Abstract
Gene delivery from tissue engineering scaffolds has potential to promote localized transgene expression that can induce the formation of functional tissues. Substrate-mediated delivery, an alternative delivery strategy to sustained release, is based on immobilization of DNA complexes to the polymer surface for subsequent delivery to cells cultured on the substrate. We investigate polyethylenimine (PEI)/DNA complex immobilization and subsequent cellular transfection on tissue engineering scaffolds fabricated from poly(lactide-co-glycolide) (PLG). The properties of the substrate and the complex affect both immobilization and cellular transfection. PLG promotes binding of PEI/ DNA complexes, with percent bound independent of the N/P ratio or the DNA dosage. The levels of transgene expression are similar to or greater than control studies based on bolus DNA delivery, with orders of magnitude less DNA. Immobilization also homogeneously distributes the DNA throughout the scaffold, resulting in large numbers of transfected cells (>60%) at low surface quantities (<50 ng). Importantly, this approach can be employed to transfect cells throughout a three-dimensional scaffold. Tissue engineering scaffolds that are prefabricated into various shapes from a range of materials could potentially employ this strategy for numerous applications.
Keywords: gene therapy, plasmid, non-viral vector, tissue, polyethyleneimine
INTRODUCTION
The combination of biomaterials, gene therapy, and tissue engineering has the potential to create synthetic environments that provide the necessary signals to promote the formation of functional tissues. Biomaterials serve a central role in many tissue engineering approaches by providing a physical support for cell adhesion and a template for tissue formation.1 A variety of materials, both natural (e.g., collagen, hyaluronic acid) and synthetic (e.g., poly(ethylene glycol), poly(lactide-co-glycolide)), have been employed as scaffolds for new tissue formation. The bioactivity of these scaffolds may be enhanced by employing the scaffold as a vehicle for efficient gene delivery, which can induce the localized expression of tissue-inductive factors. The most frequent combination of gene therapy and tissue engineering has involved the transplantation of ex vivo genetically engineered cells, which has been applied to numerous tissues, such as bone, cartilage, and nerve.2–4 Nevertheless, approaches based on the direct delivery of viral and nonviral vectors may provide alternate approaches that avoid the need for ex vivo cell manipulation and transplantation.
Sustained release of plasmids or nonviral complexes from tissue engineering scaffolds has shown the ability to promote prolonged transgene expression and induce tissue formation. Sustained DNA release has been proposed to increase or maintain the concentration of DNA in the cellular microenvironment, which may enhance gene transfer. Plasmid or DNA complexes are encapsulated within the polymer during fabrication. Localized plasmid release from tissue engineering scaffolds can promote gene expression for over 105 days,5 and has been able to promote tissue formation, such as angiogenesis and bone formation.5,6 Plasmid-releasing scaffolds have limited functionality, in vitro involve large quantities of plasmid for in vivo applications, and require procedures for encapsulation that do not substantially decrease the activity of the vector.6–9 DNA complexes can alternatively be incorporated for localized delivery and similarly promote tissue formation.10,11 The amount of plasmid is often decreased by complexation with a cationic polymer or lipid, yet the processing strategies must be considered.
An alternative approach to biomaterial-based gene delivery involves adsorption of DNA complexes to the surface.12 Plasmid may be precomplexed with the cationic lipid or polymer before immobilization, or the plasmid and transfection reagent can be sequentially deposited.13–15 In this approach, the objective is to maintain the DNA at or near the material surface to promote internalization by cells that adhere to the scaffold. In vitro studies have shown that surface immobilization significantly reduces the quantity of DNA required relative to bolus delivery. This immobilization of the vector is dependent upon the molecular scale inter-actions between the vector and the substrate, and these interactions must be appropriately balanced to support immobilization yet allowing for cellular internalization. The potential exists to apply this delivery system to traditional biomaterials used as tissue engineering scaffolds. Numerous approaches have been developed, which enable scaffolds to be fabricated into three-dimensional structures that support tissue formation; however, DNA cannot be incorporated into all scaffolds because of degradation caused by the processing conditions. Substrate-immobilization may enable DNA delivery from these scaffolds as the DNA can be immobilized following scaffold fabrication.
The objective of this study was to investigate gene delivery by immobilization of preformed nonviral vectors to the surface of tissue engineering scaffolds. We have previously demonstrated the successful immobilization and subsequent cellular transfection, using tissue culture polystyrene as the substrate.13 Tissue engineering scaffolds were fabricated from poly(lactide-co-glycolide) (PLG), a commonly used material for tissue engineering.10,16 Plasmid DNA was complexed with polyethylenimine (PEI) and complexes were deposited directly onto the scaffold after fabrication, thus avoiding any deactivation of the complexes by the processing. The properties of the complexes, surface properties of PLG, and the deposition time were investigated for their effects on DNA immobilization, surface stability, and transfection in vitro. Furthermore, transfection throughout the three-dimensional scaffold is investigated using this surface-delivery method, which would enable DNA delivery from scaffolds prefabricated into desirable geometries.
MATERIALS AND METHODS
Fabrication of polymer scaffolds
PLG (75:25 mol ratio of d,l-lactide to glycolide, i.v. = 0.6 – 0.8 dL/g) (Alkermes, OH) disks and porous scaffolds were fabricated using a previously described gas foaming process.17 Microspheres were made with a primary emulsion technique (w/o) and used as building blocks for disk and scaffold fabrication. For disk fabrication, microspheres (10 mg) were loaded into a cylindrical stainless steel die (inner diameter 5 mm) and compression molded (10 s at 1500 psi) using a Carver laboratory press (Carver, Muncie, IN). For porous scaffold fabrication, microspheres (7 mg) were mixed with a porogen (NaCl, 220 mg), loaded into a cylindrical die, and compression molded. The molded disks and scaffolds were then equilibrated with CO2 (800 psi) for 16 h in a custom-made pressure vessel. Reducing the pressure results in microsphere fusion to make intact polymer constructs. The scaffolds were immersed in water for 4 h to leach the salt. Polymer constructs were stored in a vacuum dessicator until use.
DNA complex immobilization
DNA complexes were formed by mixing plasmid with branched PEI (25 kDa) (Aldrich Chemical Company, Milwaukee, WI). The initial amount of PEI mixed with plasmid was varied to control the ratio of nitrogen/phosphate (N/P = 6, 9, and 18). PEI was dissolved in sodium bicarbonate (1 mM, pH 8.0) and diluted (100 µL final volume) with 150 mM sodium chloride solution, which was added to 100 µL of the plasmid DNA. Complexes were subsequently incubated at room temperature for 10 min before immobilization (substrate-mediated) or addition to culture media (bolus).
Polymer disks and scaffolds were coated with 10% fetal bovine serum (FBS) in PBS by incubating polymers with FBS at 37°C for 24 h before complex adsorption. Polymer disks were attached to tissue culture plates (96-well) using autoclaved silicon grease. For two-dimensional nonporous disks (diameter = 5 mm), DNA complexes (200 µL) were incubated (1, 4, 24 h) and washed twice with PBS. The surface-bound complexes were visualized by adsorbing complexes containing fluorescently-tagged DNA (Rhodamine labeling kit, Mirus Bio, WI). HEK293T (20,000 cells/well) or NIH3T3 (10,000 cells/well) in culture media (DMEM with 10% (v/v) FBS, 1% (v/v) penicillin streptomycin, and 1% (v/v) sodium pyruvate) were seeded and cultured on the PLG at 5% CO2, 37°C, and 95% humidity for 48 h. For bolus delivery, PEI/ DNA complexes were directly added to the culture media above the adhered cells.
For three-dimensional porous scaffolds (diameter = 5 mm, height = 6.5 mm), serum-coated and PBS-washed scaffolds were dried on gauze pads before incubation with DNA complexes. PEI/DNA complexes (1 mL of 50 µg in 150 mM NaCl) were lyophilized in the presence of 20 mM sucrose. Complexes were rehydrated in 100 µL of sterile deionized water and added to the porous scaffold, which were then incubated for 1 h. After washing twice with PBS, HEK293T cell suspension (60 µL of 5 × 105 cells/mL) was subsequently added to the porous scaffold, incubated for 4 h (5% CO2, 37°C), and then immersed in media (2 mL) for 48-h culture.
Quantification of surface-bound complexes
The quantity of DNA immobilized to the polymer was determined using radiolabeled DNA. Plasmid DNA was radiolabeled with a α-32P dATP using a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NJ), according to the manufacturer’s protocol with minor modifications as described in Segura et al.14 PEI/DNA complexes were formed using 32P DNA and adsorbed onto the PLG disks. At the specified times (1, 4, and 24 h), the supernatant was removed and the disks were washed twice with PBS. Subsequently, the disks were placed in scintillation cocktail (5 mL; ScintiVerse II) and the radioactivity was measured with a scintillation counter. The amount of DNA was calculated using a calibration curve.
Transfection analysis
The extent of transgene expression and the number of transfected cells were determined using the reporter genes luciferase (transgene expression) and β-galactosidase (number transfected). After 48-h culture, luciferase levels were measured on a luminometer using the luciferase assay system (Promega, WI), with levels normalized to the initial cell seeding. Alternatively, the expression of β-galactosidase was visualized by staining with X-gal solution, and the number of transfected cells was determined by counting 5 random fields on the surface (nonporous disks). The number of metabolically active cells (MTT staining) on PLG disks was analyzed to characterize the percentage of viable cells. The number of MTT stained cells was divided by the number of Hoechst stained nuclei to calculate a percentage of viable cells. To determine the total cell number, cells were trypsinized and counted using a hemocytometer. The efficiency of cell removal from the polymer was examined by incubation with Hoechst 33258 to label the nuclei of any remaining cells, which could be manually counted. The number of remaining cells was less than 0.5% of the total cell number. The fraction of transfected cells was determined as the number of transfected cells divided by the total cell number. All experimental conditions were performed in triplicate.
RESULTS
Immobilization of PEI/DNA complexes on PLG substrate
Incubation of PEI/DNA complexes with PLG substrates resulted in adsorption across the substrate, with the size of the complexes determined by the surface properties. Complexes adsorbed directly to PLG gradually aggregated with increasing deposition time [Fig. 1(a)]. Precoating PLG with serum before complex adsorption resulted in consistent complex sizes regardless of deposition time [Fig. 1(b)]. Preliminary evaluation of gene transfer in vitro by immobilizing DNA to uncoated and serum-coated scaffolds indicated that transfection was significantly higher on serum-coated scaffolds [Fig. 1(c,d)]. Note that precoating of surfaces with fibronectin, collagen, and laminin proteins supports transfection similar to serum (data not shown). We subsequently focused on DNA complex immobilization, surface stability, and in vitro transfection from serum-coated scaffolds.
Figure 1.
Adsorption and release of DNA complexes on two-dimensional PLG disks and cellular transfection. Fluorescence photomicrograph of immobilized PEI/DNA complexes (N/P = 9, 24 h deposition) on (a) untreated PLG or (b) serum-coated PLG. The scale bar indicates 50 µm. X-gal staining of transfected cells (HEK293T) by immobilized complexes (N/P = 9, 24 h deposition) on noncoated (c), and FBS-coated PLG disks (d). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
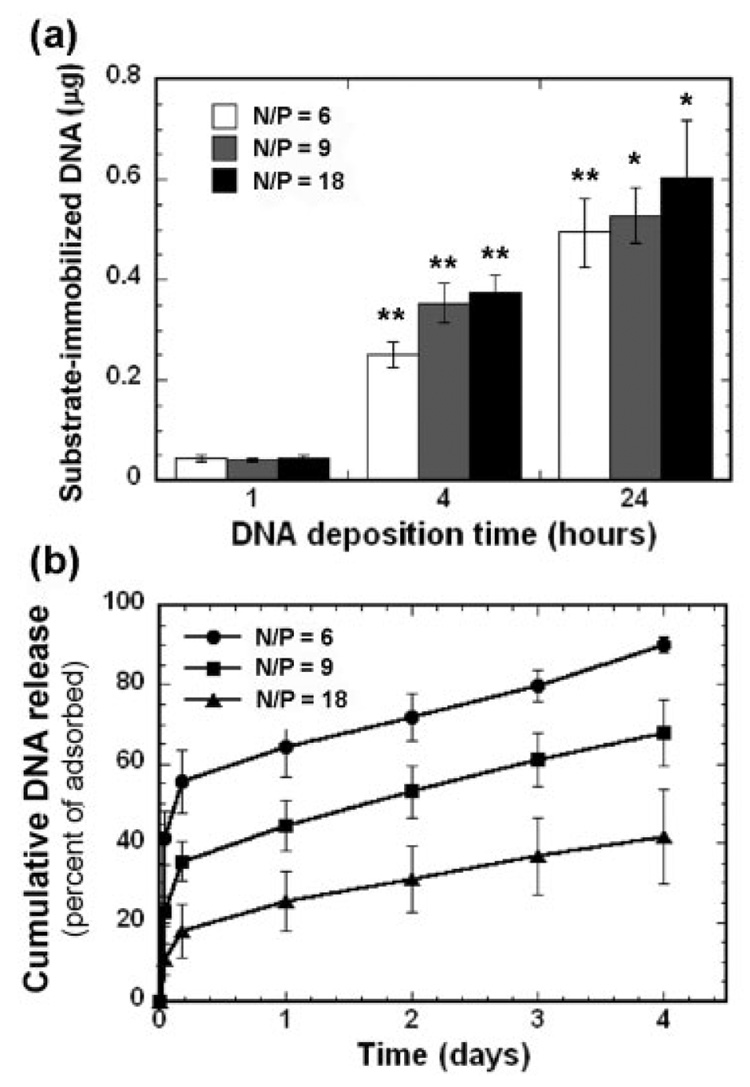
The quantity of immobilized complexes on serum-coated PLG substrate increased with the time of deposition, and were retained on the scaffold for time scales of the order of days. DNA was complexed with PEI at three N/P ratios (6, 9, 18) before adsorption, with N/P defined as the ratio of amines in PEI to phosphates in DNA. Increasing the deposition time from 1 h to 24 h increased the amount immobilized to the surface after washing, ranging from ~2.2 to 27.5% of the incubated DNA (i.e., 2 µg) [Fig. 2(a)]. For 2 µg of DNA incubated with the polymer, the mean quantity of immobilized PEI/DNA complexes for the deposition times of 1, 4, and 24 h was 45, 317, and 544 ng, respectively. Increasing the amount of complexes incubated with the scaffold to 5 µg increased the total amount immobilized, with the percent immobilized unchanged (data not shown). Although the N/P ratio did not affect adsorption, a gradual desorption was observed and dependent upon the N/P ratio of the complex. An increasing N/P ratio resulted in less dissociation of complexes from the polymer [Fig. 2(b)]. After an initial rapid desorption ranging from 17.8 to 55.5% during the first 4 h, a steady loss occurred during the subsequent 4 days, at a rate of 7.3% per day for all conditions.
Figure 2.
Quantity of immobilized PEI/DNA complexes at various N/P ratios equal to 6, 9, and 18 (a). Cumulative release of adsorbed complexes (24 h deposition) from PLG disks (b). Percentage of adsorbed complexes was defined as the ratio of dissociated DNA quantities to the initial amount of immobilized DNA. Data are presented as the average ± SD. The symbols * and ** indicate statistically significant differences relative to the previous deposition time at p < 0.05 and p < 0.001, respectively.
Cellular transfection on PLG substrate
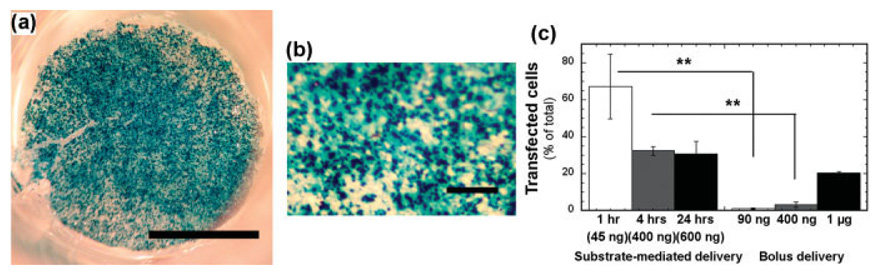
Substrate-mediated delivery substantially increased the number of transfected cells relative to bolus delivery, while requiring significantly less DNA. Large numbers of transfected cells were seen homogeneously across the PLG [Fig. 3(a,b)]. The percentage of transfected HEK293T cells by surface-bound complexes (N/P = 9) decreased from (67.1 ± 17.5)% to (30.7 ± 6.7)%, as the deposition time increased from 1 to 24 h [Fig. 3(c)]. Maximal numbers of transfected cells were observed with the lowest quantity of surface-bound complexes (45 ng), which is an order of magnitude less DNA required to achieve the highest levels of transfection by bolus delivery. For bolus delivery, addition of 1 µg of DNA was able to achieve transfection of ~20% of the cell population.
Figure 3.
Cellular transfection of HEK293T by immobilized PEI/DNA complexes. (a) X-gal staining of transfected cells by immobilized complexes (N/P = 9, 1 h deposition). The scale bar indicates 2.5 mm. (b) Enlarged view of transfected cells with high magnification. (c) Percentage of transfected cells by substrate-mediated and bolus delivery of complexes (N/P = 9). For substrate-mediated delivery, the deposition time and approximate DNA quantities (in parentheses) are shown. For bolus delivery, PEI/DNA complexes were formed under the same conditions (2 µg/200 µL) used for substrate-mediated delivery, and then the specific quantity (90 ng, 400 ng, and 1 µg) was directly added to the media above cultured cells. The symbol ** indicates statistical significance at p < 0.001. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
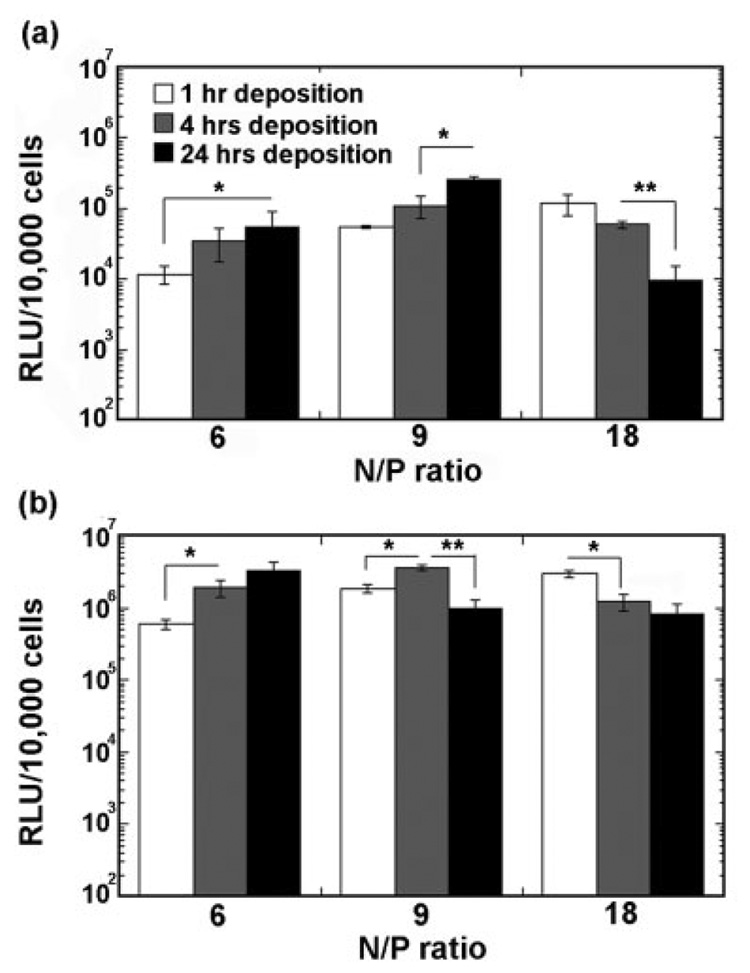
Maximal transgene expression was dependent upon an appropriate balance between the N/P ratio and the amount of immobilized DNA, which is regulated by the deposition time [Fig. 4(a,b)]. At the lowest N/P ratio, transgene expression increased with the deposition time. However, for N/P equal to 18, transgene expression decreased with an increase in deposition time. At the intermediate N/P ratio of 9, the relationship between transgene expression and deposition time was a function of cell type. For NIH/3T3 cells, transgene expression increased with an increasing deposition time, whereas HEK293T cells demonstrated a biphasic relationship between transgene expression and deposition time. At short incubation times, increasing the N/P ratio increased transgene expression. However, for the intermediate and long deposition times, transgene expression was maximal for N/P ratio equal to 9. The N/P ratio of the complex, which affected the stability at the surface, also influenced the transgene expression.
Figure 4.
Transgene expression by immobilized PEI/DNA complexes and by bolus delivery. (a) and (b) Transgene expression by substrate-mediated delivery for NIH/3T3 and HEK293T, respectively, as a function of the N/P ratio and deposition time. The symbols * and ** indicate significant differences at p < 0.05 and p < 0.001, respectively.
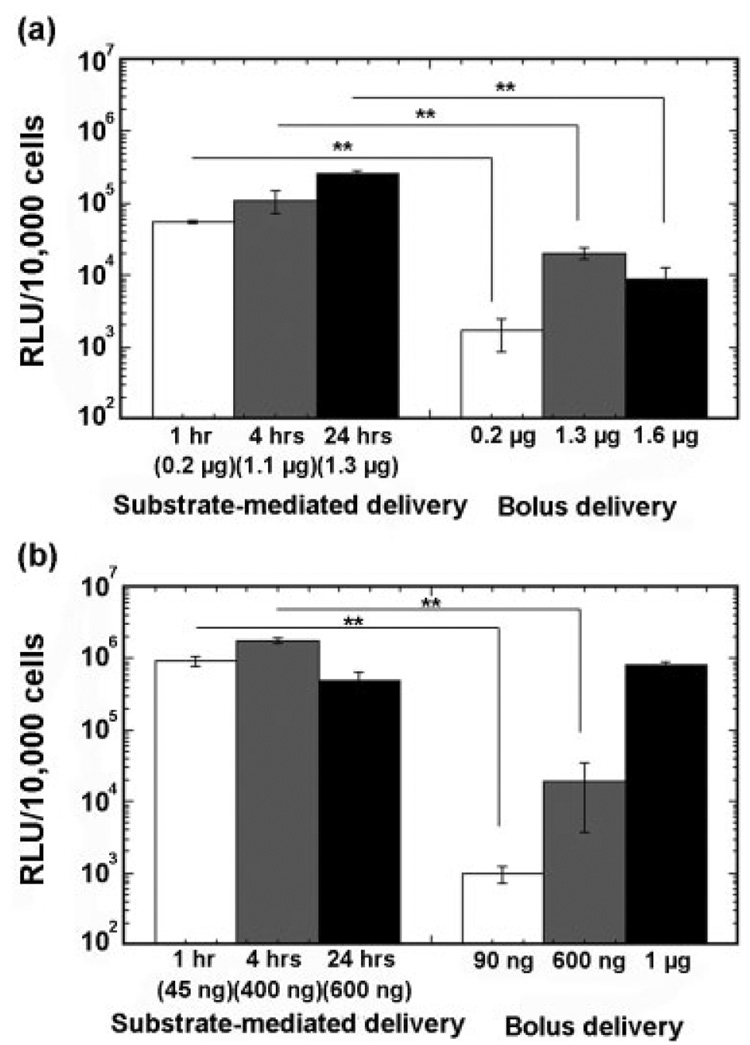
Delivery by immobilization to the substrate enhanced transgene expression by 1–3 orders of magnitude relative to bolus delivery of similar DNA quantities (Fig. 5). For NIH/3T3 cells, transfection by bolus delivery of complexes at quantities that are comparable with surface quantities resulted in approximately an order of magnitude less expression relative to delivery from the substrate [Fig. 5(a)]. Larger quantities of PEI/DNA complexes delivered in solution can produce comparable transgene expression to those observed with surface delivery. For HEK293T cells, the highest doses of DNA delivered as a bolus produced transgene expression that was comparable with transfection from the surface [Fig. 5(b)]. An important issue for the ultimate use of DNA immobilized scaffolds is the maintenance of activity during long-term storage. Scaffolds with immobilized DNA complexes maintained their transgene expression levels following lyophilization in the presence of sugars, such as sucrose or lactose (data not shown).
Figure 5.
Comparison of transgene expression by bolus and substrate-mediated delivery at similar DNA quantities (N/P = 9) for NIH/3T3 (a) and HEK293T (b), respectively. The symbol ** indicates significant differences at p < 0.001.
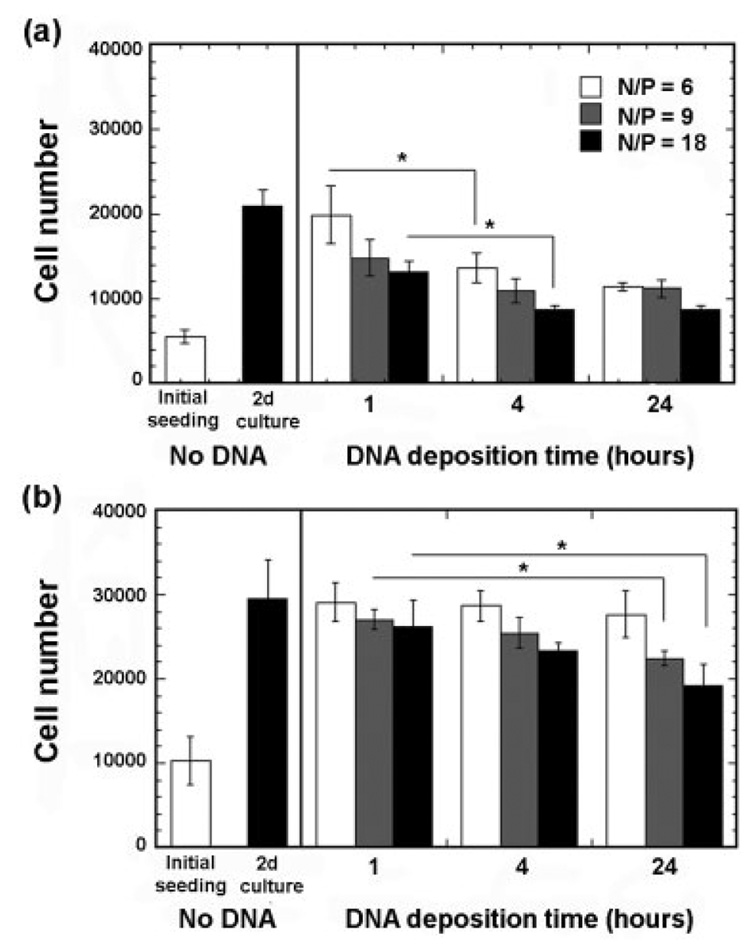
Cell growth with surface-bound PEI/DNA complexes
The quantity of surface-immobilized DNA was regulated through the amount incubated and the deposition time of complexes, and increasing doses led to a decrease in the percentage of transfected cells [Fig. 3(c)]. To examine this phenomenon, the cell number on the polymer was examined. The seeding efficiency, which was defined as the ratio of cells remaining after 3-h culture to cells initially seeded, was 55.8 and 51.7% for NIH/3T3 and HEK293T, respectively [Fig. 6(a,b)]. The percentage of viable cells on the disk was determined using an MTT stain, which indicated that more than 90% of the adhered cells were viable (not shown). For all conditions, the number of cells on the polymer increased relative to the initial number seeded during the 48-h culture; however, longer deposition times and increased N/P ratios produced smaller increases in cell number.
Figure 6.
Cellular proliferation after 48-h culture on PLG disks with immobilized complexes. (a) NIH/3T3 cells were seeded at a density of 10,000 cells/well. (b) HEK293T cells were seeded at a density of 20,000 cells/well. The symbol * indicates significant differences at p < 0.05.
Transfection on three-dimensional tissue engineering scaffolds
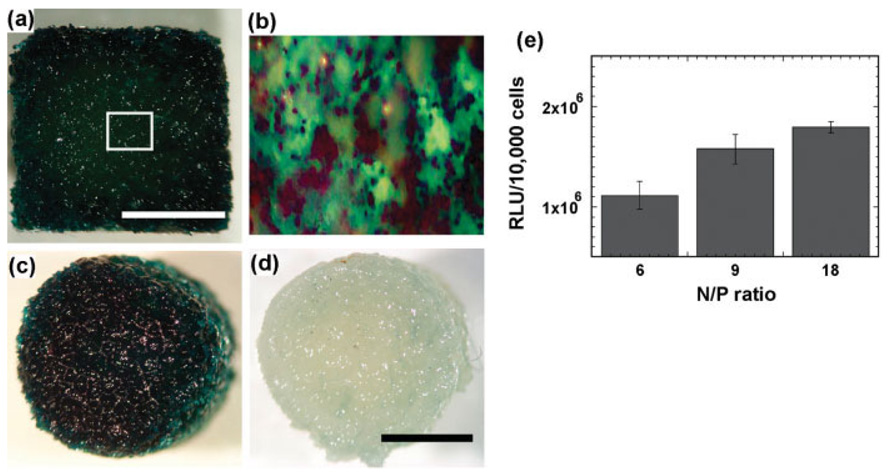
Substrate-mediated delivery can be readily applied to highly porous three-dimensional tissue engineering scaffolds. The volume in which the complexes were prepared, the quantity of DNA, and the deposition time (1h) were manipulated to achieve a homogeneous distribution of DNA immobilized throughout the scaffold. Seeding of HEK293T cells resulted in transfected cells throughout the scaffold [Fig. 7(a–c)], which was not observed on control scaffolds (no plasmid) [Fig. 7(d)]. Transgene expression on the porous scaffold was comparable with that observed for cells cultured on two-dimensional disks. Also consistent with the disk culture, average levels of transgene expression increased with an increasing N/P ratio [Fig. 7(e)].
Figure 7.
Transgene expression by complexes adsorbed within porous PLG scaffolds. Complexes were formed at N/P ratios of 6, 9, 18, and then were deposited on the scaffolds for 1 h before seeding HEK293T cells. X-gal staining of HEK293T cells seeded within the scaffold containing (a), (b), and (c) DNA encoding for β-galactosidase or (d) no DNA. Panels (a) and (b) provide a cross-sectional view, whereas panels (c) and (d) indicate a top view of the scaffold. Scale bars indicate 2.5 mm. (e) Transgene expression by immobilized complexes (initial loading: ~50 µg) formed at N/P ratios = 6, 9, 18. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
This report describes the application of a substrate-mediated DNA delivery approach to tissue engineering scaffolds. Tissue engineering scaffolds fabricated from various synthetic polymers are fabricated into highly porous structures, with the pore structure enabling cell seeding and transplantation and also cellular infiltration from the surrounding tissue for integration with the host. The pore structure also provides a large surface area for adsorption of DNA complexes, and complexes can readily be distributed throughout the scaffold. Since complex adsorption was performed after completing scaffold processing, this delivery strategy can be applied to scaffolds fabricated by various methods, such as electrospinning, gas foaming, solvent casting, and phase separation, without damaging the activity of the complexes. Adsorption of DNA complexes can increase the bioactivity of these scaffolds, by inducing transgene expression within the scaffold.
Substrate-mediated delivery from PLG scaffolds achieved similar or higher levels of transgene expression as bolus delivery with lower quantities of DNA, and was able to increase the number of transfected cells relative to bolus. Immobilization of DNA to the biomaterial places DNA directly in the cellular microenvironment, which has been shown to enhance gene transfer.18,19 The amount of surface immobilized complexes is relatively low, and the release of complexes into the media is undesirable as these complexes are unable to transfect cells in vitro.14 Cell adhesion to the substrate on which the complexes are adsorbed does promote transgene expression, likely by overcoming limitations associated with mass transport or complex aggregation. Polymeric release strategies have been successfully employed with both naked DNA and DNA complexes, yet require processing strategies that do not reduce activity of the complexes.5,6 Transgene expression does require large quantities of DNA (>100 µg), yet can induce transgene expression for months in vivo. In this report, substrate-mediated delivery requires low quantities of DNA on the surface that are lost over the course of days, which would not be expected to provide long-term expression in vivo without additional strategies for increasing the quantity bound and maintaining complexes at the surface. Alternatively, these scaffolds could be used for cell transplantation, with the immobilized DNA targeting the transplanted cells, which could reduce the ex vivo manipulation of the cells.
A variety of materials have been employed as substrates for complex immobilization, including TCPS, hydrogels, and synthetic polymers such as PLG13,14,20 and the properties of the substrate influence DNA adsorption and transfection. DNA complexes adsorbed directly to PLG were found to aggregate with increasing deposition time [Fig. 1(a)]; however, complexes incubated with tissue culture polystyrene did not aggregate.13 Additionally, the percentage of DNA immobilized to the PLG scaffold was substantially less than that previously observed on polystyrene surfaces (nearly 80% after 24 h).13 Deposition times longer than 24 h are expected to increase the amount of immobilized DNA, as higher densities of immobilized DNA have been achieved using larger doses that were incubated with polymer (data not shown). These differences in immobilization likely reflect differences in the surface chemistry of the polystyrene and the polymer, and the extent to which protein adsorbs to each. For both polystyrene and PLG, transfection was improved by surface coating with proteins. Importantly, the substrate delivery approach is applicable to a variety of ECM proteins, such as collagen, fibronectin, and laminin, which may be necessary for appropriate cell adhesion and signaling. The protein coating prevents aggregation of the polyplexes, likely through reducing the available sites for interaction with the substrate or through the complexes directly interacting with the proteins. The stability of complexes at the surface [Fig. 2(b)] can generally be described by a low affinity site that leads to rapid loss from the surface, and a high affinity site that leads to a more gradual loss. Incubation of scaffolds in buffer at various deposition times are expected to exhibit these two affinities; however, the relative quantity of the complexes associated with each site may vary with the deposition time.
Substrate-mediated delivery is also applicable to a variety of transfection reagents, such as cationic polymers,14,15,20 cationic lipids,13 and calcium phosphate nanocomposites.21 Several strategies have been investigated for the immobilization of DNA to polymeric biomaterials (e.g., PLA and PLG). Cationic polymers, such as PEI, were adsorbed to polymer substrates13 or covalently attached.20 Alternatively, complexes can be preformed in solution for subsequent immobilization to the polymer substrate. Preformed complexes can be dried onto polymer scaffolds,15 linked to the scaffold,20 or be specifically immobilized through complementary functional groups (e.g., biotin-avidin).14 This strategy of preforming the complexes may allow greater control over the complex properties (e.g., size) before substrate immobilization. For PEI/DNA complexes, the N/P ratio is a critical property that significantly affects gene transfer from the surface, through affecting complex size, zeta potential, substrate binding and stability, and intracellular trafficking. PEI supports the adsorption of complexes to the substrate and retention at the surface is improved at higher N/P ratios [Fig. 2(b)]. Higher N/P ratios have been proposed to enhance transfection by facilitating complex association with plasma membrane and promoting endosomal escape.22 However, relative to low N/P ratios, complexation at high N/P ratios can result in the presence of free PEI in quantities reaching 86% of the initial PEI amounts,23 which be toxic due to permeabilization of the plasma membrane.22,24 Free PEI can be removed from PEI/DNA complexes through ultrafiltration22 or size exclusion chromatography (SEC).24 Free PEI should not be a significant issue for substrate-mediated delivery given the multiple washing steps that are performed.
Transgene expression on the substrate was maximized through balancing the concentration of DNA in the cellular microenvironment and the N/P ratio. The amount immobilized and the fraction lost from the surface determine the concentration in the cellular microenvironment. The amount immobilized is controlled through the dose incubated and the incubation time, whereas the retention at the surface was affected by the N/P ratio. Note that the N/P ratio can also affect the potency of the complexes. Immobilization of PEI/DNA complexes to PLG increases transgene expression by two orders of magnitude and enhances the percentage of transfected cells as compared with bolus delivery (Fig. 3). Increasing the incubation time of complexes with the substrate results in greater deposition of complexes. As with bolus delivery, the quantity of complexes must be modulated to provide sufficient complexes for transfection (Fig. 5). However, excessive quantities can produce lower levels of transgene expression, either through limiting cell proliferation (Fig. 6) or becoming cytotoxic.25 Increasing doses given as a bolus have been associated with decreasing metabolic activity and increasing cytotoxicity,25 consistent with the reduction in cell number that is observed for the scaffold at higher DNA loadings. Substrate-mediated delivery is expected to produce similar trends in transgene expression and transfection efficiency as bolus delivery; however, note that the amount of DNA required will differ.
Substrate-mediated delivery can be extended to transfection within three-dimensional tissue engineering scaffolds, which are central to many tissue engineering applications. Scaffolds implanted into injury sites are typically designed to support cellular infiltration from the surrounding tissue to enable integration with the host. The pore structure that allows cellular infiltration can also be employed to deposit DNA complexes on the surface of the polymer. The ability to transfect cells throughout the structure suggests that complexes can be adsorbed to all surfaces to which the DNA complex solution is exposed. The surface area of the scaffolds thus becomes a key design parameter for regulating transgene expression,5 as the surface area regulates both complex adsorption and cell adhesion. Additionally, the three-dimensional scaffolds may limit the escape of complexes to the bulk media, and thereby extending the opportunity for the complex to be internalized. Surface immobilization may also allow complexes to be patterned within the scaffold, thus creating spatial patterns in transgene expression that could potentially mimic the expression patterns observed in developing tissues. Transgene expression from a tissue engineering scaffold, with a geometry that orients tissue formation, provides a powerful tool to stimulate the in vivo development of functional tissue replacements.
In conclusion, nonspecific adsorption of DNA complexes to tissue engineering scaffolds can achieve localized and enhanced gene transfer. This delivery approach is applicable to multiple cell types and transfection reagents, achieves similar or greater levels of transfection as bolus delivery, yet can reduce the total amount of DNA required relative to delivery as a bolus. Immobilization also homogeneously distributes the DNA throughout the scaffold, resulting in large numbers of transfected cells at these low quantities. Gene transfer is dependent upon the complex and substrate properties, which must balance substrate binding and stability with cellular internalization and trafficking. The substrate can be prefabricated in an appropriate geometry, for the subsequent immobilization of complexes. This approach may be widely applicable to numerous existing materials and a variety of geometries (three-dimensional). Alternatively, novel materials may ultimately be designed that specifically adsorb DNA complexes. The nano-scale control of chemical and physical properties of biomaterial substrates combined with the development of strategies to regulate vector immobilization and surface stability will enable numerous applications. Precisely controlling the substrate chemistry, architecture, and patterning may enhance the delivery efficiency and control gene transfer, which would increase its therapeutic application.
Acknowledgments
The authors would like to acknowledge Brian Anderson for helpful discussions.
Contract grant sponsor: Christopher Reeve Paralysis Foundation
Contract grant sponsor: NIH; contract grant number: RO1 GM066830
References
- 1.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Lee JY, Peng H, Usas A, Musgrave D, Cummins J, Pelinkovic D, Jankowski R, Ziran B, Robbins P, Huard J. Enhancement of bone healing based on ex vivo gene therapy using human muscle-derived cells expressing bone morphogenetic protein 2. Hum Gene Ther. 2002;13:1201–1211. doi: 10.1089/104303402320138989. [DOI] [PubMed] [Google Scholar]
- 3.Hidaka C, Goodrich LR, Chen CT, Warren RF, Crystal RG, Nixon AJ. Acceleration of cartilage repair by genetically modified chondrocytes over expressing bone morphogenetic protein-7. J Orthop Res. 2003;21:573–583. doi: 10.1016/S0736-0266(02)00264-4. [DOI] [PubMed] [Google Scholar]
- 4.Cao L, Liu L, Chen ZY, Wang LM, Ye JL, Qiu HY, Lu CL, He C. Olfactory ensheathing cells genetically modified to secrete GDNF to promote spinal cord repair. Brain. 2004;127(Pt 3):535–549. doi: 10.1093/brain/awh072. [DOI] [PubMed] [Google Scholar]
- 5.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: Transgene expression and cellular transfection. Mol Ther. 2005;12:475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: Prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5:753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 7.Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17:551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 8.Meilander NJ, Pasumarthy MK, Kowalczyk TH, Cooper MJ, Bellamkonda RV. Sustained release of plasmid DNA using lipid microtubules and agarose hydrogel. J Control Release. 2003;88:321–331. doi: 10.1016/s0168-3659(03)00007-5. [DOI] [PubMed] [Google Scholar]
- 9.Eliaz RE, Szoka FC., Jr. Robust and prolonged gene expression from injectable polymeric implants. Gene Ther. 2002;9:1230–1237. doi: 10.1038/sj.gt.3301786. [DOI] [PubMed] [Google Scholar]
- 10.Huang YC, Connell M, Park Y, Mooney DJ, Rice KG. Fabrication and in vitro testing of polymeric delivery system for condensed DNA. J Biomed Mater Res A. 2003;67:1384–1392. doi: 10.1002/jbm.a.20036. [DOI] [PubMed] [Google Scholar]
- 11.Huang YC, Riddle K, Rice KG, Mooney DJ. Long-term in vivo gene expression via delivery of PEI-DNA condensates from porous polymer scaffolds. Hum Gene Ther. 2005;16:609–617. doi: 10.1089/hum.2005.16.609. [DOI] [PubMed] [Google Scholar]
- 12.Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol Ther. 2004;10:19–26. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 13.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotechnol Bioeng. 2005;90:290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid-collagen hydrogels via a substrate-mediated approach. Biomaterials. 2005;26:1575–1584. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bielinska AU, Yen A, Wu HL, Zahos KM, Sun R, Weiner ND, Baker JR, Jr, Roessler BJ. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21:877–887. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- 16.Murphy WL, Mooney DJ. Controlled delivery of inductive proteins, plasmid DNA and cells from tissue engineering matrices. J Periodontal Res. 1999;34:413–419. doi: 10.1111/j.1600-0765.1999.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 17.Jang JH, Shea LD. Controllable delivery of non-viral DNA from porous scaffolds. J Control Release. 2003;86:157–168. doi: 10.1016/s0168-3659(02)00369-3. [DOI] [PubMed] [Google Scholar]
- 18.Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18:893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 19.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13:621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, Manuel WS, Hornsby PJ. Transfection of cells mediated by biodegradable polymer materials with surface-bound polyethyleneimine. Biotechnol Prog. 2000;16:254–257. doi: 10.1021/bp990150h. [DOI] [PubMed] [Google Scholar]
- 21.Shen H, Tan J, Saltzman WM. Surface-mediated gene transfer from nanocomposites of controlled texture. Nat Mater. 2004;3:569–574. doi: 10.1038/nmat1179. [DOI] [PubMed] [Google Scholar]
- 22.Godbey WT, Wu KK, Hirasaki GJ, Mikos AG. Improved packing of poly(ethylenimine)/DNA complexes increases transfection efficiency. Gene Ther. 1999;6:1380–1388. doi: 10.1038/sj.gt.3300976. [DOI] [PubMed] [Google Scholar]
- 23.Clamme JP, Azoulay J, Mely Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 25.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]