Abstract
Cyclic lipopeptides produced by Pseudomonas species exhibit potent surfactant and broad-spectrum antibiotic properties. Their biosynthesis is governed by large multimodular nonribosomal peptide synthetases, but little is known about the genetic regulatory network. This study provides, for the first time, evidence that the serine protease ClpP regulates the biosynthesis of massetolides, cyclic lipopeptides involved in swarming motility, biofilm formation, and antimicrobial activity of Pseudomonas fluorescens SS101. The results show that ClpP affects the expression of luxR(mA), the transcriptional regulator of the massetolide biosynthesis genes massABC, thereby regulating biofilm formation and swarming motility of P. fluorescens SS101. Transcription of luxR(mA) was significantly repressed in the clpP mutant, and introduction of luxR(mA) restored, in part, massetolide biosynthesis and swarming motility of the clpP mutant. Site-directed mutagenesis and expression analyses indicated that the chaperone subunit ClpX and the Lon protease are not involved in regulation of massetolide biosynthesis and are transcribed independently of clpP. Addition of Casamino Acids enhanced the transcription of luxR(mA) and massABC in the clpP mutant, leading to a partial rescue of massetolide production and swarming motility. The results further suggested that, at the transcriptional level, ClpP-mediated regulation of massetolide biosynthesis operates independently of regulation by the GacA/GacS two-component system. The role of amino acid metabolism and the putative mechanisms underlying ClpP-mediated regulation of cyclic lipopeptide biosynthesis, swarming motility, and growth in P. fluorescens are discussed.
Cyclic lipopeptides are versatile metabolites produced by a variety of bacterial genera, including Pseudomonas and Bacillus (54, 55, 60). They are composed of a short cyclic oligopeptide linked to a fatty acid tail and exhibit potent surfactant properties (60). Cyclic lipopeptides have received considerable attention for their antibiotic activities against a range of human- and plant-pathogenic organisms, including enveloped viruses, mycoplasmas, trypanosomes, bacteria, fungi, and oomycetes (60). For plant-associated Pseudomonas species, cyclic lipopeptides play important roles in swarming motility, biofilm formation, and virulence (2, 4, 14, 15, 18, 34, 45, 60, 61). Cyclic lipopeptide biosynthesis is governed by large, multimodular nonribosomal peptide synthetases via a thiotemplate process (23, 60). Compared to the understanding of cyclic lipopeptide biosynthesis in Pseudomonas and other bacterial genera, however, relatively little is known about the genetic network involved in the perception of external factors and the signal transduction pathways that drive transcription of the cyclic lipopeptide biosynthesis genes.
For pathogenic and saprophytic Pseudomonas species, only a few regulatory genes and mechanisms have been identified. The GacA/GacS two-component system functions as a master switch, as a mutation in either one of the two genes results in loss of cyclic lipopeptide production (14, 15, 20, 41, 42). For pathogenic Pseudomonas syringae pv. syringae, regulatory genes identified downstream of the Gac system include salA and syrF, two LuxR-type transcriptional regulators involved in syringomycin and syringopeptin biosynthesis (41, 47, 48, 66). For saprophytic Pseudomonas putida strain PCL1445, DnaK and DnaJ were also shown to regulate putisolvin biosynthesis (20). Although the exact roles of these heat shock proteins are not yet resolved, the authors speculated that they might be required for proper folding or activity of other regulators of the putisolvin biosynthesis gene psoA or that DnaK is required for proper assembly of the peptide synthetase complex (20). In addition, cell density plays a role in cyclic lipopeptide biosynthesis in some Pseudomonas strains. For plant-pathogenic Pseudomonas fluorescens strain 5064, Cui et al. (12) provided evidence that N-acyl homoserine lactone (N-AHL)-mediated quorum sensing is required for viscosin biosynthesis. Also, for P. putida strain PCL1445, it was shown that putisolvin production was regulated by the quorum-sensing system composed of ppuI, rsaL, and ppuR (22). In many other pathogenic and saprophytic Pseudomonas species and strains, however, cyclic lipopeptide production is not regulated via N-AHL-mediated quorum sensing (2, 14, 15, 40, 59). In this context, Nybroe and Sørensen (54) emphasized that although cyclic lipopeptide production is affected by the growth phase and nutritional conditions, the specific impacts of these factors and the underlying molecular mechanisms in relation to cyclic lipopeptide biosynthesis are still unknown and may differ considerably among species and strains.
This study focuses on the regulation of cyclic lipopeptide biosynthesis in the plant growth-promoting strain P. fluorescens SS101. Strain SS101 produces massetolide A, which consists of a 9-amino-acid cyclic peptide moiety linked to 3-hydroxydecanoic acid (14). Massetolide A was first identified in a marine Pseudomonas species isolated from Masset Inlet, BC, Canada (31), and showed surfactant and broad-spectrum antimicrobial activities. Massetolide A inhibits the growth of Mycobacterium tuberculosis and Mycobacterium avium-M. intracellulare (31) and has destructive effects on zoospores of multiple oomycete plant pathogens (15, 17). Furthermore, massetolide A induces a systemic resistance response in tomato plants and contributes to root colonization by strain SS101 (64). Massetolide A is produced in the early exponential growth phase and is essential for swarming motility and biofilm formation of strain SS101 (14). Its biosynthesis is governed by three nonribosomal peptide synthetases, designated MassA, MassB, and MassC, and is not regulated via N-AHL-based quorum sensing (14). Due to flexibility in amino acid selection by the nonribosomal peptide synthetases, strain SS101 produces several massetolide A derivatives that differ in the amino acid composition of the peptide moiety (14). To begin to identify the genetic networks and mechanisms underlying the regulation of cyclic lipopeptide biosynthesis, P. fluorescens strain SS101 was subjected to random mutagenesis. Among the massetolide-deficient mutants obtained, one mutant harbored a Tn5 insertion in the caseinolytic protease gene clpP. The clpP gene of strain SS101 was cloned and sequenced, and its genomic context was assessed by primer walking. Site-directed mutagenesis, genetic complementation, and phenotypic and transcriptional analyses were performed to assess the functions of the ClpP protease in the regulation of massetolide biosynthesis and other bacterial traits, including swarming motility, growth, and biofilm formation. The effects of the clpP mutation on the expression of two LuxR-type transcriptional regulators, as well as the role of amino acids in ClpP-mediated regulation of massetolide biosynthesis, were investigated in detail.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. fluorescens SS101 was grown on Pseudomonas agar F (Difco) plates or in liquid King's medium B (KB) at 25°C. The transposon mutants were obtained as described by De Souza et al. (17), and plasposon mutants were obtained with plasmid pTnModOKm (16). Escherichia coli strain DH5α was used as a host for the plasmids for site-directed mutagenesis and complementation. E. coli strains were grown on Luria-Bertani (LB) plates or in LB broth amended with the appropriate antibiotics.
Identification of the clpP cluster.
clpP was identified by sequencing the regions flanking the transposon insertions as described by De Sousa et al. (17). The flanking regions of clpP were sequenced by primer walking, and open reading frames (ORFs) were identified with the Softberry FGENESB program. The ORFs were analyzed using Blastx in the NCBI database, PseudoDB (http://xbase.bham.ac.uk/pseudodb/), and Pseudomonas.com.
Site-directed mutagenesis.
Site-directed mutagenesis of the lon and tig genes was performed with the pKnockout-G suicide vector (67) as described by De Bruijn et al. (14). The primers used for site-directed mutagenesis are listed in Table 1. Site-directed mutagenesis of the clpP and clpX genes was performed based on the method described by Choi and Schweizer (10). For each mutant construct, three fragments were amplified: a 5′ fragment, a Gm cassette flanked by FRT sites (FRT-Gm-FRT cassette), and a 3′ fragment. In the first-round PCR, the FRT-Gm-FRT cassette and the 5′ and 3′ fragments were amplified. In the second-round PCR, these three fragments were coupled by overlap extension PCR. The 5′ and 3′ fragments were chosen in such a way that, after homologous recombination in Pseudomonas, the FRT-Gm-FRT cassette was inserted around the 170-bp position of the clpP or clpX gene. For amplification of the FRT-Gm-FRT cassette, pPS854-GM, a derivative of pPS854 (37), was used as a template in the PCR with primers FRT-F and FRT-R. The first-round PCR was performed with KOD polymerase (Novagen) according to the manufacturer's protocol, but with the addition of 1 to 10% dimethyl sulfoxide for the clpP and clpX fragments. The program used for the PCR consisted of 2 min of denaturation at 95°C, followed by 5 cycles of 95°C, 55°C, and 68°C, each for 20 s. The PCR amplification was preceded by 25 cycles of 95°C, 60°C, and 68°C, each for 20 s. The last step of the PCR was 68°C for 7 min. All fragments were separated on a 1% (wt/vol) agarose gel and purified with a NucleoSpin kit (Macherey-Nagel). The second-round PCR was performed by mixing equimolar amounts of the 5′, FRT-Gm-FRT, and 3′ fragments with milliQ, deoxynucleotide triphosphates, KOD buffer, and KOD polymerase to a total of 47 μl. The PCR was started by 2 min of denaturation at 95°C, followed by 3 cycles of 95°C, 55°C, and 68°C for 20, 30, and 60 s, respectively. In the third extension cycle, 1.5 μl each of the Up forward and Dn reverse primers (10 μM stock) was added. The PCR amplification was preceded by 25 cycles of 95°C, 58°C, and 68°C for 20, 20, and 120 s, respectively. The last step of the PCR was 68°C for 7 min. All fragments were separated on a 1% agarose gel, and bands of the right size were purified with a NucleoSpin kit. The fragments were digested with BamHI and cloned into pEX18Tc. E. coli DH5α was transformed with a pEX18Tc-clpP or pEX18Tc-clpX plasmid by heat shock transformation according to the method of Inoue et al. (39), and transformed colonies were selected on LB supplemented with 25 μg/ml gentamicin (Sigma). Integration of the inserts was verified by PCR analysis with pEX18Tc primers and by restriction analysis of the isolated plasmids. The plasmid inserts were verified by sequencing (BaseClear, Leiden, The Netherlands). The correct pEX18Tc-clpP and pEX18Tc-clpX constructs were subsequently electroporated into P. fluorescens strain SS101. Electrocompetent cells were obtained according to the method of Choi et al. (9), and electroporation occurred at 2.4 kV and 200 μF. After incubation in SOC medium (2% Bacto tryptone [Difco], 0.5% Bacto yeast extract [Difco], 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose [pH 7]) for 2 h at 25°C, the cells were plated on KB supplemented with gentamicin (25 μg/ml) and rifampin (50 μg/ml). The colonies obtained were grown in LB for 1 h at 25°C and plated on LB supplemented with gentamicin (25 μg/ml) and 5% sucrose to accomplish the double crossover. The plates were incubated at 25°C for at least 48 h, and colonies were restreaked on LB supplemented with gentamicin plus 5% sucrose and on LB supplemented with tetracycline (25 μg/ml). Colonies that grew on LB with gentamicin plus sucrose, but not on LB with tetracycline, were selected and subjected to colony PCR to confirm the presence of the gentamicin resistance cassette and the absence of the tetracycline resistance cassette. Positive colonies were confirmed by sequencing the PCR fragments obtained with the Up forward and Dn reverse primers. The clpP and clpX mutants obtained were tested for massetolide production in a drop collapse assay and by high-performance liquid chromatography (HPLC) analysis. HPLC analyses were performed as described previously (14) with the exception that in this study, samples of the crude surfactant extract (1 mg/ml) were analyzed isocratically (flow rate, 0.5 ml/min) using a solution of 45% acetonitrile and 15% milliQ, both containing 0.1% trifluoroacetic acid, and 40% methanol as eluents.
TABLE 1.
Primers used in this study
| Fragment | Orientation | Primer sequencea | |
|---|---|---|---|
| Site-directed mutagenesis | |||
| lon | Forward | 5′-GAGCAGATGAAGGCCATTCAG-3′ | |
| Reverse | 5′-GCCACATCCGGCAAGGGCTC-3′ | ||
| tig | Forward | 5′-TCTGTGCAACGAGGAATATCC-3′ | |
| Reverse | 5′-CTTGTTCTTCCAGCACAACCG-3′ | ||
| FRT | Forward | 5′-CGAATTAGCTTCAAAAGCGCTCTGA-3′ | |
| Reverse | 5′-CGAATTGGGGATCTTGAAGTTCCT-3′ | ||
| clpP-Up | Forward | 5′-TCAAGCAAGCGGATCCTGACTACCAGAACCTGGAC-3′ | |
| Reverse | 5′-tcagagcgcttttgaagctaattcgGGAATTACGGAACATGCTCTG-3′ | ||
| clpP-Dn | Forward | 5′-aggaacttcaagatccccaattcgGTGATCTTTCTGGTTGGCC-3′ | |
| Reverse | 5′-TCAAGCAAGCGGATCCGATGTCATTGCACAGGTCGAC-3′ | ||
| clpX-Up | Forward | 5′-TCAAGCAAGCGGATCCGTGCGCAGTTGCTGTTCCTT-3′ | |
| Reverse | 5′-tcagagcgcttttgaagctaattcgCCTCACGGATGATGTCATTGC-3′ | ||
| clpX-Dn | Forward | 5′-aggaacttcaagatccccaattcgTCGATCACTCGGGACGTTTC-3′ | |
| Reverse | 5′-TCAAGCAAGCGGATCCTTGGACTTGCCTTCGATAACG-3′ | ||
| pEX18Tc | Forward | 5′-CCTCTTCGCTATTACGCCAG-3′ | |
| Reverse | 5′-GTTGTGTGGAATTGTGAGCG-3′ | ||
| Complementation | |||
| clpP | Forward | 5′-tttttttgagctcCCGCACCGAAGTTCGCAAG-3′ | |
| Reverse | 5′-aaaaaaaggatccCCTGCTGCACGCCTTCAC-3′ | ||
| luxR(mA) | Forward | 5′-TGCTCCAGGGCGCTGTAGAG-3′ | |
| Reverse | 5′-CATGCCGAGGGTGCACAG-3′ | ||
The 5′ end of the Up reverse and Dn forward primers for site-directed mutagenesis contain a 25-bp sequence (lowercase letters) complementary to the FRT-F and FRT-R primers for overlap extension in the second-round PCR. The 5′ end of the Up forward and Dn reverse primers contain a restriction site (underlined) for BamHI, which is required for cloning into pEX18Tc.
Construction of pME6031-based vectors for genetic complementation.
A fragment of approximately 2 kb containing the clpP gene, including the promoter and terminator, was obtained by PCR with specific primers (Table 1) and the KOD polymerase. The pME6031-luxR(mA) construct was generated as follows: a 1,817-bp fragment was obtained by PCR with specific primers (Table 1) and Phusion DNA polymerase (Finnzymes). The PCR fragments were subcloned in pGEM-T Easy (Promega), and the plasmids obtained were digested with EcoRI. The clpP and luxR(mA) (see below) fragments were obtained from gels with the NucleoSpin kit and cloned into the shuttle vector pME6031 (36), which was digested, dephosphorylated (shrimp alkaline phosphatase; Promega), and purified with the NucleoSpin kit according to the manufacturer's instructions. E. coli DH5α was transformed with the plasmid obtained, pME6031-clpP or pME6031-luxR(mA), by heat shock transformation (39), and transformed colonies were selected on LB agar plates supplemented with tetracycline (25 μg/ml). Correct integration of the fragments was verified by PCR analysis and restriction analysis of isolated plasmids. The pME6031-clpP and pME6031-luxR(mA) constructs were subsequently electroporated into the clpP mutant and the wild-type strain SS101. Transformed cells were plated on KB supplemented with tetracycline (25 μg/ml), and the presence of pME6031-clpP or pME6031-luxR(mA) was verified by PCR analysis with primers specific for pME6031.
Surface tension measurements and transcriptional analysis.
Cells were grown at 25°C (220 rpm) in a 24-well plate with 1.25 ml KB broth per well. At specific time points during growth, 100 μl culture was transferred to a 96-well plate, and cell density was measured at 600 nm with a microplate reader (Bio-Rad). Subsequently, 1 ml of cell culture was collected and spun down. The cells were frozen in liquid N2 and stored at −80°C. For the RNA isolations and cDNA synthesis, four biological replicates were used for each time point. Massetolide production was measured qualitatively by the drop collapse assay and quantitatively by tensiometric analysis of the cell supernatant (K6 tensiometer; Krüss GmbH, Hamburg, Germany) at room temperature. To get sufficient volume for the tensiometric analysis, the supernatants of four biological replicates were collected and pooled for each time point. The surface tension of each sample was measured in triplicate.
For the transcriptional analyses, RNA was isolated from the frozen bacterial cells with Trizol reagent (Invitrogen), followed by DNase I (GE Healthcare) treatment. One μg RNA was used for cDNA synthesis with Superscript III (Invitrogen) according to the manufacturer's protocol. For the real-time quantitative PCR (Q-PCR), conducted with the 7300SDS system from Applied Biosystems, the SYBR Green Core kit (Eurogentec) with a final concentration of 3.5 mM MgCl2 was used according to the manufacturer's protocol. The concentration of the primers was optimized (400 nM final concentration for the mass genes and rpoD; 500 nM for clpP and clpX), and a melting curve was performed to check the specificity of the primers. The primers used for the Q-PCR are listed in Table S1 in the supplemental material. To correct for small differences in the template concentration, rpoD was used as the housekeeping gene. The cycle in which the SYBR green fluorescence crossed a manually set cycle threshold (CT) was used to determine transcript levels. For each gene, the threshold was fixed based on the exponential segment of the PCR curve. The CT value of clpP was corrected for the housekeeping gene rpoD as follows: ΔCT = CT(clpP) − CT(rpoD); the same formula was used for the other genes investigated. The relative quantification (RQ) values, were calculated by the following formula: RQ = 2−[ΔCT(mutant) − ΔCT (wild type)]. If there was no difference in transcript levels between the mutant and the wild type, then RQ was equal to 1 (20) and log RQ was equal to 0. Q-PCR analysis was performed in duplicate (technical replicates) on four independent RNA isolations (biological replicates). Statistically significant differences were determined for log-transformed RQ values by analysis of variance (P < 0.05), followed by Bonferroni post hoc multiple comparisons.
Swarming motility and biofilm formation.
The swarming and swimming motility of the wild-type strain SS101, the massetolide-deficient mutants, and several transformants was assessed on soft (0.6% and 0.25% agar [wt/vol], respectively) standard succinate agar medium (SSM) consisting of 32.8 mM K2HPO4, 22 mM KH2PO4, 7.6 mM (NH4)2SO4, 0.8 mM MgSO4, and 34 mM succinic acid and adjusted to pH 7 with NaOH. After being autoclaved, the SSM was cooled down in a water bath to 55°C and kept at 55°C for 1 h. Twenty milliliters of SSM was pipetted into a 9-cm-diameter petri dish, and the plates were kept for 24 h at room temperature (∼20°C) prior to inoculation with the bacterial suspensions. For all swarming assays, the same conditions (the agar temperature, the temperature at which the plates were stored, and the time between pouring the plates and inoculation) were kept constant to maximize reproducibility. Overnight cultures of the wild-type SS101, mutants, and transformants were washed three times with 0.9% NaCl, and 5 μl of the washed cell suspension (1 × 1010 cells/ml) was spot inoculated in the center of the soft SSM plate and incubated for 48 to 72 h at 25°C. For the assays with Casamino Acids (CAA), a filter-sterile stock solution of 20% CAA (Difco, Becton Dickinson and Co.) was prepared and diluted in SSM to obtain final concentrations of 0.1, 0.4, 1, and 4%. To test each amino acid present in the CAA separately, the amounts used were equivalent to those present in 1% CAA (see Table S2 in the supplemental material). Also, the effects of citric acid (citrate, 0.4%), CaCl2 (14.7 μM), and FeCl3 (0.24 μM) on the swarming motility of strain SS101 were tested. Biofilm formation was assessed according to the method described by De Bruijn et al. (14) and O'Toole et al. (57) using flat-bottom 96-well plates made of transparent polystyrene (Greiner) with 200 μl KB broth per well. Statistically significant differences were determined by Student's t test (P <0.05).
Nucleotide sequence accession number.
The sequences of clpP and its flanking genes have been deposited in GenBank under accession number FJ403110.
RESULTS
Role of clpP in regulation of massetolide biosynthesis.
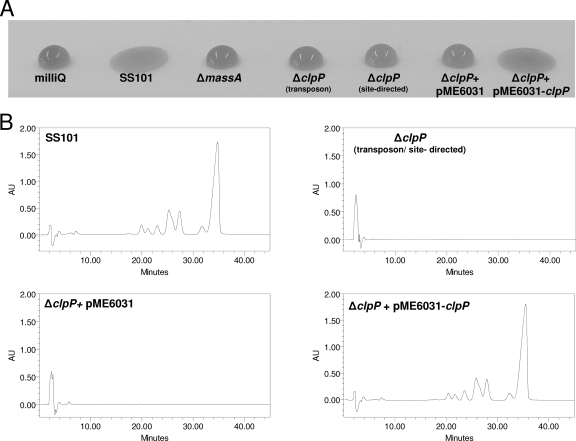
Screening of an initial 520 random transposon mutants of P. fluorescens SS101 for loss of massetolide production by a drop collapse assay (Fig. 1A) resulted in the selection of six putative mutants. All six mutants contained a single Tn5 transposon insertion, as determined by Southern blot analysis of their genomic DNAs with the km gene as a probe (data not shown). The regions flanking the Tn5 transposon insertion were cloned and sequenced for all six massetolide-deficient mutants. In five mutants, the Tn5 insertion was located in the massA, massB, or massC gene (14). In the sixth mutant, designated mutant 13.3, the transposon was inserted in the caseinolytic protease gene, clpP. The complete clpP gene comprised 636 bp, and Blastx analysis showed 80 to 98% identity to clpP in other Pseudomonas genomes and 72% identity to clpP in E. coli. To confirm the role of clpP in the regulation of massetolide biosynthesis, site-directed mutagenesis of clpP was performed. Consistent with the phenotype of transposon mutant 13.3, the site-directed clpP mutant also lacked the ability to collapse a droplet of water (Fig. 1A). HPLC analysis confirmed that the clpP mutants obtained by random or site-directed mutagenesis did not produce detectable levels of massetolide A or its derivatives (Fig. 1B). Complementation of the clpP transposon mutant with the stable vector pME6031-clpP restored massetolide production to the wild-type level, whereas the empty-vector control had no effect (Fig. 1B). Taken together, these results indicate that clpP is required for massetolide biosynthesis in P. fluorescens SS101.
FIG. 1.
Phenotypic and biochemical analyses of massetolide biosynthesis in P. fluorescens strain SS101 and in several mutants obtained by random or site-directed mutagenesis. (A) Drop collapse assay with cultures of the wild-type strain SS101 and different mutants. Bacterial cells grown for 2 days at 25°C were resuspended in sterile water (1 × 1010 cells/ml), and 10-μl droplets were spotted on parafilm; crystal violet was added to the droplets to facilitate visual assessment. A flat droplet was indicative of massetolide production. (B) HPLC chromatograms of cell-free culture extracts of wild-type SS101, clpP mutants obtained by transposon or site-directed mutagenesis, clpP+pME6031 (empty-vector control), and clpP+pME6031-clpP. The wild-type strain SS101 produces massetolide A (retention time of approximately 35.5 min) and various other derivatives of massetolide A (peaks with retention times ranging from 21 to 33 min) differing in the amino acid composition of the peptide moiety (14).
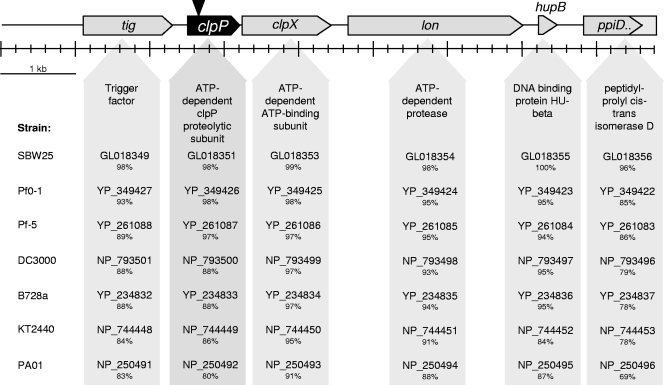
Genomic context of clpP in P. fluorescens SS101.
By primer walking up- and downstream of the transposon insertion, a total 8,670-bp sequence was obtained from the regions flanking the clpP gene in strain SS101. Several ORFs were identified (Fig. 2), including the chaperone and protein-folding trigger factor (tig); the ATPase chaperone, clpX; the lon protease; the DNA binding and -bending hupB; and a partial sequence of ppiD isomerase, a gene involved in protein folding (3, 19, 24, 35). The organization of these genes in strain SS101 is identical to that found in various other fully sequenced Pseudomonas species and strains (Fig. 2). ClpX is known to act as a chaperone in the proteolytic complex with clpP (28) and is responsible for the recognition, unfolding, and translocation of substrates into the ClpP degradation chamber (51). Furthermore, Tig and Lon were shown to be substrates for Clp-dependent proteolysis (25, 32). To determine if the genes flanking clpP also play roles in the regulation of massetolide biosynthesis, site-directed mutagenesis was performed for tig, clpX, and lon. Drop collapse assays and HPLC analyses showed that disruptions of these three genes did not affect massetolide production (data not shown), suggesting that clpP acts independently of clpX in regulating massetolide biosynthesis.
FIG. 2.
Genomic organization of clpP and flanking genes in P. fluorescens SS101 and other fully sequenced Pseudomonas strains. For each of the genes of strain SS101, the percentages of amino acid identity with their corresponding genes in other Pseudomonas strains are given. The reference strains used were P. fluorescens strains SBW25 (15), Pf0-1 (15), Pf-5 (34), P. syringae pv. tomato strain DC3000 (4), P. syringae pv. syringae strain B728a (62, 70), P. putida strain KT2440, and P. aeruginosa strain PAO1. The codes for the genes of these reference strains correspond to those in the PseudoDB and NCBI databases.
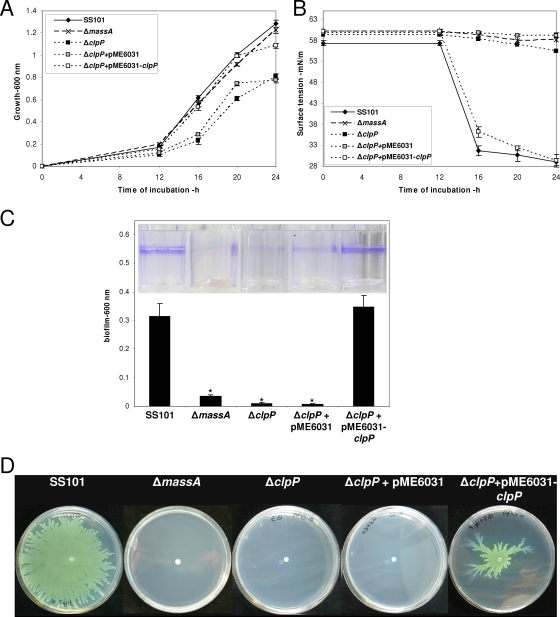
Phenotypic characterization of the clpP mutant of P. fluorescens.
Consistent with observations made previously for E. coli, Staphylococcus aureus, and Pseudomonas aeruginosa (13, 63, 65), a mutation in clpP adversely affected the growth of P. fluorescens SS101 (Fig. 3A). This reduced growth of the clpP mutant was not due to a lack of massetolide production, because the massA biosynthesis mutant showed growth comparable to that of the wild-type strain SS101 (Fig. 3A). Complementation of the clpP mutant with pME6031-clpP restored growth to the wild-type level, whereas the empty-vector control had no effect (Fig. 3A). Tensiometric analysis of cell-free culture supernatants of strain SS101 and mutants indicated that the wild-type strain SS101 started producing the massetolide surfactants at between 12 h and 16 h of incubation (Fig. 3B). A reduction in the surface tension of the growth medium was not observed for the massA mutant or for the clpP mutant, but surface tension was restored by complementation with pME6031-clpP (Fig. 3B).
FIG. 3.
Phenotypic characteristics of P. fluorescens strain SS101, the massA mutant, the clpP mutant, clpP+pME6031 (empty-vector control), and clpP+pME6031-clpP. (A) Growth at 25°C. At each time point, the cell density was measured spectrophotometrically (600 nm), and the mean values for four replicates are given; the error bars represent the standard errors of the mean. (B) Surface tension of cell-free culture supernatant of strain SS101 and the different mutants shown in panel A. (C) Biofilm formation by strain SS101 and the different mutants in a polystyrene 96-well plate containing 200 μl growth medium; cells firmly attached to the walls of the wells were stained with crystal violet, and their density was quantified spectrophotometrically at 600 nm. The mean values of four replicates are given. For each of the mutants, asterisks indicate statistically significant (P < 0.05) differences relative to wild-type SS101. (D) Swarming motility of wild-type strain SS101 and mutants on soft (0.6% [wt/vol]) agar plates. Five microliters of washed overnight cultures of wild-type SS101 and mutants was spot inoculated in the center of a soft agar plate and incubated for 48 to 72 h at 25°C.
Massetolide biosynthesis is essential for biofilm formation and swarming motility in strain SS101 (14). The capacity to form a biofilm was strongly reduced in the clpP mutant to a level similar to that observed for the massA biosynthesis mutant (Fig. 3C). Biofilm formation was fully restored to the wild-type level by complementation of the clpP mutant with pME6031-clpP (Fig. 3C). The clpP mutant also lost its ability to swarm on a soft agar surface (Fig. 3D). Swarming motility was restored in the clpP mutant by introduction of pME6031-clpP, although the extent of complementation, as well as the swarming pattern, was not identical to that of the wild-type strain (Fig. 3D). Introduction of pME6031-clpP into wild-type SS101 also resulted in reduced swarming (data not shown), suggesting that the altered swarming pattern of the complemented clpP mutant may have resulted from multiple copies of the clpP gene. In contrast to a complete loss of swarming motility, the clpP mutant was still able to swim on soft (0.25% [wt/vol]) agar plates (see Fig. S1 in the supplemental material). The observation that the swimming motility was similar to that of the massA biosynthesis mutant but reduced compared to the wild-type strain SS101 and the complemented clpP mutant (see Fig. S1 in the supplemental material) indicates that massetolide production also plays a (partial) role in swimming motility.
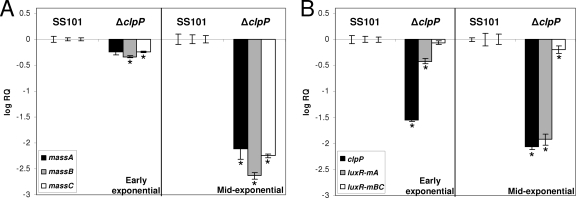
Transcriptional analysis of the clpP mutant of P. fluorescens.
Q-PCR analyses were performed to study the effects of the clpP mutation on the expression of a range of genes, including the biosynthesis genes massA, massB, and massC. To prevent differences in growth rates between the wild-type strain SS101 and the clpP mutant from interfering with gene expression measurements, cells used for RNA isolation were collected when the wild type and mutant reached a specific density, i.e., early exponential (OD600, ∼0.2) and mid-exponential (OD600, ∼0.6) phases. Consistent with previous results (14), massA, massB, and massC were expressed in the wild-type strain SS101 during the early exponential and mid-exponential growth phases (data not shown). The transcript levels of all three mass genes were significantly decreased in the clpP mutant, especially in the mid-exponential growth phase (Fig. 4A). Mutations in massA, massB, or massC did not affect transcription of clpP (data not shown). Collectively, these results indicate that clpP regulates the transcription of the mass biosynthesis genes.
FIG. 4.
(A) Transcript levels of massA, massB, and massC in cells of P. fluorescens SS101 and the clpP mutant obtained from the early and mid-exponential growth phases. (B) Transcript levels of clpP, luxR(mA), and luxR(mBC) in cells of P. fluorescens SS101 and the clpP mutant obtained from the early and mid-exponential growth phases. The transcript levels of each of the genes was corrected for the transcript level of the housekeeping gene rpoD [ΔCT = CT (gene x) − CT(rpoD)] and is presented relative to the transcript levels in wild-type SS101 (log RQ), with RQ equal to 2−[ΔCT(mutant) − ΔCT (wild type)]. For each time point, mean values of four biological replicates are given; the error bars represent the standard errors of the mean. The asterisks indicate statistically significant (P < 0.05) differences relative to wild-type SS101.
A mutation in clpP had only a minor effect on expression of clpX (Table 2), and also, a mutation in clpX only slightly reduced clpP transcript levels (data not shown), suggesting that under these conditions, clpX and clpP are transcribed independently. Moreover, the clpP mutation did not result in major or consistent changes in tig, lon, hupB, and ppiD transcript levels (Table 2). Since it was reported that dnaK regulates cyclic lipopeptide biosynthesis in P. putida (20) and that DnaK can influence proteolysis by ClpP (30), we also determined dnaK, dnaJ, and gprE transcript levels in the clpP mutant. No changes in transcript levels were observed (Table 2), indicating that clpP does not affect dnaK expression.
TABLE 2.
Transcript levels of various genes in the clpP mutant of P. fluorescens strain SS101
| Gene | Transcript levela
|
|||
|---|---|---|---|---|
| Early exponential
|
Mid-exponential
|
|||
| SS101 | ΔclpP | SS101 | ΔclpP | |
| tig | 0.00 ± 0.03 | −0.29 ± 0.03b | 0.00 ± 0.04 | −0.36 ± 0.04b |
| clpX | 0.00 ± 0.04 | −0.30 ± 0.03b | 0.00 ± 0.08 | 0.09 ± 0.11 |
| lon | 0.00 ± 0.02 | 0.06 ± 0.03 | 0.00 ± 0.02 | −0.08 ± 0.02 |
| hupB | 0.00 ± 0.03 | 0.05 ± 0.03 | 0.00 ± 0.03 | −0.46 ± 0.02b |
| ppiD | 0.00 ± 0.08 | −0.19 ± 0.03 | 0.00 ± 0.02 | −0.61 ± 0.03b |
| dnaK | 0.00 ± 0.03 | 0.12 ± 0.02b | 0.00 ± 0.01 | 0.06 ± 0.02 |
| dnaJ | 0.00 ± 0.04 | 0.13 ± 0.02 | 0.00 ± 0.03 | 0.28 ± 0.05b |
| grpE | 0.00 ± 0.04 | −0.01 ± 0.03 | 0.00 ± 0.02 | −0.02 ± 0.03 |
The transcript level of each of the genes was corrected for the transcript levels of the housekeeping gene rpoD [ΔCT = CT (gene x) − CT(rpoD)] and is presented relative to the transcript level in wild-type SS101 (log RQ ± standard errors of the mean), with RQ equal to 2−[ΔCt(mutant) −ΔCt(wild type)]. For each time point, the mean values of four biological replicates are given.
Significantly different from SS101 transcript levels (P < 0.05).
Effect of ClpP on expression of the transcriptional regulator luxR(mA).
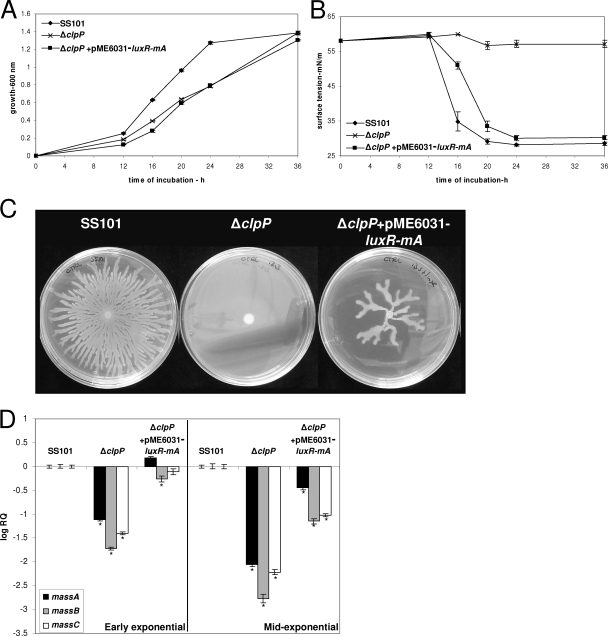
To further unravel the role of ClpP in transcriptional regulation of the massABC biosynthesis genes, we determined the transcript levels of two LuxR-type transcriptional-regulatory genes located upstream of massA (designated luxR(mA)) and downstream of massBC [luxR(mBC)] (14). The results showed that transcript levels of luxR(mA) were significantly decreased in the clpP mutant, whereas luxR(mBC) transcript levels were not or were only marginally reduced (Fig. 4B). Introduction of extra copies of luxR(mA) in the clpP mutant via pME6031-luxR(mA) restored massetolide production, based on the results of tensiometric analyses (Fig. 5B), drop collapse assays, and HPLC analysis (see Fig. S2 in the supplemental material). However, growth deficiency of the clpP mutant was not restored by pME6031-luxR(mA) (Fig. 5A), which in turn may explain why massetolide production was slightly delayed in the clpP+pME6031-luxR(mA) strain compared to the wild-type strain SS101 (Fig. 5B). Also, swarming motility was restored for the clpP+pME6031-luxR(mA) strain (Fig. 5C), but not to the same extent as in the wild type, most likely due to reduced growth. Gene expression analysis further showed that massABC transcript levels were partly restored in clpP+pME6031-luxR(mA), especially during early exponential growth (Fig. 5D). Collectively, these results strongly suggest that ClpP affects expression of the transcriptional-regulatory gene luxR(mA), thereby regulating massetolide biosynthesis and swarming motility in P. fluorescens SS101.
FIG. 5.
(A) Growth of P. fluorescens strain SS101, the clpP mutant, and clpP+pME6031-luxR(mA) at 25°C. At each time point, the cell density was measured spectrophotometrically (600 nm), and the mean values of four replicates are given; the error bars represent the standard errors of the mean. (B) Surface tension of cell-free culture supernatant of strain SS101 and the different mutants shown in panel A. (C) Swarming motility of wild-type strain SS101 and mutants on soft (0.6% [wt/vol]) agar plates. Five microliters of washed overnight cultures of wild-type SS101 and mutants was spot inoculated in the center of a soft agar plate and incubated for 48 to 72 h at 25°C. (D) Transcript levels of the massA, massB, and massC genes in wild-type strain SS101, the clpP mutant, and clpP+pME6031-luxR(mA) in early and mid-exponential growth phases. The transcript level of each of the genes was corrected for the transcript level of rpoD [ΔCT = CT(gene x) − CT(rpoD)] and is presented relative to the transcript level in wild-type SS101 (log RQ), with RQ equal to 2−[ΔCT(mutant) − ΔCT (wild type)]. For each time point, the mean values of four biological replicates are given; the error bars represent standard errors of the mean. The asterisks indicate statistically significant (P < 0.05) differences relative to wild-type SS101.
Influence of amino acids on clpP expression and massetolide biosynthesis.
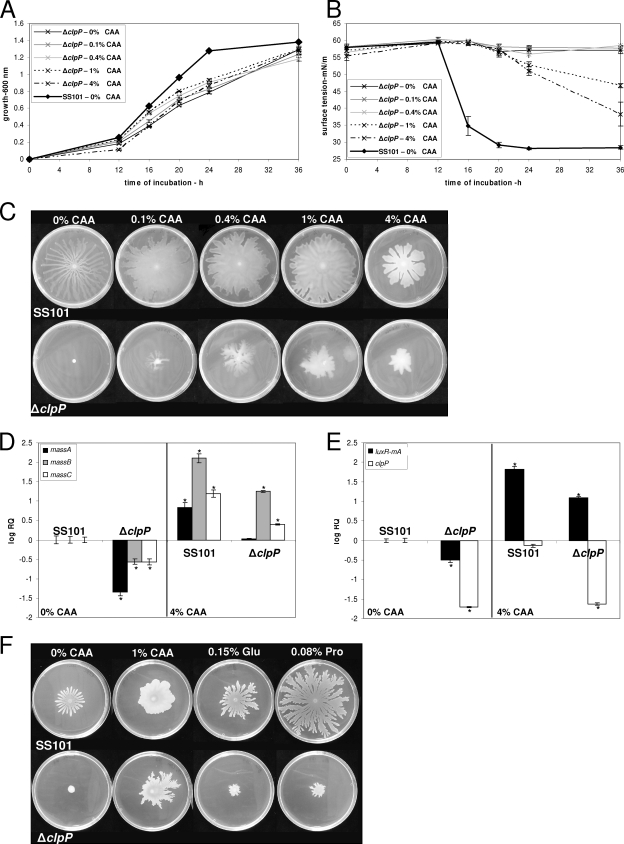
Previous studies showed that various nutritional conditions, including specific sugars and amino acids, affect cyclic lipopeptide production in P. fluorescens and P. putida (21, 31, 54, 60). Furthermore, CAA, citrate, glutamate, and iron were shown to rescue biofilm and growth defects of a range of surface attachment (sad) mutants of P. fluorescens strain WCS365, including a clpP mutant (56). Based on these observations, swarming assays, Q-PCR, and tensiometric and HPLC analyses were performed to assess the effects of specific nutrients on growth, massetolide production, and gene expression in strain SS101 and the clpP mutant. The results showed that addition of CAA did not rescue the growth defect of the clpP mutant (Fig. 6A) but did restore, at concentrations of 1% and 4% (wt/vol), massetolide production, as evidenced by a reduction in surface tension (Fig. 6B) and by HPLC analysis (see Fig. S3 in the supplemental material). Consistent with this partial recovery of massetolide production, swarming motility of the clpP mutant was also partly restored when the mutant was grown on CAA-supplemented agar medium (Fig. 6C). In contrast, swarming motility of the massA mutant was not restored by addition of CAA to the growth medium (data not shown). For wild-type strain SS101, swarming motility increased with increasing CAA concentrations; however, growth was not affected by the addition of CAA to liquid KB (see Fig. S4 in the supplemental material). With increasing CAA concentrations, the motility patterns of wild-type SS101 changed from typical dendritic to more confluent (Fig. 6C). Moreover, compared to the other CAA concentrations, the drop in surface tension was delayed when 4% CAA was added to liquid KB (see Fig. S4 in the supplemental material). Gene expression analyses showed that addition of CAA led to an increase in mass transcript levels in wild-type SS101 (Fig. 6D). In the clpP mutant, addition of CAA restored transcription of massA to the wild-type level and led to an increase in massBC transcript levels (Fig. 6D), providing support at the transcriptional level to the idea that CAA restore, at least in part, massetolide biosynthesis in the clpP mutant. In the wild-type strain SS101 and the clpP mutant, addition of CAA increased luxR(mA) transcript levels but did not affect transcription of clpP (Fig. 6E). Addition of CAA to cultures of the clpP mutant modified with pME6031-luxR(mA) completely restored swarming motility (see Fig. S5 in the supplemental material). Taken together, these results show that CAA restore and enhance transcription of the luxR(mA) and massABC biosynthesis genes, leading to a partial rescue of massetolide biosynthesis and swarming motility in the clpP mutant. Expression of the clpP gene, however, was not affected by CAA.
FIG. 6.
(A) Growth of the clpP mutant of P. fluorescens strain SS101 at 25°C in growth medium supplemented with 0 to 4% (wt/vol) CAA. At each time point, the cell density was measured spectrophotometrically (600 nm); the mean values of four replicates are given, and the error bars represent the standard errors of the mean. (B) Surface tension of cell-free culture supernatant of strain SS101 and the clpP mutant grown in media supplemented with different concentrations of CAA. (C) Swarming motility of strain SS101 and the clpP mutant on soft (0.6% [wt/vol]) agar plates supplemented with different concentrations of CAA. Five microliters of washed overnight cultures of wild-type SS101 and the mutants was spot inoculated in the center of a soft agar plate and incubated for 48 to 72 h at 25°C. (D and E) Transcript levels of the massA, massB, and massC genes (D) and luxR(mA) and clpP (E) in wild-type SS101 and the clpP mutant when grown on soft-agar plates supplemented with 0 or 4% CAA. Cells were collected from the periphery of the swarming colony. The transcript levels were corrected for the transcript levels of rpoD [ΔCT = CT(gene x) − CT(rpoD)] and are presented relative to the transcript level in wild-type SS101 grown at 0% CAA (log RQ), with RQ equal to 2−[ΔCT(sample X) − ΔCT (wild type 0% CAA)]. For each sample, the mean values of four biological replicates are given, and the error bars represent the standard errors of the mean. The asterisks indicate statistically significant (P < 0.05) differences relative to wild-type SS101. (F) Swarming motility of strain SS101 and the clpP mutant on soft agar medium supplemented with glutamic acid and proline.
To identify which amino acid is responsible for the partial complementation of the swarming motility of the clpP mutant, each amino acid present in the CAA was tested separately at concentrations identical to their respective concentrations in 1% CAA (see Table S2 in the supplemental material). The results show that the amino acids proline and glutamic acid can partially complement the deficiency in swarming motility of the clpP mutant, but not to the same extent as provided by addition of 1% CAA (Fig. 6F). When proline and glutamic acid were combined, no significant additional effects were observed (data not shown). The other amino acids, as well as calcium, iron, and citrate, did not stimulate the swarming motility of the clpP mutant. In fact, addition of several amino acids (valine, isoleucine, and leucine) inhibited the swarming motility of the wild-type strain SS101 (data not shown).
Interplay between GacA/GacS and ClpP.
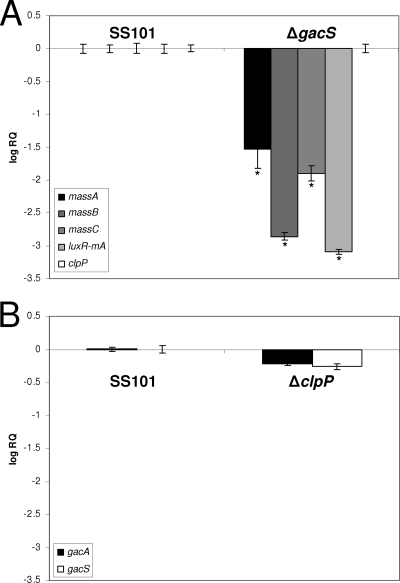
For P. fluorescens SS101, a mutation in the sensor kinase gene gacS significantly reduced the expression of the massABC genes (Fig. 7) and shut down massetolide production. Also, luxR(mA) transcript levels were reduced but clpP transcription was not affected in the gacS mutant of strain SS101 (Fig. 7). Furthermore, transcript levels of gacA/gacS were not affected in the clpP mutant (Fig. 7), suggesting that, at the transcriptional level, ClpP-mediated regulation of massetolide biosynthesis is independent of regulation by GacA/GacS.
FIG. 7.
(A) Transcript levels of massA, massB, massC, luxR(mA), and clpP in a gacS mutant of P. fluorescens SS101 at mid-exponential growth phase. (B) Transcript levels of gacA and gacS in the clpP mutant of P. fluorescens SS101 at mid-exponential growth phase. The transcript levels were corrected for the transcript levels of rpoD [ΔCT = CT(gene x) − CT(rpoD)] and are presented relative to the transcript level in wild-type SS101 (log RQ), with RQ equal to 2−[ΔCT(mutant) − ΔCT (wild type)]. The mean values of four biological replicates are given. The error bars represent the standard errors of the mean. The asterisks indicate statistically significant (P < 0.05) differences relative to wild-type SS101.
DISCUSSION
ClpP is a serine protease that is highly conserved in bacteria and eukaryotes (68, 69). Together with other proteases, ClpP plays a crucial role in intracellular refolding and degradation of proteins, which is an essential process for the viability and growth of cells. In this study, we cloned and sequenced clpP from plant growth-promoting P. fluorescens strain SS101 and showed that clpP plays an important role in the regulation of cyclic lipopeptide biosynthesis, swarming motility, biofilm formation, and growth. These results confirm and extend observations made for other Pseudomonas species and bacterial genera. For example, biofilm formation was reduced in clpP mutants of P. fluorescens WCS365 and S. aureus but enhanced in a clpP mutant of P. aeruginosa (26, 56, 63, 65). ClpP is also important for virulence in several bacterial pathogens, like Streptococcus pneumoniae, S. aureus, Salmonella enterica serovar Typhimurium, Yersinia enterocolitica, Listeria monocytogenes, and Porphyromonas gingivalis (7, 29, 38, 58). In Listeria, the hemolytic activity, but not the production, of the virulence factor listeriolysin O was strongly reduced in a clpP mutant (29). In Bacillus subtilis, ClpP plays a role in competence development, motility, and sporulation (52). Although specific extracellular metabolites of Pseudomonas strains are known to play roles in swarming motility and biofilm formation, the involvement of ClpP in regulation of the biosynthesis genes encoding these metabolites has, to our knowledge, not been demonstrated conclusively. This study provides for the first time evidence that the ClpP protease regulates the biosynthesis of cyclic lipopeptide surfactants that play an important role in swarming the motility, biofilm formation, and antimicrobial activity of P. fluorescens. More specifically, ClpP was shown to affect expression of the transcriptional-regulatory gene luxR(mA), thereby regulating massetolide biosynthesis and concomitantly biofilm formation and swarming motility in P. fluorescens SS101. Whether this is typical for the Pseudomonas strain under study remains to be addressed, but the observation by Nakano et al. (53) that expression of the surfactin gene srfA in B. subtilis is affected in a clpP mutant suggests that a similar role of ClpP may apply to other bacterial genera and species producing lipopeptide antibiotics.
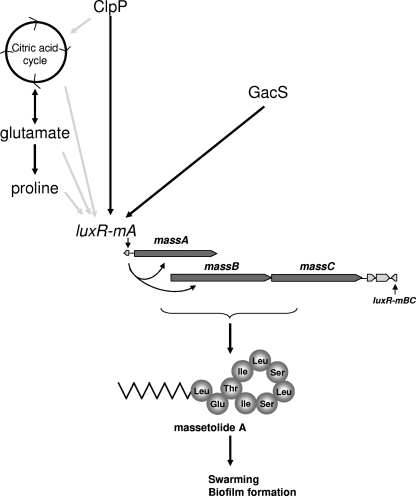
Based on the results of this and previous studies, several hypotheses can be proposed for the mechanisms underlying ClpP-mediated regulation of luxR(mA) expression, massetolide biosynthesis, and swarming motility in P. fluorescens (Fig. 8). In E. coli, ClpP consists of two heptameric rings that form a barrel-shaped core with active sites in an interior chamber (69). ClpP forms a proteolytic complex with Clp-ATPases, i.e., ClpX and ClpA, that carry one or two nucleotide binding domains (28). These ATPases belong to the Hsp100 protein family and unfold the substrates so they can be translocated to the active sites of the ClpP protease, which then leads to protein degradation and release of protein fragments (51, 69). Besides ClpXP and ClpAP, other ATP-dependent proteolytic complexes, like HslUV, Lon, and FtsH, have been identified in bacteria, particularly in E. coli (33, 68). However, based on site-directed mutagenesis and transcriptional analyses performed in this study, the chaperone subunit ClpX and also the Lon protease do not appear to be involved in regulation of massetolide biosynthesis in P. fluorescens SS101. Whether other Clp-ATPases are required as chaperones in ClpP-mediated regulation of these processes was not determined and will be investigated in more detail as soon as the whole genome of strain SS101 is sequenced. Alternatively, ClpP may also act as a peptidase in the absence of the Clp-ATPases, thereby hydrolyzing short peptides of up to 6 amino acids (5). Studies of B. subtilis further showed that in addition to its function in the degradation of misfolded and defective proteins, ClpP is also involved in targeted proteolysis of specific protein substrates, including key regulators and transcriptional factors involved in competence and developmental programs (5, 26, 32, 43, 52). Based on these observations in B. subtilis, we postulate that in P. fluorescens strain SS101 ClpP may degrade, alone or in concert with a Clp-ATPase, proteins that repress or interfere with transcription of the massetolide-regulatory gene luxR(mA). To identify the cellular substrates and target proteins of the ClpP protease in P. fluorescens, an extensive proteomic analysis, as was performed previously for E. coli (25), will be required to support this hypothesis.
FIG. 8.
Proposed model for ClpP-mediated regulation of massetolide biosynthesis and swarming motility in P. fluorescens strain SS101. The darkly shaded arrows indicate interactions based on observations in this study; the lightly shaded arrows indicate putative interactions based on previous findings in other studies.
Another scenario for how ClpP may regulate massetolide biosynthesis is by influencing the citric acid cycle and amino acid metabolism (Fig. 8). In E. coli, ClpAP plays a role in the degradation of l-glutamate dehydrogenase (49), and ClpXP associates with the two principal enzymes (AceA and GlcB) of the glyoxylate shunt, which replenishes the pool of citric acid cycle intermediates (25). The results of other studies showed that the degradation rate of enzymes involved in amino acid metabolism was significantly reduced in a clpP mutant of B. subtilis (32). More specifically, one of the ClpP substrates in B. subtilis was PycA, a pyruvate carboxylase that catalyzes the conversion of pyruvate into oxaloacetate, which replenishes the citric acid cycle (32). For P. fluorescens SS101, preliminary results of Q-PCR analyses showed that the transcript levels of a pycA homologue are indeed significantly reduced (log RQ = −1.76) in the clpP mutant (data not shown). However, the role of this gene and other enzymes involved in the amino acid metabolism of P. fluorescens SS101, as well as their effects on massetolide biosynthesis and swarming motility, remain to be investigated. Assuming that ClpP adversely affects the citric acid cycle and amino acid metabolism in P. fluorescens SS101, it also may provide an explanation for the reduced growth observed for the clpP mutant. At higher temperatures, a condition known to increase the levels of misfolded proteins (33), growth was reduced in clpP mutants of Campylobacter jejuni, L. monocytogenes, and B. subtilis (11, 29, 52), but at regular temperatures, growth deficiencies were also observed for clpP mutants of E. coli, S. aureus, and P. aeruginosa (13, 63, 65). In this context, Chandu and Nandi (8) suggested that the ClpP protease degrades proteins, resulting in the release of amino acids that are subsequently recycled in the cellular pool and used for growth. For example, in E. coli, the growth deficiency of clpP mutant colonies was restored by the addition of CAA (13). For P. fluorescens SS101, however, growth of the clpP mutant was not restored by addition of CAA, suggesting that this effect may be strain specific. When the effects of individual amino acids were analyzed, the results of our study showed that glutamic acid and proline restored, in part, the swarming deficiency of the clpP mutant of strain SS101. The possibility that these amino acids may have served as building blocks for the nonribosomal peptide synthetases MassABC to synthesize the peptide moieties of the massetolide compounds seems unlikely. Although glutamic acid is a constituent of the massetolide compounds, proline is not (14). Furthermore, valine, leucine, and isoleucine, three amino acids in the peptide moieties of massetolides (14), did not complement the swarming deficiency in the clpP mutant and even adversely affected swarming in the wild-type strain SS101. Alternatively, glutamic acid and proline may have served as chemical signals that triggered, directly or indirectly, the expression of luxR(mA) and the mass biosynthesis genes, leading to a partial rescue of massetolide biosynthesis and swarming motility in the clpP mutant (Fig. 8). It is well known that specific amino acids, including glutamate and proline, can promote swarming in P. aeruginosa (44) and Proteus mirabilis (1) and act as a chemoattractant (1). Moreover, glutamine can serve as a signal for the cellular nitrogen state; in E. coli, glutamine is sensed by enzymes that trigger a signal transduction cascade that activates the glutamine synthase gene glnA (46). Also, exogenously provided proline can release the transcriptional repressor PutA from the proline utilization (put) genes (6, 71). These studies demonstrate that these amino acids can induce gene transcription.
Finally, we looked into the possible interplay between ClpP and the two-component regulatory system GacA/GacS (Fig. 8). In other systems, ClpP affects global regulation. For example, in S. aureus, the global regulator agr was repressed in the clpP mutant, which resulted in reduced alpha-toxin and extracellular protease activities (27, 50). Also in Bacillus, ClpP-dependent proteolysis is regulated in response to environmental signals (nutrients) and transmitted via the two-component signal transduction system ComK/ComS (28). For P. fluorescens SS101, gacS regulates transcription of the massABC and luxR(mA) genes and thereby massetolide production, but clpP transcription is not affected. Furthermore, expression of gacA/gacS was not affected in the clpP mutant, suggesting that, at the transcriptional level, ClpP-mediated regulation of massetolide biosynthesis is independent of regulation by GacA/GacS.
Supplementary Material
Acknowledgments
This work was funded by the Dutch Technology Foundation (STW), the applied science division of NWO, and Productschap Tuinbouw.
We thank Jorge de Souza, who initially isolated the clpP mutant of P. fluorescens. We are very grateful to Teresa Sweat and Joyce Loper (USDA, Corvallis, OR) and Dimitri Mavrodi (USDA, Pullman, WA) for providing plasmids (pEX18Tc and pPS854-Gm), protocols, and advice for the site-directed mutagenesis.
Footnotes
Published ahead of print on 29 December 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allison, C., H. C. Lai, D. Gygi, and C. Hughes. 1993. Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol. Microbiol. 853-60. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., B. Koch, T. H. Nielsen, D. Sorensen, M. Hansen, O. Nybroe, C. Christophersen, J. Sorensen, S. Molin, and M. Givskov. 2003. Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology 14937-46. [DOI] [PubMed] [Google Scholar]
- 3.Bartels, F., S. Fernandez, A. Holtel, K. N. Timmis, and V. de Lorenzo. 2001. The essential HupB and HupN proteins of Pseudomonas putida provide redundant and nonspecific DNA-bending functions. J. Biol. Chem. 27616641-16648. [DOI] [PubMed] [Google Scholar]
- 4.Berti, A. D., N. J. Greve, Q. H. Christensen, and M. G. Thomas. 2007. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 1896312-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brotz-Oesterhelt, H., D. Beyer, H. P. Kroll, R. Endermann, C. Ladel, W. Schroeder, B. Hinzen, S. Raddatz, H. Paulsen, K. Henninger, J. E. Bandow, H. G. Sahl, and H. Labischinski. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 111082-1087. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. D., and J. M. Wood. 1993. Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J. Biol. Chem. 2688972-8979. [PubMed] [Google Scholar]
- 7.Capestany, C. A., G. D. Tribble, K. Maeda, D. R. Demuth, and R. J. Lamont. 2008. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 1901436-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandu, D., and D. Nandi. 2004. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res. Microbiol. 155710-719. [DOI] [PubMed] [Google Scholar]
- 9.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64391-397. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. H., and H. P. Schweizer. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohn, M. T., H. Ingmer, F. Mulholland, K. Jorgensen, J. M. Wells, and L. Brondsted. 2007. Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 737803-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui, X., R. Harling, P. Mutch, and D. Darling. 2005. Identification of N-3-hydroxyoctanoyl-homoserine lactone production in Pseudomonas fluorescens 5064, pathogenic to broccoli, and controlling biosurfactant production by quorum sensing. Eur. J. Plant Pathol. 111297-308. [Google Scholar]
- 13.Damerau, K., and A. C. St John. 1993. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J. Bacteriol. 17553-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Bruijn, I., M. J. de Kock, P. de Waard, T. A. van Beek, and J. M. Raaijmakers. 2008. Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 1902777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Bruijn, I., M. J. D. de Kock, M. Yang, P. de Waard, T. A. van Beek, and J. M. Raaijmakers. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 63417-428. [DOI] [PubMed] [Google Scholar]
- 16.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 642710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Souza, J. T., M. De Boer, P. De Waard, T. A. Van Beek, and J. M. Raaijmakers. 2003. Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 697161-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 1492005-2013. [DOI] [PubMed] [Google Scholar]
- 19.Dougan, D. A., A. Mogk, and B. Bukau. 2002. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol. Life Sci. 591607-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubern, J.-F., E. L. Lagendijk, B. J. J. Lugtenberg, and G. V. Bloemberg. 2005. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J. Bacteriol. 1875967-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubern, J. F., and G. V. Bloemberg. 2006. Influence of environmental conditions on putisolvins I and II production in Pseudomonas putida strain PCL1445. FEMS Microbiol. Lett. 263169-175. [DOI] [PubMed] [Google Scholar]
- 22.Dubern, J. F., B. J. J. Lugtenberg, and G. V. Bloemberg. 2006. The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol. 1882898-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finking, R., and M. A. Marahiel. 2004. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58453-488. [DOI] [PubMed] [Google Scholar]
- 24.Fischer, G., T. Tradler, and T. Zarnt. 1998. The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett. 42617-20. [DOI] [PubMed] [Google Scholar]
- 25.Flynn, J. M., S. B. Neher, Y. I. Kim, R. T. Sauer, and T. A. Baker. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11671-683. [DOI] [PubMed] [Google Scholar]
- 26.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 541445-1462. [DOI] [PubMed] [Google Scholar]
- 27.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 481565-1578. [DOI] [PubMed] [Google Scholar]
- 28.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 631285-1295. [DOI] [PubMed] [Google Scholar]
- 29.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 351286-1294. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Fruitos, E., M. Martinez-Alonso, N. Gonzalez-Montalban, M. Valli, D. Mattanovich, and A. Villaverde. 2007. Divergent genetic control of protein solubility and conformational quality in Escherichia coli. J. Mol. Biol. 374195-205. [DOI] [PubMed] [Google Scholar]
- 31.Gerard, J., R. Lloyd, T. Barsby, P. Haden, M. T. Kelly, and R. J. Andersen. 1997. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60223-229. [DOI] [PubMed] [Google Scholar]
- 32.Gerth, U., H. Kock, I. Kusters, S. Michalik, R. L. Switzer, and M. Hecker. 2008. Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis. J. Bacteriol. 190321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30465-506. [DOI] [PubMed] [Google Scholar]
- 34.Gross, H., V. O. Stockwell, M. D. Henkels, B. Nowak-Thompson, J. E. Loper, and W. H. Gerwick. 2007. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 1453-63. [DOI] [PubMed] [Google Scholar]
- 35.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 2951852-1858. [DOI] [PubMed] [Google Scholar]
- 36.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13232-237. [DOI] [PubMed] [Google Scholar]
- 37.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 38.Ibrahim, Y. M., A. R. Kerr, N. A. Silva, and T. J. Mitchell. 2005. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect. Immun. 73730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 9623-28. [DOI] [PubMed] [Google Scholar]
- 40.Kinscherf, T. G., and D. K. Willis. 2002. Global regulation by gidA in Pseudomonas syringae. J. Bacteriol. 1842281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitten, T., T. G. Kinscherf, J. L. McEvoy, and D. K. Willis. 1998. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 28917-929. [DOI] [PubMed] [Google Scholar]
- 42.Koch, B., T. H. Nielsen, D. Sorensen, J. B. Andersen, C. Christophersen, S. Molin, M. Givskov, J. Sorensen, and O. Nybroe. 2002. Lipopeptide production in Pseudomonas sp. strain DSS73 is regulated by components of sugar beet seed exudate via the Gac two-component regulatory system. Appl. Environ. Microbiol. 684509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kock, H., U. Gerth, and M. Hecker. 2004. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 511087-1102. [DOI] [PubMed] [Google Scholar]
- 44.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 1825990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuiper, I., E. L. Lagendijk, R. Pickford, J. P. Derrick, G. E. M. Lamers, J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2004. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 5197-113. [DOI] [PubMed] [Google Scholar]
- 46.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61349-377. [DOI] [PubMed] [Google Scholar]
- 47.Lu, S. E., B. K. Scholz-Schroeder, and D. C. Gross. 2002. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 1543-53. [DOI] [PubMed] [Google Scholar]
- 48.Lu, S. E., N. Wang, J. Wang, Z. J. Chen, and D. C. Gross. 2005. Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol. Plant-Microbe Interact. 18324-333. [DOI] [PubMed] [Google Scholar]
- 49.Maurizi, M. R., and F. Rasulova. 2002. Degradation of l-glutamate dehydrogenase from Escherichia coli: allosteric regulation of enzyme stability. Arch. Biochem. Biophys. 397206-216. [DOI] [PubMed] [Google Scholar]
- 50.Michel, A., F. Agerer, C. R. Hauck, M. Herrmann, J. Ullrich, J. Hacker, and K. Ohlsen. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 1885783-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mogk, A., R. Schmidt, and B. Bukau. 2007. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 17165-172. [DOI] [PubMed] [Google Scholar]
- 52.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27899-914. [DOI] [PubMed] [Google Scholar]
- 53.Nakano, M. M., Y. Zhu, J. Liu, D. Y. Reyes, H. Yoshikawa, and P. Zuber. 2000. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase alpha can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol. 37869-884. [DOI] [PubMed] [Google Scholar]
- 54.Nybroe, O., and J. Sorensen. 2004. Production of cyclic lipopeptides by fluorescent pseudomonads, p. 147-172. In J.-L. Ramos (ed.), Pseudomonas, vol. 3. Biosynthesis of macromolecules and molecular metabolism. Kluwer Academic Publishers, New York, NY. [Google Scholar]
- 55.Ongena, M., and P. Jacques. 2008. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16115-125. [DOI] [PubMed] [Google Scholar]
- 56.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 57.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 31091-109. [DOI] [PubMed] [Google Scholar]
- 58.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32449-458. [DOI] [PubMed] [Google Scholar]
- 59.Quinones, B., G. Dulla, and S. E. Lindow. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant-Microbe Interact. 18682-693. [DOI] [PubMed] [Google Scholar]
- 60.Raaijmakers, J. M., I. De Bruijn, and M. J. D. De Kock. 2006. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant-Microbe Interact. 19699-710. [DOI] [PubMed] [Google Scholar]
- 61.Roongsawang, N., K. Hase, M. Haruki, T. Imanaka, M. Morikawa, and S. Kanaya. 2003. Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chem. Biol. 10869-880. [DOI] [PubMed] [Google Scholar]
- 62.Scholz-Schroeder, B. K., J. D. Soule, and D. C. Gross. 2003. The sypA, sypB, and sypC synthetase genes encode twenty-two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant-Microbe Interact. 16271-280. [DOI] [PubMed] [Google Scholar]
- 63.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 725027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran, H., A. Ficke, T. Asiimwe, M. Hofte, and J. M. Raaijmakers. 2007. Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens. New Phytol. 175731-742. [DOI] [PubMed] [Google Scholar]
- 65.Wang, C., M. Li, D. Dong, J. Wang, J. Ren, M. Otto, and Q. Gao. 2007. Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microbes Infect. 91376-1383. [DOI] [PubMed] [Google Scholar]
- 66.Wang, N., S. E. Lu, A. R. Records, and D. C. Gross. 2006. Characterization of the transcriptional activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J. Bacteriol. 1883290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193201-205. [DOI] [PubMed] [Google Scholar]
- 68.Wong, P., and W. A. Houry. 2004. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J. Struct. Biol. 14679-89. [DOI] [PubMed] [Google Scholar]
- 69.Yu, A. Y., and W. A. Houry. 2007. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 5813749-3757. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, J. H., N. B. Quigley, and D. C. Gross. 1995. Analysis of the syrB and syrC genes of Pseudomonas syringae pv. syringae indicates that syringomycin is synthesized by a thiotemplate mechanism. J. Bacteriol. 1774009-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, Y., W. Zhu, P. S. Bellur, D. Rewinkel, and D. F. Becker. 2008. Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli. Amino Acids 35711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.