis C104 and type 5Gn RPS of strain SK144 were previously distinguished by an antigenic difference noted in immunodiffusion studies (8). NMR spectra recorded for type 5Gn RPS at that time suggested that this polysaccharide and previously characterized type 4Gn RPS (3) were identical, except for the linkage between Gal and ribitol-5-phosphate. The more refined NMR techniques and analyses used in the present study confirmed this suggestion but indicated that the linkage between Gal and ribitol-5-phosphate in type 5Gn RPS was α1-2 rather than α1-3, as previously suggested (8). We also established the molecular basis of the structural difference between these polysaccharides in the present study by genetic complementation of a wefM mutant of S. oralis C104 with wcrC from S. oralis SK144 or S. pneumoniae serotype 10A (Fig. 5). The expression of wcrC rather than wefM in strain C104 resulted in the production of type 5Gn RPS, thereby associating wcrC with the α1-2 linkage between Gal and ribitol-5-phosphate in type 5Gn RPS and 10A CPS (1) and wefM with the α1-1 linkage between the same residues in type 4Gn RPS (Fig. 3).
The 75% sequence identity noted between WefM of strain C104 and WcrC of strain SK144 (Fig. 3) is not distributed evenly. Instead, the N-terminal regions of these proteins are approximately 50% identical, whereas the C-terminal regions are more than 90% identical. WefM and WcrC are members of CAZy (a carbohydrate-active enzyme database) glycosyltransferase family 4 and thus are expected to have similar GT-B folds consisting of two Rossman-like β/α/β domains (10). Structural studies of different CAZy family 4 members, including recently characterized MshA of Corynebacterium glutamicum (31) have identified the binding site for nucleotide sugar donors in the C-terminal Rossman-like domains of these proteins and shown that donor binding induces a rotational reorientation of the two domains, creating the acceptor binding site, primarily from features in the N-terminal domain. Thus, the homology seen between WefM and WcrC may reflect similar donor binding sites for UDP-Gal in the C-terminal regions of these proteins and different acceptor binding sites for ribitol-5-phosphate in the N-terminal regions. This insight and the availability of multiple wefM and wcrC sequences provide a much improved basis for distinguishing these genes in strains of S. oralis, S. pneumoniae, and other species.
We suspect that wefE of S. oralis C104 (Fig. 3) and wcrH of S. pneumoniae serotype 10F (1) are the same gene, as both encode similar proteins (Table 3) that appear to catalyze the β1-6 transfer of Galf. However, based on the available structure of 10F CPS (30), WcrH was predicted to act on the α-Gal moiety in 4GalNAcβ1-3Galα1, creating a Galf branch at this site (1), whereas our findings indicate that WefE transfers Galf to the β-GalNAc moiety of GalNAcβ1-3Galα1 (Fig. 3). At present, it is unclear whether these findings indicate a subtle difference in the acceptor specificities of WefE and WcrH or alternatively, an error in the available structure of 10F CPS. To distinguish between these alternatives, we are currently determining the structure of this polysaccharide by NMR and are comparing wcrH of S. pneumoniae and wefE of S. oralis for genetic complementation of type 4Gn RPS production. We expect that the results will precisely define the structural and molecular differences that exist between these types of CPS and RPS and provide insight into the role of β1-6-linked Galf in exposing adjacent GalNAcβ1-3Gal for adhesin-mediated recognition.
We recently suggested (37) that the evolution of recognition motifs in different types of RPS may depend on the selective advantage gained from adhesin-mediated recognition of RPS-bearing streptococci by other commensal species, leading to the establishment of mutualism in biofilm communities. Other recent results (17) suggest that the commensal species Streptococcus mitis, which is closely related to S. oralis, evolved from pneumococcus-like pathogens through the loss of genes for essential virulence factors, including capsule production. In view of this proposal, the molecular similarities noted between different types of RPS and CPS in the present study, as well as those noted between Glc- and Rha-containing types of RPS and serotype 21 CPS (20), suggest the evolution of coaggregation receptors on modern-day commensal species from the antiphagocytic capsules of ancestral pathogens. Further comparative molecular, structural, and functional studies of RPS and CPS are under way to examine this intriguing possibility.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Program of the NIH, NIDCR.
We thank Daron Freedberg of the Center for Biologics Evaluation and Research of the FDA, Bethesda, MD, for facilities and assistance with 31P NMR experiments.
Footnotes
Published ahead of print on 16 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aanensen, D. M., A. Mavroidi, S. D. Bentley, P. R. Reeves, and B. G. Spratt. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 1897856-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abeygunawardana, C., and C. A. Bush. 1993. Determination of the chemical structure of complex polysaccharides by heteronuclear NMR spectroscopy. Advan. Biophysical Chem. 3199-249. [Google Scholar]
- 3.Abeygunawardana, C., C. A. Bush, and J. O. Cisar. 1991. Complete structure of the cell surface polysaccharide of Streptococcus oralis C104: a 600-MHz NMR study. Biochemistry 308568-8577. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cisar, J. O., A. L. Sandberg, C. Abeygunawardana, G. P. Reddy, and C. A. Bush. 1995. Lectin recognition of host-like saccharide motifs in streptococcal cell wall polysaccharides. Glycobiology 5655-662. [DOI] [PubMed] [Google Scholar]
- 8.Cisar, J. O., A. L. Sandberg, G. P. Reddy, C. Abeygunawardana, and C. A. Bush. 1997. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect. Immun. 655035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cisar, J. O., A. E. Vatter, W. B. Clark, S. H. Curl, S. Hurst-Calderone, and A. L. Sandberg. 1988. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 562984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328307-317. [DOI] [PubMed] [Google Scholar]
- 11.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 1471383-1391. [DOI] [PubMed] [Google Scholar]
- 12.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 7761-68. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 436073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans streptococcal species. Microb. Ecol. Health Dis. 7125-137. [Google Scholar]
- 15.Jiang, S. M., L. Wang, and P. R. Reeves. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 691244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, C. 1995. Full assignment of the NMR spectrum of the capsular polysaccharide from Streptococcus pneumoniae serotype 10A. Carbohydr. Res. 269175-181. [DOI] [PubMed] [Google Scholar]
- 17.Kilian, M., K. Poulsen, T. Blomqvist, L. S. Havarstein, M. Bek-Thomsen, H. Tettelin, and U. B. Sorensen. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS ONE 3e2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 1815004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain wicky. J. Bacteriol. 1785831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavroidi, A., D. M. Aanensen, D. Godoy, I. C. Skovsted, M. S. Kaltoft, P. R. Reeves, S. D. Bentley, and B. G. Spratt. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 1897841-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntire, F. C., L. K. Crosby, A. E. Vatter, J. O. Cisar, M. R. McNeil, C. A. Bush, S. S. Tjoa, and P. V. Fennessey. 1988. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 1702229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntire, F. C., A. E. Vatter, J. Baros, and J. Arnold. 1978. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect. Immun. 21978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95369-380. [DOI] [PubMed] [Google Scholar]
- 24.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 1884117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 1853400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 695794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stingele, F., J. W. Newell, and J. R. Neeser. 1999. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J. Bacteriol. 1816354-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120105-110. [DOI] [PubMed] [Google Scholar]
- 30.van Dam, J. E., A. Fleer, and H. Snippe. 1990. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie van Leeuwenhoek 581-47. [DOI] [PubMed] [Google Scholar]
- 31.Vetting, M. W., P. A. Frantom, and J. S. Blanchard. 2008. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis. J. Biol. Chem. 28315834-15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L., D. Liu, and P. R. Reeves. 1996. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J. Bacteriol. 1782598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, D. Q., J. Thompson, and J. O. Cisar. 2003. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J. Bacteriol. 1855419-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, Q., and C. A. Bush. 1996. Molecular modeling of the flexible cell wall polysaccharide of Streptococcus mitis J22 on the basis of heteronuclear NMR coupling constants. Biochemistry 3514521-14529. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida, Y., S. Ganguly, C. A. Bush, and J. O. Cisar. 2005. Carbohydrate engineering of the recognition motifs in streptococcal coaggregation receptor polysaccharides. Mol. Microbiol. 58244-256. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, Y., S. Ganguly, C. A. Bush, and J. O. Cisar. 2006. Molecular basis of l-rhamnose branch formation in streptococcal coaggregation receptor polysaccharides. J. Bacteriol. 1884125-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, Y., J. Yang, P. E. Peaker, H. Kato, C. A. Bush, and J. O. Cisar. 2008. Molecular and antigenic characterization of a Streptococcus oralis coaggregation receptor polysaccharide by carbohydrate engineering in Streptococcus gordonii. J. Biol. Chem. 28312654-12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







