Abstract
Expression of the multidrug efflux pump ttgDEF and ttgGHI operons is modulated in vivo mainly by the TtgV repressor. TtgV is a multidrug recognition repressor that exhibits a DNA binding domain with a long interaction helix comprising residues 47 to 64. The pattern of expression of the two pumps is different in Pseudomonas putida: in the absence of effectors, the promoter for the ttgD gene is silent, whereas the ttgG gene is expressed at a high basal level. This correlates with the fact that TtgV exhibits a higher affinity for the ttgD operator (KD = 10 ± 1 nM) than for the ttgG (KD = 19 ± 1 nM) operator. Sequence analysis revealed that both operators are 40% identical, and mutational analysis of the ttgD and ttgG operators combined with electrophoretic mobility shift assays and in vivo expression analysis suggests that TtgV recognizes an inverted repeat with a high degree of palindromicity around the central axis. We generated a collection of alanine substitution mutants with substitutions between residues 47 and 64 of TtgV. The results of extensive combinations of promoter variants with these TtgV alanine substitution mutants revealed that TtgV modulates expression from ttgD and ttgG promoters through the recognition of both common and different sequences in the two promoters. In this regard, we found that TtgV mutants at residues 48, 50, 53, 54, 60, and 61 failed to bind ttgG but recognized the ttgD operator. TtgV residues R47, R52, L57, and T49 are critical for binding to both operators. Based on three-dimensional models, we propose that these residues contact nucleotides within the major groove of DNA.
Multidrug efflux pumps are widely distributed among prokaryotic and eukaryotic organisms and are responsible for resistance to a number of antibiotics, flavonoids, organic solvents, superoxide-generating agents, dyes, anticancer compounds, and other drugs (2, 14, 15, 16, 24).
The Pseudomonas putida DOT-T1E strain (Table 1) has the extraordinary capacity to withstand and even grow in the presence of high concentrations of organic solvents such as toluene, styrene, and xylenes (19). The toxicity of these aromatic hydrocarbons derives from their preferential partitioning in the cell membrane, which leads to collapse of the cell membrane potential and eventually causes cell death. The main mechanism underlying solvent resistance in this and other gram-negative bacteria lies in the action of RND (resistance-nodulation-cell division) efflux pumps (17, 18). In the DOT-T1E strain, three of these pumps, called TtgABC, TtgDEF, and TtgGHI (13, 18, 22), are involved in the concerted extrusion of organic solvents, although the TtgGHI pump is chiefly responsible, from a quantitative point of view, for the extrusion of toluene and other solvents from the cell membranes (22, 23).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. putida strains | ||
| DOT-T1E | Rifr | 17 |
| DOT-T1EVT | Rifr Kmr TelrttgV::Km ttgT::kilA/telAB | 27 |
| E. coli strains | ||
| BL21(DE3) | Carries T7 RNA polymerase under the control of the lacUV5 promoter | Novagen |
| DH5α | supE44 Δ(lacZYA-argF) U169 deoR (φ80lacZΔM15) hsdR17(rK− mK−) recA1 endA1 gyrA (Nalr) thi-1 relA1 Δ(lacIZYΔ-argF) | 28 |
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14 (McrA−) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17(rK mK) relA1 supE44 recA1 | 29 |
| Plasmids | ||
| pET28b(+) | Kmr; protein expression vector | Novagen |
| pANA126 | Kmr; pET28b(+) derivative vector used to produce His6-TtgV | 22 |
| pMP220 | Tcr; promoterless lacZ expression vector | 26 |
| pMPD1 | Tcr; ttgD::′lacZ fusion cloned in pMP220 | 27 |
| pANA96 | Tcr; ttgG::′lacZ fusion cloned in pMP220 | 22 |
| pGEM-T | Apr; cloning vector | Promega |
| pMBL-T | Apr; cloning vector | Promega |
| pGG1 | Apr; pUC18 bearing an 8-kb BamHI fragment with ttgGHI and ttgVW | 22 |
| pT1-B6 | Apr; pUC18 bearing an 6.8-kb BamHI fragment with ttgDEF and ttgT | 22 |
| pBBRN | pBBR-MCS5 derivative with an additional NdeI restriction site | This work |
| pBBRN::ttgV | Gmr; pBBRN derivative vector expressing TtgV | This work |
| pBBRN::R47A | Gmr; pBBRN derivative vector expressing TtgV(R47A) | This work |
| pBBRN::V50A | Gmr; pBBRN derivative vector expressing TtgV(V50A) | This work |
| pBBRN::Q51A | Gmr; pBBRN derivative vector expressing TtgV(Q51A) | This work |
| pBBRN::E60A | Gmr; pBBRN derivative vector expressing TtgV(E60A) | This work |
| pET28b(+)::R47A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(R47A) | This work |
| pET28b(+)::S48A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(S48A) | This work |
| pET28b(+)::T49A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(T49A) | This work |
| pET28b(+)::V50A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(V50A) | This work |
| pET28b(+)::Q51A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(Q51A) | This work |
| pET28b(+)::R52A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(R52A) | This work |
| pET28b(+)::I53A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(I53A) | This work |
| pET28b(+)::I54A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(I54A) | This work |
| pET28b(+)::N55A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(N55A) | This work |
| pET28b(+)::L57A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(L57A) | This work |
| pET28b(+)::E58A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(E58A) | This work |
| pET28b(+)::E59A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(E59A) | This work |
| pET28b(+)::E60A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(E60A) | This work |
| pET28b(+)::F61A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(F61A) | This work |
| pET28b(+)::L62A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(L62A) | This work |
| pET28b(+)::V63A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(V63A) | This work |
| pET28b(+)::E64A | Kmr; pET28b(+) derivative vector used to produce His6-TtgV(E64A) | This work |
| pMBLT::pDEF | Apr; pMBLT derivative plasmid bearing the ttgD promoter | This work |
| pMBLT::pDEF box 1 | Apr; pMBLT derivative plasmid bearing a mutant ttgD promoter in box 1 | This work |
| pGEMT::pDEF box 3 | Apr; pGEMT derivative plasmid bearing a mutant ttgD promoter in box 3 | This work |
| pMBLT::pDEF box 4 | Apr; pMBLT derivative plasmid bearing a mutant ttgD promoter in box 4 | This work |
| pGEMT::pDEF box 5 | Apr; pGEMT derivative plasmid bearing a mutant ttgD promoter in box 5 | This work |
| pMBLT::pGHI | Apr; pMBLT derivative plasmid bearing the ttgG promoter | This work |
| pGEMT::pGHI box 1 | Apr; pGEMT derivative plasmid bearing a mutant ttgG promoter in box 1 | This work |
| pMBLT::pGHI box 3 | Apr; pMBLT derivative plasmid bearing a mutant ttgG promoter in box 3 | This work |
| pGEMT::pGHI box 4 | Apr; pGEMT derivative plasmid bearing a mutant ttgG promoter in box 4 | This work |
| pGEMT::pGHI box 5 | Apr; pGEMT derivative plasmid bearing a mutant ttgG promoter in box 5 | This work |
Apr, Gmr, Kmr, Rifr, Tcr, and Telr stand for resistance to ampicillin, gentamicin, kanamycin, rifampin, tetracycline, and tellurite, respectively.
Control of the expression of the ttgABC operon is mediated by TtgR, a member of the TetR family of regulators (1, 20), whereas expression of ttgDEF and ttgGHI is mediated mainly by the TtgV repressor, a member of the IclR family of regulators (12). Expression of the last two efflux pumps is also under the side control of TtgT, another repressor that exhibits 63% identity to TtgV (27); we found that in vivo the ttgV gene was expressed at a higher level than the ttgT gene and that the TtgV protein exhibited a higher affinity than TtgT for its operators. Consequently, the TtgV protein turned out to be the main regulator of in vivo expression of the ttgDEF and ttgGHI efflux pumps (27). In a ttgV-deficient background, the role of TtgT as a repressor of the ttgD and ttgG promoters became measurable, as the expression of these promoters was still modulated in response to effectors. This constituted the first example of cross-regulation of two RND efflux pumps involved in the defense against toxic compounds (27).
Due to the dominant role in vivo of TtgV, its interaction with the ttgG promoter has been documented more widely than that of TtgT. Footprint assays revealed that TtgV protected a 42-bp region that covers the −10/−35 regions of the ttgG promoter and the −10 region of the divergently oriented ttgV promoter (5, 23). Isothermal titration calorimetry analyses showed that TtgV recognition specificity is restricted within the ttgG operator to a 34-nucleotide stretch, and it was proposed that TtgV recognized intercalated inverted repeats that share no significant DNA sequence similarities (4) (Fig. 1). In addition, atomic force microscopy studies of TtgV-ttgG operator complexes showed that TtgV induced a 57° convex bend in the DNA (4). It was therefore proposed that the mechanism of TtgV repression was based on steric occlusion of the RNA polymerase binding site, reinforced by DNA bending of the ttgV-ttgG promoter region. Early studies also showed that TtgV exhibits multidrug effector specificity and recognizes mono- and biaromatic compounds. The most efficient in vivo inducers of TtgV were 1-naphthol, 2,3-dihydroxynaphthalene, and indole (3, 5). Stimulation of transcription from the ttgD, ttgG, and ttgV promoters occurs through a derepression mechanism such that in the absence of effectors, TtgV is bound to the target operators and represses transcription. Upon effector binding, it dissociates from the target DNA, and RNA polymerase subsequently transcribes the ttgV, ttgG, and ttgD promoters (3, 27).
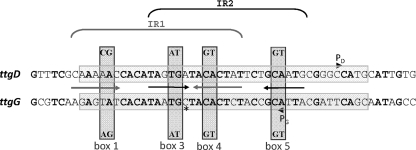
FIG. 1.
Alignment of the ttgD and ttgG operators recognized by TtgV. The transcription initiation points, marked PG and PD, were determined in vivo by primer extension (3). The sequences shadowed in gray correspond to protected regions in the footprint. The transcription start points are indicated by arrowheads above (ttgD) and below (ttgG) the DNA sequence. The indicated overlapping inverted repeats (IR) were proposed by Guazzaroni et al. (4), based on EMSAs and isothermal titration calorimetry assays using variants of the ttgG operator and the TtgV protein. The dinucleotides marked boxes 1 and 4 and boxes 3 and 5 were called IR1 and IR2, respectively. The same nomenclature is used in this study to refer to the corresponding aligned sequences in ttgD. The dinucleotides above the ttgD sequence and below the ttgG sequence indicate the changes introduced to create mutants in the marked boxes.
This study was undertaken to better understand ttgD repression by TtgV. Initially, we expected that TtgV would recognize similar sequences in both the ttgD and ttgG operators; however, the operators share only 40% identity, as deduced from the alignment of the regions protected by TtgV in both operators (Fig. 1). It was surprising that the lowest identity was at the 5′-terminal end zone (Fig. 1, box 1), a segment that was proposed to be critical for the recognition of the ttgG promoter by TtgV (4) (Fig. 1). We then hypothesized that TtgV might recognize ttgD and ttgG differentially depending on the precise composition of the binding site. Our results support the hypothesis that TtgV modulates expression from ttgD and ttgG through the recognition of common and distinct sequences in both operators and that the importance of some residues of the TtgV DNA recognition helix also differs depending on the promoter under scrutiny.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture medium.
The bacterial strains and plasmids used in this study are shown in Table 1. These strains were routinely grown in liquid Luria-Bertani (LB) medium at 30°C (P. putida) or 37°C (Escherichia coli) with shaking on an orbital platform operating at 200 rpm. Escherichia coli DH5α and E. coli JM109 were used as host strains to construct and maintain different plasmids. Escherichia coli BL21(DE3) was used to overproduce TtgV and its mutant variants. When required, the following antibiotics were added to the cultures to the following final concentrations: 100 μg/ml ampicillin, 10 μg/ml gentamicin, 50 μg/ml kanamycin, 20 μg/ml rifampin, and 20 μg/ml tetracycline. K2TeO3 (30 μg/ml) was also used to select some of the mutants.
Construction of TtgV mutants.
TtgV mutants in which amino acid residues at positions 47 to 64 were replaced by alanine were generated by overlapping PCR mutagenesis, using the pANA126 plasmid (22) as a source of the ttgV wild-type allele. For each mutant, three PCRs were carried out. The first two PCR runs involved amplifications using an upstream primer (corresponding to the ttgV coding sequence), a mismatched primer that included the segment to be mutated as well as a PCR amplification using the downstream primer, and an oligonucleotide complementary to the mismatched primer. The resulting overlapping PCR products were annealed, supplemented with upstream and downstream primers, and subjected to the third PCR. The final PCR product was cloned into the pMBL-T or pGEM-T vector, which was subsequently digested with NdeI and BamHI enzymes. The digestion product was cloned into the pET28b(+) vector (Novagen).
Mutant ttgD and ttgG promoters were generated by overlapping PCR mutagenesis, using plasmids pT1-B6 and pGG1, respectively, as templates (13, 22). We used 38-bp primers mutated in each box (1 to 5) for amplification. The PCR products were cloned into pGEM-T or pMBL-T. In all cases, the introduction of site-specific changes was confirmed by DNA sequencing.
β-Galactosidase assays.
Cultures were inoculated with bacterial cells from fresh LB agar plates supplemented with the appropriate antibiotics and grown overnight at 30°C on LB medium with appropriate antibiotics. Cultures were diluted to an initial optical density at 660 nm of 0.05 in the same medium supplemented or not with 1-naphthol (1 mM) dissolved in dimethyl sulfoxide (note that the latter did not interfere with the induction assays in this study). β-Galactosidase activity was determined in triplicate for permeabilized cells when cultures reached a turbidity at 660 nm of 0.5 (11). The results are reported as the means for nine different experiments.
Overexpression and purification of His-tagged TtgV and mutants.
The pANA126 plasmid is a pET28b(+) derivative that was transformed into E. coli BL21(DE3) and used to overproduce a His6-TtgV-tagged protein. ttgV mutant alleles were cloned into the same plasmid, which was then used to overexpress ttgV mutant variants.
Electrophoretic mobility shift assays (EMSAs).
The DNA probes were 295-bp fragments containing the ttgT-ttgDEF and ttgV-ttgGHI intergenic regions obtained from plasmid DNA (pT1-B6 or derivatives and pGG1 or derivatives, respectivley) by PCR with primers D5′E (5′-NNNNNNGAATTCCCTTCTGATCCAGGCCACCG-3′) and D3′P (5′-NNNNNNCTGCAGTAACTGTCTCGCACGCAAAG-3′) and with primers G5′E (5′-NNNNNNGAATTCGTTCATATCTTTCCTCTGCG-3′) and G3′P (5′-NNNNNNCTGCAGGGGGATTACCCGTAATGCAC-3′), respectively.
Cycling parameters were 2 min at 95°C followed by 30 cycles at 95°C for 1 min, 50°C for 30 s, and 72°C for 30 s, ending with 10 min at 72°C. PCR products were isolated from agarose gel by use of a Qiaquick gel extraction kit (Qiagen) and radiolabeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. A 1 nM concentration (∼104 cpm) of the labeled probe was then incubated with the indicated concentrations of purified proteins in 10 μl STAD (25 mM Tris-acetate, pH 8.0, 10 mM KCl, 8 mM magnesium acetate, 3.5% [wt/vol] polyethylene glycol 8000, and 1 mM dithiothreiol) supplemented with 15 μg/ml poly(dI-dC) and 200 μg/ml bovine serum albumin. Reaction mixtures were then incubated for 10 min at 30°C, and samples were run in 4.5% (wt/vol) native polyacrylamide gels (Bio-Rad Mini-Protean II) for 2 h at 50 V at room temperature in Tris-glycine buffer (25 mM Tris-HCl, pH 8.0, 200 mM glycine). The results were analyzed with Personal FX equipment and Quantity One software (Bio-Rad).
RESULTS
Mutational analysis of the extended DNA recognition helix of TtgV: differential effect on operator binding.
Molina-Henares et al. (12) reported that the helix-turn-helix (HTH) DNA binding domain of the IclR family was relatively extended with respect to other regulators. Based on the sequence alignment of 53 IclR family members (10) and α-helix secondary structure predictions, TtgV residues 34 to 43 might form the first helix of the HTH DNA binding motif, whereas residues 47 to 64 might represent the binding helix of this domain (Fig. 2). To determine whether these residues are involved in the binding of TtgV to its operators and to explore whether TtgV interacts with the ttgD and ttgG promoters differentially depending on the precise base composition of the target sites, we mutated all residues between positions 47 and 64 to alanine. All alleles encoding mutant proteins were cloned into pET28b(+), and the proteins were purified as N-terminally His-tagged variants. Homogeneously purified mutant variants were used for EMSAs with similarly sized ttgD and ttgG operator regions and with the wild-type protein as a control (see Fig. 3 and 4 for examples with some of the mutants). Densitometric analysis of the amount of DNA shifted by TtgV revealed that TtgV shifted the ttgD operator better than the ttgG fragment (Fig. 3B). We also tested all mutant proteins in EMSAs with ttgD and ttgG and distinguished three types of mutant proteins. (i) Some TtgV mutants had changes that had no effect on binding to either promoter (Q51A, N55A, E58A, E59A, L62A, V63A, and E64A) (Fig. 3A and Q51A EMSA in Fig. 3B). Further support for this lack of effect was obtained when we determined that the ttgD/ttgG ratio of retarded DNA at a fixed concentration of TtgV (i.e., 50 nM) was close to 1 (Fig. 4). Also identified were (ii) TtgV mutants that did not bind to the ttgG or ttgD operator (R47A, T49A, R52A, and L57A) (Fig. 3A and B, R47A mutant) and (iii) TtgV variants with mutations with a more severe effect on binding to the ttgG operator than to the ttgD operator (S48A, V50A, I53A, I54A, E60A, and F61A) (Fig. 3A and E60A and V50A EMSAs in Fig. 3B). This was further supported by the fact that at a fixed concentration of TtgV mutant variants, the ratio of the amount of ttgD- to ttgG-shifted DNA was >4 (Fig. 4). We found that S48A, V50A, I53A, and I54A mutants were more severely affected in binding to ttgG, with ratios of 5.8- to 8.5-fold with respect to the wild-type TtgV (Fig. 4).
FIG. 2.
Amino acid sequence at the N-terminal end of TtgV, where an extended HTH DNA binding domain is located. The proposed TtgV DNA binding protein includes a position helix (residues 34 to 43) followed by a turn and an extended recognition helix that covers residues 47 to 64. Below is the physical organization of the members of the IclR family, consisting of two domains, one involved in DNA recognition and another that includes a central segment of the proteins and extends toward the C-terminal end. The second domain seems to be involved in effector recognition and multimerization (5, 10). PFAM refers to the HMM algorithm identifying IclR members in that region.
FIG. 3.
TtgV mutant variants grouped according to their binding to the ttgD and ttgG operators. (A) Three groups of mutants were defined according to their ability to bind and retard (+) the ttgD and/or ttgG operator. (B) EMSA of 295-bp DNA fragments incubated in the absence (−) or presence of 10, 50, or 100 nM TtgV (WT) or the indicated mutant variant. F, free DNA; B, retarded DNA.
FIG. 4.
Effects of mutations in the HTH DNA binding domain of TtgV on binding to the ttgG and ttgD operators. EMSAs were carried out as described in Materials and Methods and in the legend for Fig. 3, using a fixed amount of TtgV (50 nM) or its mutants. Densitometric analysis was carried out to determined the amount of protein DNA bound to each operator. The ttgD/ttgG ratio shown at the top distinguishes mutant variants that bind and retard both operators or one preferentially over the other.
To quantify these effects, we carried out EMSA with a wide range of wild-type and mutant regulator concentrations and a 295-bp ttgG or ttgD promoter fragment, which allowed us to determine the apparent dissociation constant (KD). We found that TtgV had an affinity of around 10 ± 1 nM for ttgD (Table 2), whereas the affinity for ttgG was around 19 ± 1 nM (Table 2). We also found that mutants in group 1, e.g., the Q51A mutant, bound ttgD and ttgG DNAs with KD values similar to those of the parental protein and that TtgV mutants in the third group (e.g., V50A, I53A, I54A, E60A, and F61A mutants) exhibited reduced affinities for their target operators, with the reductions being in the range of 2.5- to 4.9-fold in the case of the ttgD promoter and over 35-fold in the case of ttgG.
TABLE 2.
Determination of apparent dissociation constants of wild-type and TtgV mutants for the operators at the ttgG and ttgD promotersa
| TtgV protein variant | KD (nM) for ttgD operator | KD (nM) for ttgG operator |
|---|---|---|
| Wild type | 10 ± 1 | 19 ± 1 |
| R47A | >750 | >750 |
| V50A | 49 ± 11 | >750 |
| Q51A | 8 ± 1 | 19 ± 1 |
| I53A | 42 ± 3 | ND |
| I54A | 27 ± 2 | >750 |
| E60A | 26 ± 1 | >750 |
| F61A | 25 ± 2 | >750 |
Data were obtained from densitometric analyses of EMSAs in the presence of concentrations of 1, 5, 10, 20, 30, 40, 50, 100, 500, and 750 nM. Results shown are the means for at least four independent assays done in duplicate. The DNA probe was 1 nM of a 295-bp fragment comprising the entire ttgG or ttgD promoter region. ND, not determined.
To analyze the physiological relevance of the above mutations in TtgV, we decided to test their effects on expression levels from the ttgD and ttgG promoters in vivo. To this end, we constructed fusions of the promoters to a promoterless ′lacZ gene (PttgG::′lacZ and PttgD::′lacZ). The ttgV gene (or an allele encoding a mutant variant [we chose the Q51A and ES8A mutants from group 1, the R47A mutant from group 2, and the V50A and E60A mutants from group 3]) was cloned into the low-copy-number compatible pBBRN plasmid (see Materials and Methods). Expression from PttgG and PttgD was tested in a ttgV/ttgT-deficient background, and β-galactosidase activity in the absence and presence of 1-naphthtol was determined (Table 3). In this background, we found that expression from the promoter was similar regardless of the presence of 1-naphthol and that the expression from ttgG was higher (1,900 ± 160 Miller units) than that from PttgD (around 600 Miller units). When, in addition, the cells expressed ttgV, we found that basal expression of ttgD (30 ± 5 Miller units) was lower than that of ttgG (750 ± 40 Miller units). As expected, in the presence of 1-naphthol, expression from the two promoters increased dramatically (550 ± 120 Miller units for ttgD and around 2,000 Miller units for ttgG) (Table 3). The V50A and E60A mutants in group 3 were not able to repress the ttgG promoter; consequently, high levels of β-galactosidase were seen regardless of the presence of 1-naphthol (Table 3). However, the behavior of the group 3 mutants with the ttgD promoter was similar to that observed for the wild-type TtgV protein, namely, basal expression seen in the absence of effector molecules increased in the presence of 1-naphthol. This indicates that the mutation at the DNA binding domain did not interfere with effector recognition. The R47A mutant, which belongs to group 2, was not able to repress the ttgD or ttgG promoter, and no significant induction in the presence of 1-naphthol was therefore detected. In contrast, the Q51A and E58A group 1 mutants seemed to be able to bind to both promoters tightly, since basal levels of expression of ttgD and ttgG were lower than those achieved in the presence of TtgV. However, in the presence of 1-naphthol, the Q51A mutant was able to induce the expression of ttgG to maximum levels, but expression from ttgD was fourfold lower than that with the wild-type protein, which suggests that the Q51A mutant may not be as efficient as TtgV in derepressing the ttgD promoter.
TABLE 3.
In vivo effects of TtgV and its mutant variants on the expression of ttgD (pMPD) and ttgG (pMPG) promoters fused to ′lacZa
| Strain | Presence of 1-naphthol | β-Galactosidase expression (Miller units) | Mutant group |
|---|---|---|---|
| TIEVT(pMPD) | − | 600 ± 60 | Not relevant |
| + | 600 ± 100 | Not relevant | |
| TIEVT(pMPG) | − | 1,900 ± 160 | Not relevant |
| + | 2,100 ± 160 | Not relevant | |
| TIEVT(pBBRN::ttgV, pMPD) | − | 30 ± 5 | Not relevant |
| + | 550 ± 120 | Not relevant | |
| TIEVT(pBBRN::ttgV, pMPG) | − | 750 ± 40 | Not relevant |
| + | 1,850 ± 110 | Not relevant | |
| TIEVT(pBBRN-Q51A, pMPD) | − | 10 ± 5 | 1 |
| + | 150 ± 50 | 1 | |
| TIEVT(pBBRN-Q51A, pMPG) | − | 100 ± 20 | 1 |
| + | 2,400 ± 210 | 1 | |
| T1EVT(pBBRN-E58A, pMPD) | − | 10 ± 10 | 1 |
| + | 130 ± 5 | 1 | |
| T1EVT(pBBRN-E58A, pMPG) | − | 30 ± 20 | 1 |
| + | 1,560 ± 80 | 1 | |
| TIEVT(pBBRN-R47A, pMPD) | − | 600 ± 80 | 2 |
| + | 900 ± 30 | 2 | |
| TIEVT(pBBRN-R47A, pMPG) | − | 2,500 ± 200 | 2 |
| + | 2,800 ± 390 | 2 | |
| TIEVT(pBBRN-V50A, pMPD) | − | 50 ± 5 | 3 |
| + | 750 ± 60 | 3 | |
| TIEVT(pBBRN-V50A, pMPG) | − | 2,250 ± 150 | 3 |
| + | 2,600 ± 470 | 3 | |
| TIEVT(pBBRN-E60A, pMPD) | − | 100 ± 10 | 3 |
| + | 800 ± 100 | 3 | |
| TIEVT(pBBRN-E60A, pMPG) | − | 2,200 ± 300 | 3 |
| + | 2,800 ± 580 | 3 |
Pseudomonas putida strains deficient in ttgV and ttgT were transformed with plasmid pMPD1 or pANA96 and pBBRN::ttgV or its mutant derivatives. Cells were grown on LB medium as described in Materials and Methods with rifampin, tetracycline, and kanamycin in the absence (−) and in the presence (+) of 1 mM 1-naphthol. β-Galactosidase activity was assayed in permeabilized cells as described in Materials and Methods.
Identification of sequences in ttgD important for TtgV recognition.
Guazzaroni et al. (4) suggested two overlapping inverted repeats in the ttgG promoter (Fig. 1) as the TtgV target. Their results showed that mutations within the so-called IR1 inverted repeat (boxes 1 and 4) and IR2 inverted repeat (boxes 3 and 5) had a significant effect on recognition by TtgV. These results were corroborated in this study, as we observed that the amount of target DNA retarded by a fixed amount of TtgV decreased with the mutant operators compared to the amount of wild-type DNA (Fig. 5).
FIG. 5.
Effects of nucleotide changes in the operator sequences of ttgD (left) and ttgG (right) on the binding of TtgV. EMSAs were carried out with 1 nM of the indicated wild-type or mutant operator variant (295-bp fragments) and 50 nM TtgV. Other conditions were the same as those described in the legends for Fig. 3 and 4. F and B, free and bound DNAs, respectively.
Based on the alignment of the ttgG and ttgD promoters (Fig. 1), we introduced similar double mutations in the ttgD operator to correspond to the inverted repeats in ttgG, although these sequences did not always correspond to potential inverted repeats in ttgD. We maintained the box 1, box 3, box 4, and box 5 nomenclature for the mutant promoters. We used EMSA to test the role of these boxes in recognition by TtgV and found that mutations in boxes 3 and 4 had a significant effect on TtgV binding to ttgD, since only 56% and 72%, respectively, of the DNA was retarded (Fig. 5), compared to nearly 95% of the DNA bound by the wild-type ttgD sequence (Fig. 5). Therefore, a clear difference between the ttgD and ttgG promoters is that boxes 1 and 5 are irrelevant for TtgV binding at ttgD but are important at ttgG, suggesting that the base composition of the binding sites is relevant.
Amino acids important for ttgG and ttgD operator recognition in the HTH DNA binding motif of TtgV.
We analyzed the effects of mutations in the ttgG and ttgD operators on the binding of the whole collection of TtgV mutants. TtgV mutants in group 2 failed to bind any of the mutant promoters tested (not shown), and this was expected since the mutant proteins did not bind wild-type DNA (Fig. 3B).
TtgV mutants in group 3 (V50A, I53A, I54A, E60A, and E61A mutants) did not bind the ttgG wild-type operator or its mutant variants (Fig. 6B); however, these mutants recognized ttgD operator variants (Fig. 6A). This indicates that TtgV can establish differential contacts with its target DNA between the two operators. Although all group 3 mutants interacted with all ttgD operator variants, it should be noted that in general, the amount of shifted DNA was lower than that retarded by the wild-type protein. This was particularly clear for the V50A and I53A mutants, since only approximately 30% of the total DNA was retarded (Fig. 6A).
FIG. 6.
Effects of nucleotide changes in the operators of ttgD (A) and ttgG (B) on the binding of TtgV and its mutants. EMSAs were carried out with 1 nM of the indicated wild-type or mutant operator variant (295-bp fragments) and 50 nM TtgV. Densitometric analysis were done to determine the amount of shifted DNA with respect to the total DNA. Data are averages for at least three independent assays plus standard errors.
When DNA shifts of the ttgG or ttgD operator mutant variants were assayed with TtgV mutants in group 1, e.g., the Q51A, E58A, and E59A mutants, the patterns of interactions were similar to those found with the wild-type TtgV protein (Fig. 6A and B). We also found that the group 1 L62A, V63A, and E64A mutants also bound to the ttgG and ttgD promoters and their variants. However, although none of these TtgV mutants affected binding to the ttgD box 1 and box 5 variants (the amount of shifted DNA was >90%), binding of these mutant proteins to ttgG box 1 and box 5 variants was weaker (Fig. 6A and B).
This set of results suggests that differential recognition of the promoters is influenced by specific and distinct contacts of the TtgV HTH with the two operators. This set of interactions requires resolution of the three-dimensional (3D) structure of TtgV in complex with both operators, and the nature of this structure is currently under investigation at our laboratories.
DISCUSSION
Multidrug resistance often involves the concerted action of efflux pumps, membrane permeability barriers, drug inactivation, and detoxification via metabolic conversion to less toxic molecules. The high level of tolerance to toluene in Pseudomonas putida DOT-T1E is mediated mainly by three efflux pumps, two of which—TtgGHI and TtgDEF—are under cross-regulation by the IclR family multidrug recognition regulators TtgV and TtgT (10, 12). Of these two repressors, TtgV has been shown to play a dominant role in the transcriptional control of these two pumps in vivo (27).
An earlier observation made by our research group was that the patterns of expression of the ttgDEF and ttgGHI operons are different, in the sense that expression of the ttgDEF operon is silent in the absence of aromatic hydrocarbons, whereas the ttgGHI operon is expressed at a relatively high basal level (13, 22). When an effector molecule is added to the culture medium, both operons are expressed at higher levels, although the maximal level of expression of the ttgGHI operon is superior to that of ttgDEF (27) (Table 3). Why does this marked difference in the expression patterns of the two operons exist if they are under the transcriptional control of the same repressors? The answer to this question is twofold. (i) The physical organizations of the genes encoding the repressors and the efflux pumps are different, and thus the access of RNA polymerase to the promoters may differ. The ttgT gene is adjacent to and transcribed divergently from ttgDEF, and the corresponding promoters are separated by 80 nucleotides (the TtgV binding site is located between positions −41 and −16 with respect to ttgD), whereas not only do the divergent PttgV and PttgG promoters fully overlap with each other, but the TtgV operator lies at the −10/−35 regions of these promoters. (ii) The DNA sequences of the ttgG and ttgD operators show only 40% identity (Fig. 1 and 7). TtgV recognizes the ttgD operator with a higher affinity (twofold) than that for ttgG, which correlates with the lower basal expression from the ttgD promoter. Therefore, in vivo the different expression patterns of the two efflux pump operons seem to result from the combination of the affinity of TtgV for its target operators and the physical organization of these operators.
FIG. 7.
Proposed inverted repeat targets for TtgV, based on mutational analyses of the interactions of PttgG and PttgD with wild-type TtgV and its mutant variants. The conserved sequences in footprints of the ttgG and ttgD promoters are shown. The inverted repeat within the sequences protected in previous footprint assays (3, 27) is based on sequence alignment and our current mutational assays. Nucleotides in the consensus sequence were identical in the alignment, and palindromic nucleotides are shown in bold in the consensus sequence.
Previous work using TtgV and mutant variants of the ttgG operator revealed that TtgV binds to a region spanning about 34 nucleotides. Guazzaroni et al. (4) suggested the existence of four subsites for this interaction, called boxes 1, 3, 4, and 5, which were proposed to be organized as a set of intercalated inverted repeats. Alignment of the ttgG and ttgD operators revealed high sequence conservation at the central boxes 3 and 4, with less conservation at box 5 and large differences at box 1. In the present study, we evaluated the importance of these boxes in the binding of TtgV to the ttgD operator through construction of the appropriate mutants. We found that boxes 3 and 4 were critical for the recognition of ttgD by TtgV; however, to our surprise, boxes 1 and 5 in this promoter were dispensable (Fig. 5). To gain more insight into these observations, we reanalyzed the alignment of the ttgD and ttgG operators at the region corresponding to the footprint (Fig. 7) and found a weak inverted repeat (26 nucleotides long) centered between boxes 3 and 4 that may serve as the potential TtgV target sequence (Fig. 7). Of note is the fact that the palindromic order around the axis is higher for ttgD than for ttgG and defines a 15-bp inverted repeat. If this set of nucleotides were those directly bound by TtgV, this may explain why the affinity of TtgV for the ttgD operator is higher and correlates with the lower expression of the ttgDEF operon in vivo. Long palindromic DNA targets for IclR family members have been described recently for Streptomyces CcaR, a regulator involved in the control of β-lactam antibiotics in this microorganism (25), and Acinetobacter PcaU, a regulator involved in the metabolism of aromatic carboxylic acids (7).
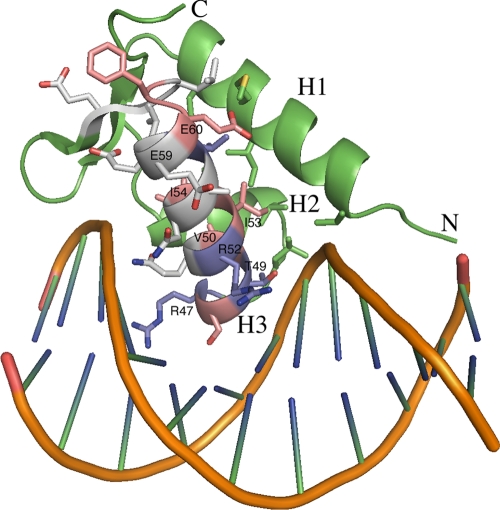
To test the hypothesis that TtgV can establish different contacts in each of the two promoters, we created mutants in the recognition helix of the HTH binding domain predicted to contact DNA and searched for differential effects on the repression of the ttgG and ttgD promoters and their variants. EMSA revealed that residues R47, T49, and R52 are critical for binding to ttgD and ttgG operators, whereas residues 48, 50, 53, 54, 60, and 61 in TtgV are critical for TtgV binding to the ttgG promoter but not to the ttgD promoter (compare Fig. 6A and B). To learn more about the potential role of the different amino acids, we modeled the DNA binding domain of TtgV, using the program Modeler, with Thermotoga maritima IclR as the template (PDB code 1MKM) (30). The DNA binding domain was then modeled onto a B-DNA, using a lambda repressor-operator complex (PDB code 1LMB). Residues R47, T49, and R52, which when mutated to A significantly reduced DNA binding, were proposed to be involved directly in DNA interaction (Fig. 8). For PobR, the positive regulator of the pobA gene for p-hydroxybenzoate metabolism and a member of the IclR family of regulators, an R56S mutant (a position that aligns with R47 in TtgV) was also unable to bind to its target sequence (9), in support of our results. Residues N55, A56, E58, and E59 (white in Fig. 8), although within the recognition helix from our model, do not appear to be involved directly in DNA interaction. Our results support this conclusion, since mutating these residues had little effect on DNA binding affinity. Residues V50, I53, and I54 (Fig. 8) were located at the interface between helices 2 and 3. Replacement of these residues by A resulted in mutant proteins that only affected DNA binding to the ttgGHI promoter, not to the ttgDEF promoter. As such, these mutations seemed to alter the conformations of helices 2 and 3, leading to a differential effect on the binding of TtgV to the two operators. Rhee et al. (21) showed that the OmpR regulator of Salmonella spp. can adopt different orientations depending on the precise base composition of the different binding sites that this regulator recognizes. Therefore, evidence from the present study and research with other systems support the hypothesis that HTH and DNA operators should be viewed as dynamic elements, rather than static interacting elements, in order to optimize the modulation of transcription.
FIG. 8.
Representation of the recognition helix of TtgV with B-DNA. The TtgV recognition helix is represented as a ribbon modeled on the 3D structure of the recognition helix of the IclR-TM protein (30).
In summary, we provide the first detailed study of the extended DNA recognition helix of a member of the IclR family. One conclusion from our findings is the existence of a large degree of flexibility in the interactions of the TtgV regulator with its target sequences. This flexibility may be related to the extensive interface for multimerization of monomers, as deduced from the 3D structure of IclR in Thermotoga maritima (30). Detailed mutational analysis of the operators recognized by TtgV showed that the central region of the zone protected in previous footprint studies (5, 23, 27) is essential for binding, whereas the border sequences are dispensable to some degree. This leads us to propose that TtgV recognizes a single inverted repeat, as shown in Fig. 7. Crystallization of TtgV in the absence and presence of target sequences is being carried out and will help to shed light on this set of complex DNA-protein interactions.
Acknowledgments
Work in the laboratory of J. L. Ramos was supported by grants from the Consolider-C (BIO 2006-05668) program of the Spanish Ministry of Science and Technology, the Excelencia (CVI-344) program of the Junta de Andalucia, and the PSYSMO project of the ERA-NET program of the European Commission (GEN2006-27750-C5-5-E/SYS).
We thank Antonia Felipe for technical assistance, Paloma Gutiérrez for help with interpreting the atomic force microscopy images, M. Eugenia Guazzaroni for help in the early stages of this work, K. Shashok for improving the use of English in the manuscript, and M. Fandila and C. Lorente for invaluable secretarial assistance.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 391100-1106. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman, M. M., and I. Pastam. 1993. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 62385-427. [DOI] [PubMed] [Google Scholar]
- 3.Guazzaroni, M. E., T. Krell, A. Felipe, R. Ruíz, C. Meng, X. Zhang, et al. 2005. The multidrug efflux regulator TtgV recognizes a wide range of structurally different effectors in solution and complexed with target DNA. Evidence from isothermal titration calorimetry. J. Biol. Chem. 28020887-20893. [DOI] [PubMed] [Google Scholar]
- 4.Guazzaroni, M. E., T. Krell, P. Gutierrez del Arroyo, M. Vélez, M. Jiménez, G. Rivas, and J. L. Ramos. 2007. The transcriptional repressor TtgV recognizes a complex operator as a tetramer and induces convex DNA bending. J. Mol. Biol. 369927-939. [DOI] [PubMed] [Google Scholar]
- 5.Guazzaroni, M. E., W. Terán, X. Zhang, M. T. Gallegos, and J. L. Ramos. 2004. TtgV bound to a complex operator site represses transcription of the single promoter for the multidrug and solvent extrusion TtgGHI pump. J. Bacteriol. 1862921-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isken, S., and J. A. M. de Bont. 1996. Active efflux of toluene in a solvent-resistant bacterium. J. Bacteriol. 1786056-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerg, B., and U. Gerischer. 2008. Relevance of nucleotides of the PcaV binding site from Acinetobacter baylyi. Microbiology 154756-766. [DOI] [PubMed] [Google Scholar]
- 8.Kieboom, J., J. J. Dennis, J. A. M. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 27385-91. [DOI] [PubMed] [Google Scholar]
- 9.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1998. Mutation analysis of PobR and PcaU, closely related transcriptional activators in Acinetobacter. J. Bacteriol. 1805058-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krell, T., A. J. Molina-Henares, and J. L. Ramos. 2006. The IclR family of transcriptional activators and repressors can be defined by a single profile. Protein Sci. 151207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, J. H. 1972. Experiments in molecular biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Molina-Henares, A. J., T. Krell, M. E. Guazzaroni, A. Segura, and J. L. Ramos. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol. Rev. 30157-186. [DOI] [PubMed] [Google Scholar]
- 13.Mosqueda, G., and J. L. Ramos. 2000. A set of genes encoding a second toluene efflux system in Pseudomonas putida DOT-T1E is linked to the tod genes for toluene metabolism. J. Bacteriol. 182937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 1516-523. [DOI] [PubMed] [Google Scholar]
- 15.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria. Antimicrob. Agents Chemother. 442233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob. Agents Chemother. 442595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-González, A. Rojas, W. Terán, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56743-768. [DOI] [PubMed] [Google Scholar]
- 18.Ramos, J. L., E. Duque, P. Godoy, and A. Segura. 1998. Efflux pumps involved in toluene tolerance in P. putida DOT-T1E. J. Bacteriol. 1803323-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos, J. L., E. Duque, M. J. Huertas, and A. Haïdour. 1995. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 1773911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos, J. L., M. Martínez-Bueno, A. J. Molina-Henares, W. Terán, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, J. E., W. Sheng, L. K. Morgan, R. Nolet, X. Liao, and L. J. Kenney. 2008. Amino acids important for DNA recognition by the response regulator OmpR. J. Biol. Chem. 2838664-8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 1833967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas, A., A. Segura, M. E. Guazzaroni, W. Terán, A. Hurtado, M. T. Gallegos, and J. L. Ramos. 2003. In vivo and in vitro evidence that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J. Bacteriol. 1854755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier, M. J., Jr., and I. Paulsen. 2001. Phylogeny of multidrug transporters. Cell. Dev. Biol. 12205-213. [DOI] [PubMed] [Google Scholar]
- 25.Santamarta, I., M. T. López-García, R. Pérez-Redondo, B. Koekman, J. F. Martin, and P. Liras. 2007. Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the ARE(ccaR) sequence of S. clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Mol. Microbiol. 66511-524. [DOI] [PubMed] [Google Scholar]
- 26.Spaink, H. P., J. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 927-39. [DOI] [PubMed] [Google Scholar]
- 27.Terán, W., A. Felipe, S. Fillet, M. E. Guazzaroni, T. Krell, R. Ruiz, J. L. Ramos, and M. T. Gallegos. 2007. Complexity in efflux pump control: cross-regulation by the paralogues TtgV and TtgT. Mol. Microbiol. 661416-1428. [DOI] [PubMed] [Google Scholar]
- 28.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 173469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improve M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vector. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, R. G., Y. Kim, T. Skarina, S. Beasley, R. Laskowski, C. Arrowsmith, A. Edwards, A. Joachimiak, and A. Savchenko. 2002. Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family. J. Biol. 27719183-19190. [DOI] [PMC free article] [PubMed] [Google Scholar]