Abstract
Mycoplasma mobile glides on solid surfaces by use of a unique mechanism that involves two large proteins, Gli349 and Gli521. Here we isolated and analyzed two antibodies and three mutants that modified mycoplasma gliding. Mapping of the target points of antibodies and mutations currently available suggested that a 301-amino-acid region on the whole 3,138-amino-acid sequence, a C-terminal region of Gli349, and an N-terminal region of Gli521 are directly involved in the movements of the gliding machinery.
Mycoplasma mobile forms a membrane protrusion at a cell pole and glides in the direction of the protrusion on solid surfaces by use of a unique mechanism (8-10, 13, 21). It glides smoothly on glass at an average speed of 2.0 to 4.5 μm/s (3, 4, 12, 14, 19). Previously, we identified huge proteins involved in this gliding mechanism (6, 17, 20, 22, 24), visualized the putative machinery and the binding protein (1, 11), and identified the direct energy source used and the direct binding target (5, 15, 23). The force generated by the gliding machinery may be supported from the inside of the cell by a cytoskeletal “jellyfish” structure (16). We then proposed a working model, called a “centipede” model, in which cells are propelled by “legs” composed of Gli349 repeatedly catching and releasing sialic acids fixed on the glass surface (2, 8, 9) and are driven by the force exerted by P42 through Gli521 molecules, supported by the jellyfish structure, based on the energy of ATP. In the present study, we isolated antibodies and mutants which influence gliding and analyzed these and previously isolated ones.
Effects of anti-Gli349 antibodies on binding and gliding.
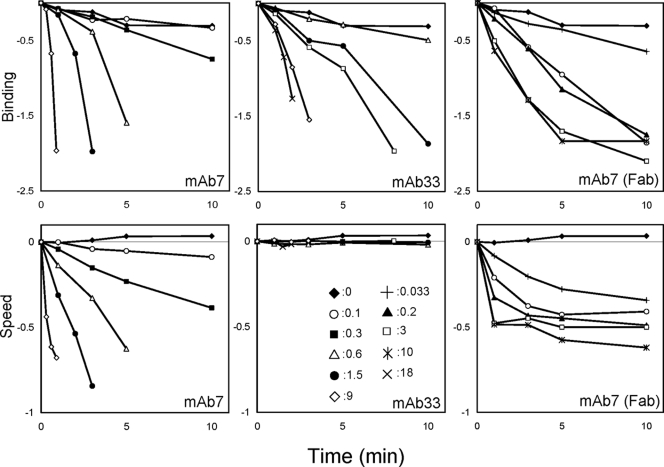
Previously, we isolated a monoclonal antibody (MAb), MAb7, against the leg protein Gli349 and analyzed its effects on mycoplasma gliding (6, 20, 22). In the present study, similar results were obtained (Fig. 1, left panels). MAb7 displaced gliding mycoplasmas from the glass in a concentration-dependent manner and also reduced the gliding speed. A mycoplasma cell generates a maximum force of 27 pN (12), which is 1,800 times larger than the force (15 fN) calculated to be necessary for the normal speed of mycoplasma movement (18). Considering this fact, the additional drag force should be considered. One possible scenario is that a conformation of Gli349 generates the drag force. Here, we isolated a new antibody, MAb33 (20); analyzed its effect on gliding, and found that this antibody displaced the gliding mycoplasmas from the glass without a reduction of speed prior to detachment (Fig. 1, center panels). This observation suggests that the putative drag in the inhibition by MAb7 is caused by Gli349, because other causes are unlikely to depend on the binding sites of anti-Gli349 MAbs.
FIG. 1.
Effects of anti-Gli349 antibodies on glass binding and gliding speed of wild-type cells. The effects on binding and speed are presented in the upper and lower panels, respectively. The antibodies and Fab were added at time zero. The numbers of bound cells in an area 280 μm square and their average gliding speeds are presented relative to the initial values on a logarithmic scale. The gliding speeds were analyzed for three consecutive intervals of 0.333 s by using Image J version 1.33v with the plug-in software MultiTracker ver1 (http://rsb.info.nih.gov/ij/plugins/multitracker.html) and are presented as the average for more than 40 cells on a logarithmic scale when sufficient numbers of cells were on glass. Antibody concentrations in mg/ml are as indicated in the central lower graph.
Since MAb7, classified as an immunoglobulin G type, has two binding sites (6), the drag by Gli349 may be caused by cross-linking effects. To examine this possibility, we prepared Fab of MAb7 using the ImmunoPure Fab preparation kit (Pierce). The Fab reduced both the binding and the gliding speed, suggesting that the reduction of speed observed for the immunoglobulin G is not caused by a cross-linking effect (Fig. 1, right panels).
Isolation and characterization of mutants resistant to MAb7.
Colonies of M. mobile have the ability to adsorb red blood cells (RBC) (hemadsorption [HA] activity), and this activity was blocked when an RBC suspension was mixed with MAb7 (22). Previously, we isolated an adhesive mutant, the gli521(P476R) mutant, by screening for a colony which can adsorb RBC in the presence of MAb7 (23). Here, we screened about 6,000 colonies for other spontaneous mutants with HA activity resistant to MAb7 and obtained 43 isolates. We sequenced the 30,469-bp DNA region encoding the gliding proteins, as described previously (20, 24), for eight strains and found that all of the mutants have a single nucleotide substitution, resulting in a single amino acid substitution (Table 1). Seven isolates had the same substitution, from serine to arginine at amino acid 859, among the entire 4,727-amino-acid sequence of the gli521 open reading frame. One of them was named the gli521(S859R) mutant and was used for further studies. Another isolate was analyzed and named the gli349(S1362W) mutant.
TABLE 1.
Antibodies and mutants
| Antibody or mutant | Binding activitya | Gliding speedb | Epitope or amino acid substitution in Gli349 or Gli521c | Nucleotide changed | Reference
|
|
|---|---|---|---|---|---|---|
| Isolation | Mapping | |||||
| Antibodies | ||||||
| MAb7 | Reduced | Reduced | 1193-1203/Gli349 | 6 | This study | |
| MAb3 | Reduced | No effect | 1477-1492/Gli349 | This study | This study | |
| MAbR19 | Reduced | Reduced | 3736-4020/Gli521 | 20 | 20 | |
| Mutants | ||||||
| m9 | None | None | E1670*/Gli521 | G19148T | 14 | 20 |
| m12 | None | None | Q523*/Gli123 | C2627T | 14 | 24 |
| m13 | None | None | Q1257*/Gli349 | C8305T | 14 | 22 |
| m23 | None | None | S2770L/Gli349 | C12845T | 14 | This study |
| m26 | None | None | 1228RPTA1229/Gli349 | 8208-8219 tandem repeate | 14 | This study |
| gli521(S859R) | 54% | 54% | S859R/Gli521 | T16717A | This study | This study |
| gli521(P476R) | 165% | 75% | P476R/Gli521 | C15567G | 23 | 23 |
| gli349(S1362W) | 59% | 108% | S1362W/Gli349 | C8621G | This study | This study |
Percentages refer to the cell density on glass compared to that of the wild-type strain.
Percentages refer to the gliding speed compared to that of the wild-type strain.
The protein fragments comprising amino acids 11 to 90, 109 to 223, 217 to 426, 421 to 821, 822 to 1021, 1024 to 1247, 1241 to 1608, 1581 to 1966, 1929 to 2380, 2374 to 2582, 2582 to 2703, 2685 to 2810, 2832 to 3066, 1159 to 1247, 1024 to 1182, 1024 to 1203, 1024 to 1212, 1024 to 1192, 1159 to 1342, 1242 to 1442, 1242 to 1522, 1242 to 1460, 1242 to 1476, 1242 to 1492, and 1242 to 1509 of the 3,183-amino-acid Gli349 protein were expressed in E. coli cells and analyzed by Western blotting. The total number of amino acids in translated Gli521 is 4,727, and processing gave 4,684. Asterisks indicate stop codons.
Numbering is as in the sequence under accession number AB084781.
An additional C18522T mutation was found but is silent in the amino acid sequence.
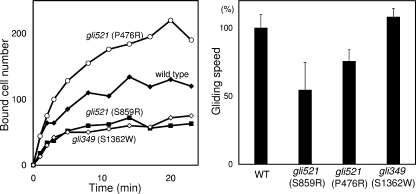
Next, we examined the binding activity and gliding speed of each strain, as described previously (15) (Fig. 2; Table 1). The cells inserted into the tunnel bound to the glass surfaces and then started gliding. The gli521(P476R) mutant showed increased binding activity, consistent with our previous observations (16, 23). M. mobile cells are known to bind to glass surfaces via sialic acids on sialoproteins fixed on the glass in the same way that they bind to animal cell surfaces (15). The observation in the present study suggests that the resistance of the HA activity of the gli521(P476R) mutant to MAb7 is caused by the elevated binding activity. In contrast, the gli521(S859R) and gli349(S1362W) mutants were less active than the wild-type strain in glass binding. The average speed of the gli521(P476R) mutant was 75% of that of the wild-type strain. The speeds of the gli521(S859R) and gli349(S1362W) mutants were 54% and 108% of that of the wild-type strain, respectively.
FIG. 2.
Binding and gliding properties of each strain. (Left) The cell suspension was inserted into a tunnel slide at time zero, and the number of bound cells in an area 280 μm square was counted at various time points. (Right) Speeds were averaged for more than 40 cells and are presented with standard deviations. WT, wild type.
The sensitivity of individual gliding mycoplasmas to MAb7 was examined for these strains. The reduction of both binding and gliding speed occurred for all strains in a similar concentration-dependent manner, showing that the resistance of HA activity to MAb7 was not caused by the reduced binding affinity of the antibody to the mutated proteins. In terms of glass binding, the gli521(P476R) mutant was more resistant to MAb7 than the wild-type strain. However, the decrease in bound cell number was more obvious in the gli521(S859R) and gli349(S1362W) mutants. Electrophoretic and Western blot analyses showed that the characteristics of the mutants are not caused by the change in amounts of gliding proteins or the change in reactivity to MAb7. What, then, causes the resistance of the HA activity of the gli521 (S859R) and gli349 (S1362W) mutants to MAb7? In the experiments described above, the effects were examined in the transient state, after the addition of MAb7. However, in the HA assay, the antibody was added before the mycoplasmas encountered RBC, and the binding was analyzed after a sufficient time for equilibrium to be reached. Therefore, we examined the behavior of individual cells in the presence of MAb7 and found that the binding of individual cells is resistant to MAb7 much more for those three mutants than for the wild type under such conditions (see Fig. S4 in the supplemental material).
Target points of antibody and nonbinding mutants on gliding proteins.
We expressed 25 protein fragments of the 3,183-amino-acid Gli349 protein in Escherichia coli cells, performed Western blotting, and determined the binding regions of MAb7 and MAb33 (Table 1).
Previously, we isolated five nonbinding and nongliding mutants, named m9, m12, m13, m23, and m26 (Table 1) (14). Mutants m9, m12, and m13 lack the Gli521, Gli123, and Gli349 proteins, respectively, due to a nonsense mutation in the N-terminal positions of their open reading frames (20, 22, 24). In the present study, we sequenced the 30,469-bp DNA region encoding the gliding proteins and found that the m23 mutant had a substitution and the m26 mutant had a tandem duplication nucleotide sequence, resulting in the insertion of the amino acid sequence RPTA at the C-terminal side of amino acid 1228, alanine. As we sequenced only the 30,469-bp region of the genome, additional mutations in other regions in those mutants cannot be ruled out. However, we assumed that the mutant phenotypes are caused by the mutations identified here, because so far we have sequenced the 30,469-bp DNA region for 14 mutants and found that only the m26 mutant possesses an additional mutation.
Hot spots on Gli349 and Gli521 molecules.
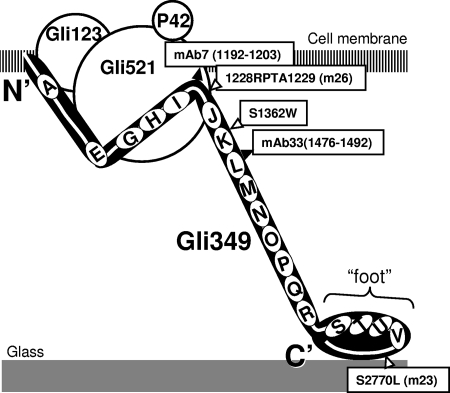
The Gli349 protein plausibly plays a role as a leg. In the present study, we mapped the binding points of two inhibitory antibodies and three mutation points affecting gliding on Gli349, and we found that four target points are located in a 301-amino-acid region comprising amino acids 1193 to 1492. Inhibition of gliding by the antibodies suggests that this region should be exposed to the outside, and the conformational changes of this domain are essential for the mechanism. Direct involvement of this region in the gliding mechanism is supported by the isolation of the m26 and gli349(S1362W) mutants. Previously, we assigned the amino acid sequence to the molecular structure of the Gli349 protein (Fig. 3) (1, 7). According to this assumption, the 301-amino-acid region including those four target points is predicted to form a 12-nm part at the proximal end of the leg structure. On the other hand, m23, a binding-defective mutant, has a mutation at amino acid 2770 of Gli349. The C-terminal region has been assigned to the foot in the schematic image, suggesting that the foot is responsible for catching the binding target, sialyllactose. This assumption is consistent with the position of the mutation in the m23 mutant.
FIG. 3.
Antibody binding sites and mutation points shown on a schematic of a Gli349 molecule. The gliding machinery is composed of the Gli123, Gli349, Gli521, and P42 proteins. The molecular shape of Gli349 was suggested from electron microscopy studies to be formed by three rods and one oval “foot,” which are tandemly connected by three hinges (1). A transmembrane segment is predicted for amino acids 9 to 31 (22). Repeat sequences of 100 amino acids with weak similarity are shown by ovals as follows: A, 118 to 222; E, 616 to 727; G, 830 to 938; H, 944 to 1047; I, 1048 to 1161; J, 1248 to 1343; K, 1344 to 1449; L, 1450 to 1546; M, 1553 to 1657; N, 1658 to 1762; O, 1765 to 1872; P, 1873 to 1972; Q, 1974 to 2080; R, 2084 to 2191; S, 2286 to 2391; T, 2396 to 2501; U, 2515 to 2608; and V, 2610 to 2720 (7). The binding sites of MAb7 and MAb33, comprised of regions 1193 to 1203 and 1477 to 1492, are shown by closed triangles. The mutation points of m26, the gli349(S1362W) mutant, and m23 at amino acids 1228, 1362, and 2770 are shown by open triangles.
We found that two mutants with HA activity resistant to MAb7 have a mutation in the N-terminal region of Gli521, at amino acids 476 (23) and 859, respectively, suggesting that Gli521 interaction with Gli349 involves this region.
Working model.
All effects caused by each antibody and mutation can be qualitatively explained by the assignment of these effectors to a single step of our working model (8, 9), as postulated in Fig. S5 in the supplemental material.
Supplementary Material
Acknowledgments
We thank George Oster and Jing Chen at UC Berkeley for helpful discussions.
This work was supported by a Grant-in-Aid for Scientific Research (A) and a Grant-in-Aid for Scientific Research on the Priority Areas “Applied Genomics” and “Structures of Biological Macromolecular Assemblies” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.M.) and by a grant from the Institution for Fermentation Osaka (to M.M.).
Footnotes
Published ahead of print on 5 January 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adan-Kubo, J., A. Uenoyama, T. Arata, and M. Miyata. 2006. Morphology of isolated Gli349, a leg protein responsible for glass binding of Mycoplasma mobile gliding revealed by rotary-shadowing electron microscopy. J. Bacteriol. 1882821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charon, N. W. 2005. Mycoplasma takes a walk. Proc. Natl. Acad. Sci. USA 10213713-13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiratsuka, Y., M. Miyata, T. Tada, and T. Q. P. Uyeda. 2006. A microrotary motor powered by bacteria. Proc. Natl. Acad. Sci. USA 10313618-13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiratsuka, Y., M. Miyata, and T. Q. P. Uyeda. 2005. Living microtransporter by uni-directional gliding of Mycoplasma along microtracks. Biochem. Biophys. Res. Commun. 331318-324. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 1864254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 1504001-4008. [DOI] [PubMed] [Google Scholar]
- 7.Metsugi, S., A. Uenoyama, J. Adan-Kubo, M. Miyata, K. Yura, H. Kono, and N. Go. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 133-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyata, M. 2008. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 166-12. [DOI] [PubMed] [Google Scholar]
- 9.Miyata, M. 2007. Molecular mechanism of mycoplasma gliding—a novel cell motility system, p. 137-175. In P. Lenz (ed.), Cell motility. Springer, New York, NY.
- 10.Miyata, M., and H. Ogaki. 2006. Cytoskeleton of mollicutes. J. Mol. Microbiol. Biotechnol. 11256-264. [DOI] [PubMed] [Google Scholar]
- 11.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 1864382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 1841827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215285-289. [DOI] [PubMed] [Google Scholar]
- 14.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 1461311-1320. [DOI] [PubMed] [Google Scholar]
- 15.Nagai, R., and M. Miyata. 2006. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J. Bacteriol. 1886469-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakane, D., and M. Miyata. 2007. Cytoskeletal “jellyfish” structure of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 10419518-19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohtani, N., and M. Miyata. 2007. Identification of a novel nucleoside triphosphatase from Mycoplasma mobile: a prime candidate for motor of gliding motility. Biochem. J. 40371-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosengarten, R., M. Fisher, H. Kirchhoff, G. Kerlen, and K.-H. Seack. 1988. Transport of erythrocytes by gliding cells of Mycoplasma mobile 163K. Curr. Microbiol. 16253-257. [Google Scholar]
- 19.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 1691891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 1873502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44431-434. [DOI] [PubMed] [Google Scholar]
- 22.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 1861537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uenoyama, A., and M. Miyata. 2005. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. USA 10212754-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 1875578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.