Abstract
Peptides can potentiate lipid-mediated gene delivery by modifying lipoplex physiochemical properties to overcome rate-limiting steps to gene transfer. The objectives of this study were to determine the regimes over which cationic peptides enhance lipofection and to investigate the mechanism of action, such as increased cellular association resulting from changes in lipoplex physical properties. Short, cationic peptides were incorporated into lipoplexes by mixing peptide, lipid and DNA. Lipoplexes were characterized using gel retardation, dynamic light scattering, and fluorescent microscopy, and the amount of surface-displayed amines was quantified by fluorescamine. Size, zeta potential, and surface amines for lipoplexes were dependent on peptide/DNA ratio. Inclusion of peptides in lipoplexes resulted in up to a 13-fold increase in percentage of cells transfected, and up to a 76-fold increase in protein expression. This transfection enhancement corresponded to a small particle diameter and positive zeta potential of lipoplexes, as well as increased amount of surface-displayed amines. Relative to lipid alone, these properties of the peptide-modified lipoplexes enhanced cellular association, which has been reported as a rate-limiting step for transfection with lipoplexes. The addition of peptides is a simple method of lipofection enhancement, as direct chemical modification of lipids is not necessary for increased transfection.
Keywords: DNA delivery, gene delivery, gene vectors, lipoplexes, cationic lipids, peptides
INTRODUCTION
Lipoplexes have been used extensively for in vitro and in vivo gene delivery applications, and are currently in clinical trials. Lipid-based vectors condense DNA and protect it from degradation, promote membrane interaction and destabilize endosomes, and can have low toxicity.1-5 Despite their widespread use, the efficiency of lipid-mediated gene transfection still lags behind that of viral vectors. Lipoplex stability, cellular internalization, and intracellular trafficking barriers can limit the efficacy of lipoplexes.6-8 Efforts to improve lipid-based gene delivery have focused on modifying the cationic lipid structure, altering lipid formulations, and adding polymers to lipid-DNA complexes to enhance transfection.9-15 In particular, polymers or peptides with specific functionalities may be added to lipids to function as modular units that can target a range of cellular processes.16 For example, bioactive peptides for receptor targeting, endosomal destabilization, and nuclear localization have been utilized to enhance lipofection.17-27
Bioactive peptides can influence lipofection through a sequence-specific response or by imparting nonspecific changes in conformation or other physiochemical properties of the lipoplex, such as zeta potential and average diameter. Increasing the amount of peptides within lipoplexes initially results in an increase of particle size from less than 500 nm to up to 2000 nm, while further increasing the peptide loading beyond a certain threshold resulted in a reduction in diameter to less than 500 nm.28,29 Despite the similar physical characteristics of peptide/DNA lipoplexes at the high peptide regime (i.e., the region in which the lipoplex size decreases from its maximum following the initial increase) compared to that of the low peptide regime, to our knowledge gene delivery in the high peptide regime has not been widely investigated. Relative to high molecular weight proteins, peptides typically have a lower affinity for DNA,30 and their inclusion in the lipoplex may be dependent on the amount of peptide added. For either bioactive or nonbioactive peptides, increasing the peptide quantity may increase the incorporation of peptides into the lipoplexes, which may facilitate cellular interactions through modified lipoplex surface properties.31
In this report, we investigate peptide-mediated lipofection to determine whether peptides can interface specifically or nonspecifically with one or more steps in the gene transfer process. We hypothesize that the physical properties of the lipoplex as well as the surface characteristics, such as the density of surface-displayed amines, may be dependent on the amount of peptide added to the lipoplexes and may affect transfection efficiency. Peptide-lipoplexes were formed at peptide/DNA weight ratios ranging from 0 to 120 by adding cationic peptides to DNA prior to complexation with lipids. The physiochemical properties of lipoplexes were characterized by gel retardation, average diameter, zeta potential, lipid and peptide content, and the amount of surface-displayed amines. Lipoplexes were visualized using fluorescence microscopy to determine the extent of peptide and DNA colocalization within lipoplexes, and to determine the extent of lipoplex localization to cells. In addition, the percentage of cells transfected, protein expression and the quantity of DNA associated with cells were quantified. This report investigates peptide addition as a simple method to manipulate the physical properties and surface chemistry of lipoplexes, which can increase cellular association and transfection efficiency. These studies add to the basic understanding of gene delivery and may contribute to the design of modular gene delivery vectors.
MATERIALS AND METHODS
Materials
Plasmids encoding for luciferase and enhanced green fluorescent fusion protein (pEGFP-Luc) with a CMV promoter were purified from bacteria culture using Qiagen (Valencia, CA) reagents and stored in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 7.4). Lipofectamine, Lipofectamine 2000, fluorescamine, Oregon Green dye, tetramethylrhodamine (TAMRA), and LysoTracker reagent were purchased from Invitrogen (Carlsbad, CA). Amino acids and resin for peptide synthesis were purchased from Novabiochem (San Diego, CA). The remaining peptide synthesis reagents were purchased from Applied Biosystems (Foster City, CA). All other reagents were obtained from Fisher Scientific (Waltham, MA) unless otherwise noted.
Peptide Synthesis and Purification
An Applied Biosystems 433A peptide synthesizer was used to synthesize the SV40 T-antigen nuclear localization sequence (NLS) and a scrambled peptide control. The SV40 peptide, EGPKKKRKVG, containing the minimal SV40 T-antigen NLS, and the sSV40 peptide, EKRGKVKPKG, a scrambled version of SV40, were synthesized using standard solid phase methods and FastMocTM chemistry. The peptide was cleaved with a mixture of 90% trifluoroacetic acid (TFA), 2.5% triisopropylsilane (TIS), 2.5% thioanosole, and 5% water for 1−2 h at room temperature, and then lyophilized. The crude peptide was analyzed for purity by reversed-phase high-pressure liquid chromatography (RP-HPLC). The peptides were purified by preparative RP-HPLC with a gradient of 0−20% acetonitrile with 0.1% TFA in water with 0.1% TFA. Peptide molecular weight was confirmed using electrospray ionization (ESI) mass spectroscopy and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry at Northwestern University's Analytical Services Laboratory. Peptide purity was analyzed using analytical RP-HPLC and the purity of both peptides was determined to be >95%.
Gel Retardation Assay
DNA condensation by peptides was analyzed by gel electrophoresis. Peptide/DNA complexes were formed by adding 22.5 μL of peptide solution containing varying amounts of peptide to 22.5 μL DNA solution (1.5 μg EGFP-Luc plasmid in TBS buffer), and the resulting solution was mixed by gentle pipetting. Complexes were incubated for 20 min at room temperature. For some samples, 5 μL of trypsin-EDTA solution was added to degrade the peptide after complex formation. After complex formation, 10 μL of each sample were loaded onto a 1% agarose gel in TBE buffer, which ran at 120 V for 30 min. DNA bands on the gel were visualized under UV light by ethidium bromide staining.
Peptide-Lipoplex Formation
Lipofectamine (Invitrogen), a 3:1 (w/w) formulation of the polycationic lipid 2,3-dioleyloxy-N-[2(sperminecarboxamido)ethyl]-N,N-dimethyl-1-propanaminium trifluoroacetate (DOSPA) and the neutral lipid dioleoyl phosphatidylethanolamine (DOPE), was used at a 10:1 (w/w) ratio with DNA, within the manufacturer's recommended usage range. Peptide-lipoplexes were formed by adding 22.5 μL of peptide solution containing varying amounts of peptide to 22.5 μL DNA solution, and the resulting solution was then mixed by gentle pipetting. The peptide/DNA mixture was allowed to incubate for 20 min at room temperature prior to the dropwise addition of 45 μL of lipid solution. The resulting solution was mixed by gentle pipetting and incubated for 20 min. Peptide-lipoplexes were formed in serum-free cell growth media [Dulbecco's Modified Eagle Medium (DMEM), Invitrogen] at pH 7.4. For studies using Lipofectamine 2000, the lipid formulation was used at a 1:3 ratio with DNA. Zeta potential and z-average diameters were measured with a Zetasizer Nano ZS (Malvern, Worcestershire, UK). All peptide/DNA ratios reported herein are by weight; the conversion between peptide/DNA (w/w) ratio and peptide/DNA N/P ratio is 1 (w/w) ratio to 1.46 N/P ratio.
Free Lipid and Peptide after Lipoplex Formation
Complexes were formed with Lipofectamine, peptide, and DNA as described above. After complex formation, complexes were filtered using a Millipore (Billerica, MA) Microcon spin filter (MWCO 3000) to separate complexes from free lipid and peptide in solution. The concentration of free lipid in the filtrate was measured by a colorimetric method using ammonium ferrothiocynate, which readily forms a colored complex with phospholipids that is soluble in chloroform.32 An aqueous ammonium ferrothiocynate solution was prepared by dissolving 2.70 g ferric chloride hexahydrate and 3.04 g ammonium thiocyanate in 100 mL deionized distilled water. Samples (20−40 μL) were added to chloroform and vigorously mixed. The ammonium ferrothiocyanate solution was then added and the resulting mixture was vigorously shaken for 1 min. After incubating at room temperature for 45 min, the absorbance of the organic phase was measured at 488 nm. A standard curve was prepared using known amounts of Lipofectamine.
Free peptide in the filtrate was measured by dialyzing the samples using a Spectrum Labs (Rancho Dominguez, CA) dialysis membrane with 1000 MWCO, lyophilizing, and then taking the absorbance of samples resuspended in water at 220 nm using a Cary 50 UV/Vis spectrophotometer (Varian, Inc., Palo Alto, CA). A standard curve was prepared using known amounts of peptide.
Cell Culture and Transfection
Transfection studies were performed with NIH/3T3 and HEK293T cells (ATCC, Manassas, VA) cultured at 37°C and 5% CO2 in DMEM supplemented with 1% sodium pyruvate, 1% penicillin–streptomycin, 1.5 g/L NaHCO3, and 10% fetal bovine serum (FBS). For bolus delivery, cells were seeded at a density of 15000 cells per well for NIH/3T3 cells and 20000 cells per well for HEK293T cells in 48-well plates (300 μL total volume) and cultured overnight before complexes were added to the culture media. Complexes were formed as described above, with 30 μL of the peptide-lipoplexes added per well. Transfection was characterized by the number of transfected cells (GFP expression) measured 24 h post-transfection and the extent of transgene expression (luciferase levels) measured 48 h post-transfection. Transfected cells were visualized and manually counted using an epifluorescence microscope (Leica, Bannockburn, IL) with a FITC filter and equipped with a digital camera. The percentage of transfected cells was calculated as the number of EGFP-positive cells divided by total cell number, which was determined by manual counting. The extent of transgene expression was quantified by measuring the luciferase activity using the Luciferase Assay System (Promega, Madison, WI). Cells were lysed and assayed for enzymatic activity 48 h after transfection. The luminometer was set for a 3 s delay with signal integration for 10 s. Luciferase activity was normalized to the amount of total protein in the sample, which was measured using a BCA assay (Pierce, Rockford, IL) following the manufacturer's instructions. Transfection by surface-bound lipoplexes was performed to determine the effect of particle density on transfection efficiency. For surface-mediated DNA delivery, complexes were formed in unsupplemented DMEM as described above, then allowed to deposit onto 48-well tissue culture polystyrene plates for 24 h, with plates being washed twice with unsupplemented DMEM before the seeding of cells to remove unbound lipoplexes.
Viability of the cell population was analyzed using the [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxyme-thoxyphenyl)-2-(4-sulfophenyl)]-2H-tetrazolium, inner salt (MTS) assay (Promega), which reports on metabolic activity. Briefly, MTS reagent was added (20% of culture volume) to the cultures, and after 1.5 h incubation, the absorbance at 490 nm was measured with a spectrophotometer. The relative cell viability was reported as the absorbance for the experimental condition divided by the absorbance for the control condition (no DNA or transfection reagent), with absorbances corrected by subtracting the absorbance of the reagents (without cells).
Colocalization of Fluorescently Labeled DNA and Peptide
EGFP-Luc plasmid was labeled with rhodamine using a Label-IT rhodamine-labeling kit (Mirus, Madison, WI) according to the manufacturer's instructions. Briefly, DNA and Label IT reagents were mixed and incubated at 37°C for 1 h. After incubation, DNA was precipitated with 70% ethanol and resuspended in 10 μL Tris-EDTA (TE) buffer. Peptides were labeled with TAMRA (5-(and-6)-carboxytetramethylrhodamine, succinimidyl ester) or Oregon Green (Invitrogen) according to the manufacturer's protocol. Briefly, peptides were mixed with Oregon Green or TAMRA dissolved in DMSO and incubated with gentle agitation for 2 h in the dark. The peptides were then dialyzed and lyophilized, and the lyophilized peptide was resuspended in water to a concentration of 10 mg/mL.
To determine if DNA and peptide colocalized, lipoplexes were formed with rhodamine-labeled DNA and Oregon-Green-labeled peptides as described above. Complexes were immobilized on tissue culture polystyrene (96-well plate; Corning, Corning, NY) immediately following complex formation by incubation of DNA complexes (50 μL) with the substrate for 2 or 24 h and were then visualized using fluorescence microscopy. To determine if DNA and peptide colocalized with cells, lipoplexes were formed with rhodamine-labeled DNA or TAMRA-labeled peptides. Lipoplexes were added to cells 24 h after cell seeding in 48-well plates. At specified time points after transfection, cells were washed twice with PBS and then incubated for 30 min with Lysotracker reagent. Cells and complexes were then visualized using fluorescence microscopy at 2, 24, and 48 h.
Cellular Association of Lipoplexes
The amount of DNA associated with cells was monitored using plasmids radiolabeled with α-32P dATP. Briefly, a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NJ) was used following the manufacturer's protocol with minor modifications. Cells were transfected with radiolabeled DNA as described above and harvested from plates by trypsinization. Following harvesting and washing with PBS, the amount of DNA associated with cells was determined by immersing samples in 5 mL scintillation cocktail (Biosafe II, Research Products International Corp., Mount Prospect, IL) for measurement with a scintillation counter. The counts were correlated to DNA concentration using a standard curve and results were reported as the amount of DNA measured per cell.
Detection of Accessible Primary Amines on Lipoplexes
A fluorescamine assay was used to determine the amount of primary amines accessible on the peptide-lipoplexes.33 Complexes were formed with Lipofectamine, peptide and α-32P-labeled DNA as described above. After complex formation, complexes were isolated by filtration using a Millipore Microcon spin filter (MWCO 3000). The retentate was recovered and diluted in 1.4 mL assay buffer (100 mM boric acid–NaOH, pH 7.4) prior to rapid addition of 500 μL 0.01% fluorescamine (prepared in acetone). Samples were rapidly inverted 4−5 times and incubated at room temperature for 10 min. Fluorescence was measured using a TBS-380 Mini-Fluorometer (Turner Biosystems, Sunnyvale, CA) using λex 365 nm and λem 440−470 nm. A standard curve was constructed using known amounts of peptide to correlate fluorescence signal to amount of primary amines. The quantity of DNA associated with the lipoplexes was determined by scintillation as previously described. The total number of primary amines detected on lipoplexes was normalized by the mass of DNA in the sample.
Statistics
Statistical analysis was performed using JMP software (SAS Institute, Inc., Cary, NC). Comparative analyses were completed using one-way ANOVA with Tukey post-tests, at a 95% confidence level. Mean values with standard deviation are reported and all experiments were performed in triplicate.
RESULTS
Physiochemical Properties of Peptide-Lipoplexes
Condensation of DNA by cationic peptides was investigated by gel retardation (Fig. 1). As the weight ratio of peptide to DNA was increased from 10 to 120, DNA migration was increasingly inhibited for both intact (SV40) and scrambled-sequence (sSV40) NLS peptides. However, even at the highest peptide/DNA ratio of 120, the peptides did not completely inhibit DNA migration. The intensity of each band was similar, indicating that complexation did not exclude ethidium bromide intercalation. Incubation of the peptide/DNA complex with trypsin, which degrades the peptide, allowed the DNA to migrate unhindered.
Figure 1.
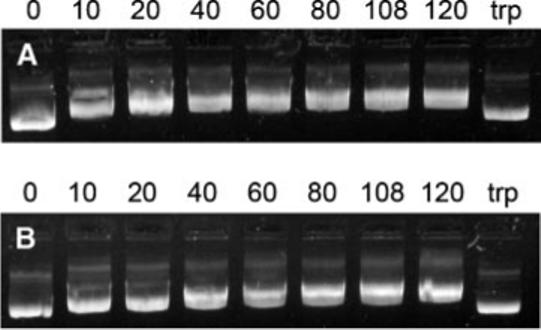
Electrophoretic mobility of DNA with SV40 peptide (A) and sSV40 peptide (B). Lane assignments correspond to peptide/DNA weight ratios. The last lane (labeled trp) corresponds to complexes formed at a peptide/DNA ratio of 120 that were incubated with trypsin.
The size and zeta potential of peptide-lipoplexes were dependent on peptide/DNA ratio. At peptide/DNA ratios of 40 and below, the peptide-lipoplexes maintained an average diameter of less than 500 nm for both the SV40 and the sSV40 peptides (Fig. 2A). For peptide/DNA ratios of 60−100, the average diameter of the peptide-lipoplexes was greater than 500 nm, increasing up to 1500 nm. Further increasing the peptide/DNA ratio above 108 resulted in a decrease in the diameter of the lipoplexes to less than 500 nm. The trends observed with particle size were comparable to zeta potential. At low and high peptide/DNA ratios (i.e., <60 and >100), the zeta potential of the peptide-lipoplexes was positive, which is desirable for cell membrane interactions, but at an intermediate peptide/DNA ratio between 60 and 100, the zeta potential of the complexes was negative (Fig. 2B). Thus, although the peptides do not completely condense DNA as seen by gel retardation, the peptides affect the size and zeta potential of the lipoplexes.
Figure 2.
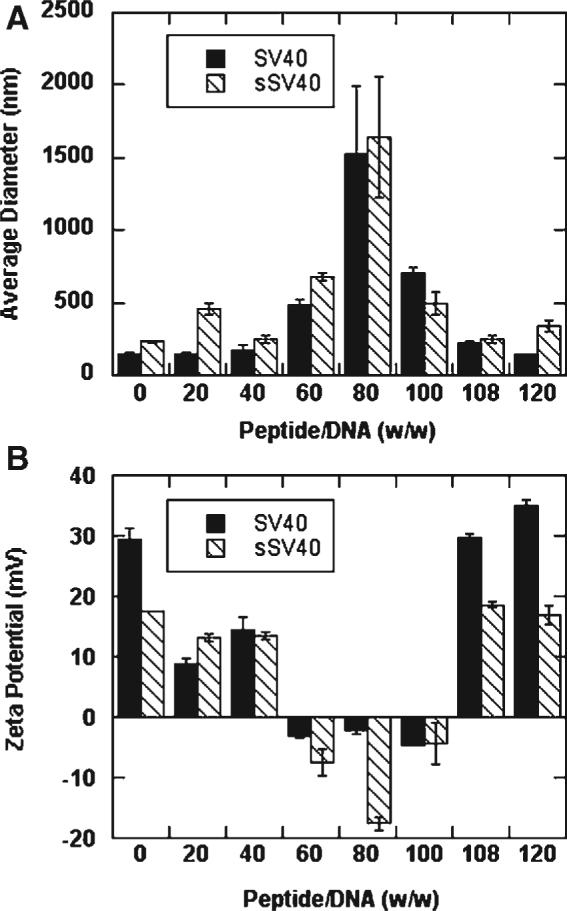
Average diameter (A) and zeta potential (B) of peptide-lipoplexes formed with SV40 and sSV40 peptides. All values are reported as mean±standard deviation.
Subsequent experiments were performed to determine if peptides were incorporated into the lipoplexes at several peptide/DNA ratios. The fluorescence images support the size results obtained by dynamic light scattering; lipoplexes formed without peptide (Fig. 3A) were noticeably smaller than lipoplexes formed at a peptide/DNA ratio of 80 (Fig. 3B). Peptides colocalized with DNA at both a peptide/DNA ratio of 80 (Fig. 3B) and 108 (Fig. 3C). Similar fluorescence images were obtained for both the sSV40 peptide and the SV40 peptide (data not shown).
Figure 3.

Fluorescence microscopy images of peptide-lipoplexes. Rhodamine-labeled DNA was mixed with Oregon Green-labeled sSV40 peptide and subsequently mixed with Lipofectamine. Images are as follows: fluorescence microscopy image of Lipofectamine/DNA complexes without peptide (A); overlaid fluorescence microscopy images of lipoplexes with sSV40 peptide, peptide/DNA ratio = 80 (B); overlaid fluorescence microscopy images of lipoplexes with sSV40 peptide, peptide/DNA ratio = 108 (C). Scale bar = 20 μm.
Free Lipid and Peptide after Complex Formation
The amount of free lipid and peptide in solution were measured to further investigate the observations for the physical properties of the peptide-lipoplexes. The quantity of free lipid increased as the peptide/DNA ratio increased from 0 to 80 (Fig. 4A). Further increasing the sSV40 peptide/DNA ratio to 120 resulted in a decrease in free lipid. At a peptide/DNA ratio of 80, the percentage of free lipid was approximately 60%, which differs significantly from other peptide/DNA ratios. Additionally, the lipoplex diameter was maximal at this condition among all the conditions tested, while the zeta potential was minimal. The percentage of free sSV40 peptide increased with an increase in the peptide/DNA ratio (Fig. 4B). Nearly all of the peptide is included in the lipoplex at low peptide/DNA ratios, while at higher ratios, an increased amount of peptide is excluded from the complex. The highest percentage of excluded peptide was approximately 20%, which occurred at a peptide/DNA ratio of 108. Similar results were obtained with the SV40 peptide (data not shown).
Figure 4.
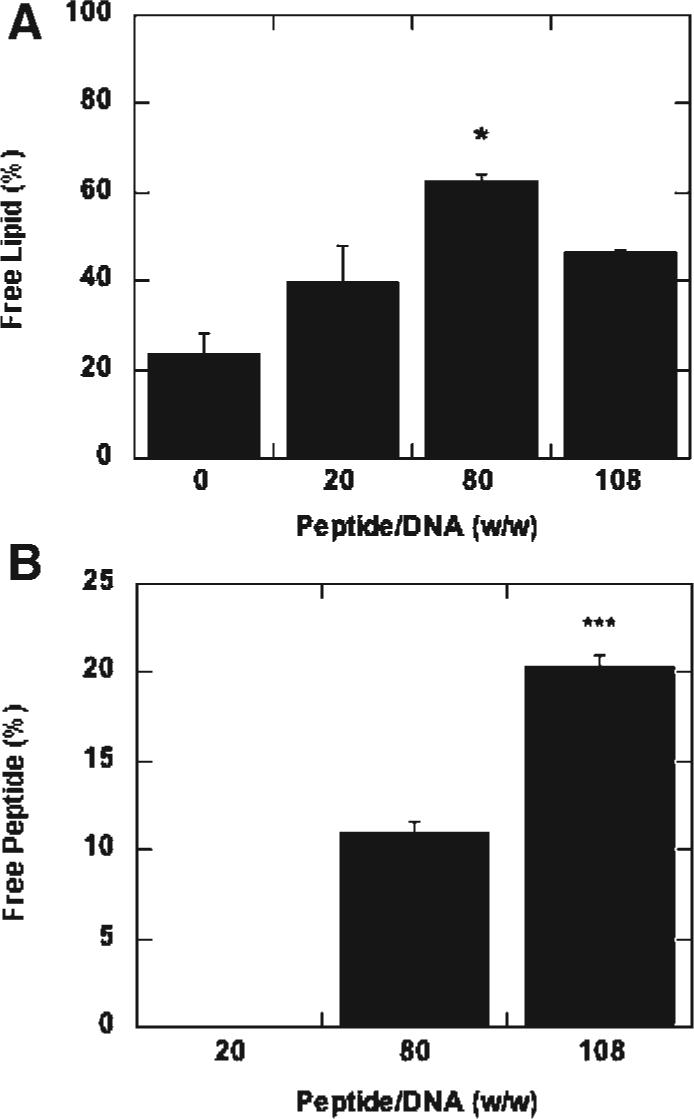
Percentage of free lipid and peptide remaining after complex formation. The percentage of free lipid (A) and peptide (B) remaining after complex formation was calculated as a function of peptide/DNA (w/w) ratio. All values are reported as mean±standard deviation (* p < 0.05, *** p < 0.001).
Peptide Content and Lipofection Efficiency
Lipoplexes formed at peptide/DNA ratios of 108 and above significantly increased both the percentage of transfected cells and reporter protein expression in NIH/3T3 mouse fibroblasts compared to lower peptide/DNA ratios with both the SV40 peptide and the sSV40 peptide (Fig. 5A and B). This trend was also observed with HEK293T human embryonic kidney cells (data not shown). The addition of cationic peptide increased the percentage of transfected NIH/3T3 cells up to 13-fold and protein expression 76-fold. Fluorescence microscopy experiments using lipoplexes formed with TAMRA-labeled peptide and rhodamine-labeled DNA indicated colocalization of peptide and DNA within cells (peptide/DNA, w/w, ratios of 20 and 100) at 2, 24, and 48 h (data not shown). For high peptide/DNA ratios, the transfection efficiency is comparable to the transfection efficiency of using lipids alone with more than twice the amount of DNA (Fig. 5C). For Lipofectamine 2000, the addition of peptide at a peptide/DNA ratio of 108 increased the percentage of transfected NIH/3T3 cells 1.9-fold and protein expression by 14.9-fold (data not shown). Although lipoplexes without any peptide or with small peptides amounts demonstrated positive zeta potential and small particle size, they did not transfect as well as the lipoplexes with peptide/DNA ratios of 108 and 120, which had similar physical characteristics independent of sequence. Thus, the size and zeta potential of the complexes are not the only factors that contribute to the increased transfection efficiency.
Figure 5.
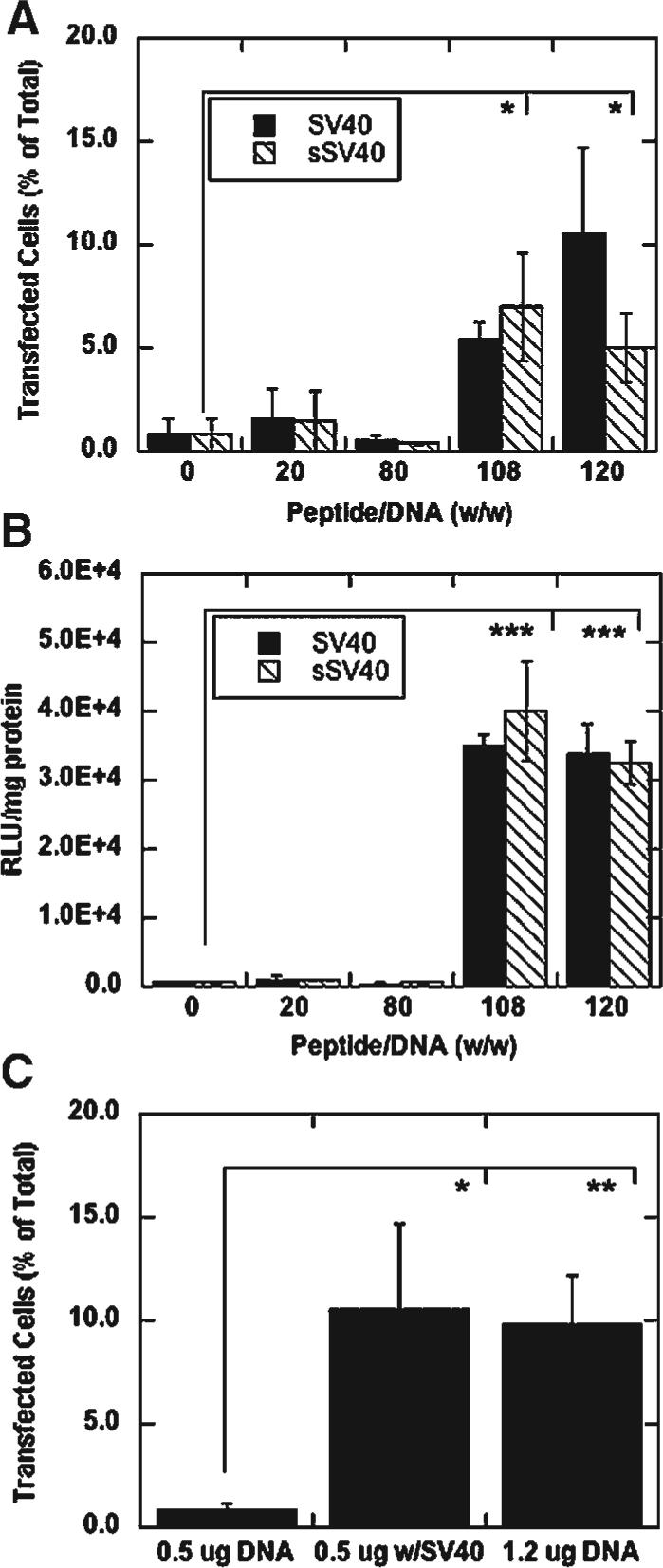
Transfection at varying peptide/DNA ratios and DNA amounts. Percentage of cells transfected (A) and luciferase expression (B) of NIH/3T3 cells transfected with peptide-lipoplexes, 0.5 μg DNA per well, 10:1 lipid/DNA ratio (w/w), with varying amounts of SV40 and sSV40 peptide added. Percentage of NIH/3T3 cells transfected as a function of DNA added per well (C); the SV40 peptide/DNA ratio was 108 (w/w) and the lipid/DNA ratio was 10 (w/w). All values are reported as mean±standard deviation (*p < 0.05, **p < 0.01, ***p < 0.001).
The order of addition of transfection reagents on transfection enhancement was also investigated. Transfection enhancement was not observed when lipids were added to DNA before the addition of SV40 peptide at any of the peptide/DNA ratios tested (Fig. 6). Similar results were obtained with the sSV40 peptide (data not shown). The preformed lipid/DNA complex may have excluded the peptides due to the peptides’ short length and low relative affinity compared to the lipids, whereas the addition of peptides before lipids to DNA resulted in a significant increase in transfection efficiency (at peptide/DNA ratios at 108 and above) presumably because the peptides were included in the lipoplexes.
Figure 6.
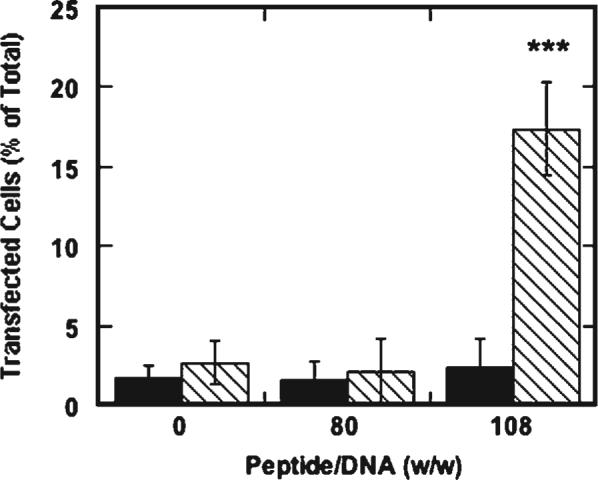
Percent transfection of NIH/3T3 cells by alternating the addition of lipid and SV40 peptide to DNA. Lipofectamine was added to DNA prior to peptide (solid bars) or after peptide (dashed bars). All values are reported as mean±standard deviation (*** p < 0.001).
Cellular Association of DNA
Cellular association of lipoplexes was subsequently quantified to investigate the mechanism of transfection enhancement. At a peptide/DNA ratios of 108, a significantly higher amount of DNA was associated to the NIH/3T3 cells compared to peptide/DNA ratios of 0, 20, and 80 for both intact NLS peptide and scrambled NLS peptide (Fig. 7). The inclusion of peptide with lipoplexes increased DNA association by up to threefold compared to lipoplexes without peptide.
Figure 7.

Cellular association of DNA. Radiolabeled DNA (0.5 μg per well) was complexed with varying amounts of SV40 and sSV40 peptides and Lipofectamine (10:1, w/w, lipid/DNA) for subsequent addition to NIH/3T3 cells. Amount of DNA associated was determined using a scintillation counter. All values are reported as mean±standard deviation (*** p < 0.05, **p < 0.01, ***p < 0.001).
Lipofection Efficiency of Surface-Immobilized Lipoplexes
We subsequently tested the hypothesis that the increased cellular quantities may result from the peptide-lipoplexes having a higher density than lipoplexes alone, which could increase delivery due to rapid settling of the lipoplexes onto the cells. The average diameter of lipoplexes without peptide was similar to the average diameters of lipoplexes with a high amount of peptide. Given that the peptide colocalizes with the lipoplexes, the peptide-containing lipoplexes may have a higher density than the lipoplexes without peptide. To minimize the influence of lipoplex density on transfection, lipoplexes were deposited onto tissue culture plates 24 h prior to the addition of NIH/3T3 cells, which reduces mass transport limitations affected by particle density.34 At a peptide/DNA ratio of 120, a significantly higher amount of luciferase expression was observed compared to lipid alone for surface-bound lipoplexes (Fig. 8). Similar results were obtained with SV40 peptide (data not shown). Thus, lipofection is enhanced with the addition of peptide when complexes are administered to cells by either bolus delivery or surface-mediated delivery, suggesting that complex density is not responsible for increased transfection.
Figure 8.
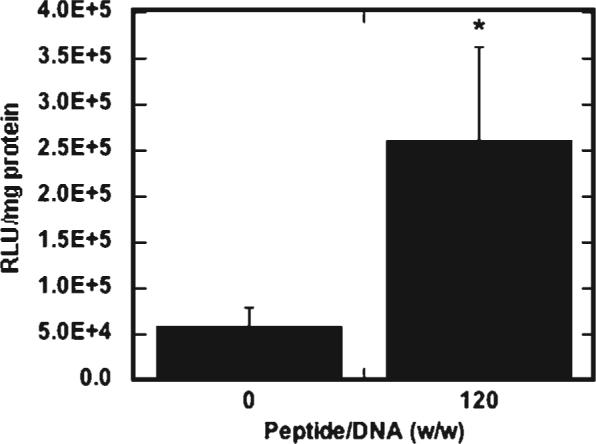
Transfection by surface-bound lipoplexes. Luciferase expression of NIH/3T3 cells transfected with peptide-lipoplexes immobilized to tissue-culture polystyrene, 1.25 μg DNA per well, 10:1 lipid/DNA ratio (w/w), with sSV40 peptide added. All values are reported as mean±standard deviation (*p < 0.05).
Quantification of Surface Amines on Peptide-Lipoplexes
We subsequently tested the hypothesis that increasing the amount of peptide added to the lipoplexes not only affects average size and zeta potential, but also increases the amount of peptide displayed on the surface, which could be observed through the number of accessible amine groups. The amount of primary amines on the lipoplex surface was quantified using a fluorescamine assay and normalized to the total DNA. As the sSV40 peptide/DNA (w/w) ratio increased above 20, the primary amines on the lipoplex surface also increased (Fig. 9). A peptide/DNA ratio of 80 had the highest amount of detectable surface amines, indicating that the number of surface amines does not directly correlate with the amount of peptide added. Similar results were obtained with SV40 peptide (data not shown).
Figure 9.
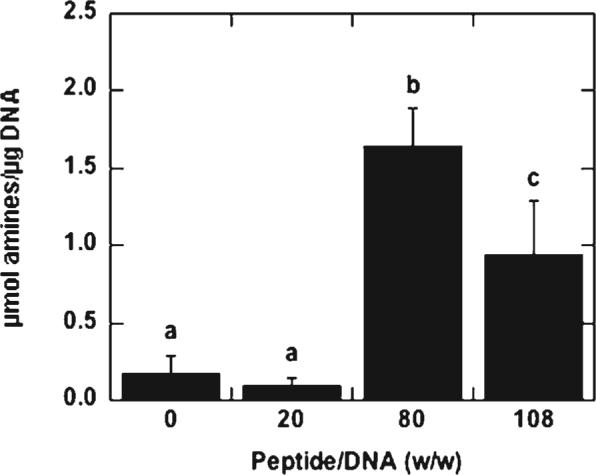
Accessible amines on peptide-lipoplexes. Amines on the lipoplex surface were quantified and normalized to the amount of DNA. All values are reported as mean±standard deviation, with different letters denoting significant difference between values (p < 0.05).
DISCUSSION
This report investigates transfection with peptide-lipoplexes formed at varying peptide/DNA weight ratios and the influence of the peptides on the lipoplex's physical properties. At peptide/DNA ratios below 60 or above 108, the average diameters of the complexes were less than 500 nm and the zeta potential was positive, while at peptide/DNA ratios between 60 and 100, the average diameters of the complexes were greater than 500 nm, increasing to up to 1500 nm with negative zeta potentials. Peptide-lipoplexes containing either intact or scrambled NLS peptides at peptide/DNA ratios of 108 and above significantly increased both the percentage of transfected cells and reporter protein expression compared to peptide/DNA ratios of 80 and below, with increases up to 13-fold for percent transfection and 76-fold for luciferase expression of NIH/3T3 cells. Comparable transfection efficiencies were achieved with less than half the amount of DNA with peptide-lipoplexes compared to lipoplexes without peptide. An increased cellular association of DNA was observed at peptide/DNA ratios above 108 compared to lower peptide/DNA ratios. In addition, inclusion of peptides in lipoplexes did not increase cytotoxicity (data not shown), consistent with literature reports on low molecular weight peptides relative to large proteins.35 Finally, the physical properties of the peptide-lipoplexes and surface primary amines on the lipoplexes contributed to increased cellular association and subsequent gene expression. The results obtained herein were similar with both the SV40 and sSV40 peptides, indicating that sequence specificity was not responsible for the observed results. The approach to form peptide-lipoplexes used herein is a more robust approach to form modular complexes relative to approaches based on synthesis of a single molecule with multiple domains, as different peptides can easily be substituted into the lipoplex.
We hypothesize that an increased quantity of peptide may be necessary to achieve significant loading of peptide in the lipoplex due to the relatively low affinity of the peptide for DNA. Earlier studies of poly-l-lysine (PLL) with lipids showed that PLL enhanced lipid-mediated gene delivery at lower polycation/DNA ratios than with peptides, most likely because PLL is a much larger molecule and has higher relative affinity to the DNA compared to the peptides.29 The short length and relatively small number of charges allows only weak association between the peptide and the DNA relative to high molecular weight proteins and polymers. At intermediate peptide amounts, the lipoplexes have a large diameter and a negative zeta potential, indicative of a disrupted structure that is perhaps due to the weakly bound peptides interfering with lipoplex formation. A disrupted structure is consistent with the free lipid data; at a peptide/DNA ratio of 80, there is significantly more free lipid than at peptide/DNA ratios of 0, 20, and 108. At higher peptide amounts, the peptide and lipids form smaller particles with positive zeta potentials, suggesting that the peptides and lipids may form ordered structures that shield the negative charge of DNA. Although the peptide-lipoplexes formed at a peptide/DNA ratio of 80 have detectable amines on the surface, these lipoplexes still have a negative zeta potential. Zeta potential does not indicate surface potential, but reflects the effective charge of the particles compared to the bulk solution. Thus, although some peptide-lipoplexes have detectable amines on the surface, the overall charge of the particle in solution may still be negative, which was observed for the peptide-lipoplexes formed at a peptide/DNA ratio of 80. The negative zeta potential and large particle diameter of the lipoplexes formed at a peptide/DNA (w/w) ratio of 80 likely contribute to the relatively low transfection. Interestingly, the addition of peptides prior to the addition of lipids did not prevent the peptides’ ability to be displayed on the surface of the lipoplexes. In addition, the order of addition of the peptides and lipids was critical to enhanced lipofection; lipid addition prior to peptides likely excludes the small peptides from the lipoplex.
The increasing quantity of peptides within the lipoplexes increases the amount of accessible amines, which is most likely from an increase in the amount of peptides on the lipoplex surface. This increase of peptides on the lipoplex surface could facilitate cellular interactions. Cellular association of DNA has been determined to be a rate-limiting step for lipofection.36 Size, zeta potential, and density have also been implicated as important factors for cellular internalization of lipoplexes;25-27 however, these factors cannot explain observations for transfection with lipoplexes containing significant quantities of peptide, as size and zeta potential were similar for low and high peptide amounts while transfection enhancement was only observed at high peptide/DNA ratios. Thus, the increased quantity of cell-associated DNA reported herein is due to both favorable lipoplex physical properties (i.e., small average diameter and positive zeta potential) and an increased amount of peptides at the surface of the lipoplex, which could promote cellular association.37 For liposomes modified by stearylated peptides, the peptide density on the surface of the liposome dictated transfection behavior; higher densities of peptides on the surface resulted in increase cellular uptake and protein expression.37 In the study presented herein, the increase in cell-associated DNA was modest compared to the increase in transfection efficiency at peptide/DNA (w/w) ratios above 80. Thus, the addition of peptide may affect cellular processes other than increased association and uptake. For example, the addition of peptides may protect the DNA from degradation or impart enhanced intracellular trafficking due to changes in endocytic uptake. Note that polyplexes, such as PEI/DNA complexes, are not limited by cellular association but by intracellular trafficking. The SV40 and sSV40 peptides added in large quantities do not enhance PEI-mediated transfection (data not shown), possibly because cell association is not the limiting step for PEI vectors.
Despite the inability to fully condense DNA, these cationic peptides contributed to the physiochemical properties, cellular association, and transfection efficiency of the lipoplexes. Complete condensation of DNA by peptides had been proposed as necessary for effective transfection,27 and cationic peptides condense DNA efficiently with a minimum of eight or more positively charged amino acids.30 However, the peptide sequences used here contained five positively charged residues and were incorporated into the lipoplex. The colocalization of the peptides with the DNA may allow for the peptides to protect the DNA from intracellular degradation, despite the fact that the DNA was not fully condensed. In addition, the short peptides may be beneficial to facilitate vector unpacking; “looser” particles may result in higher transfection efficiencies than “tighter” particles.35,38,39 Shorter polycations can have a higher probability of dissociating from DNA than longer polycations and lipids, as shown by a thermodynamic model as well as experimentally, thus permitting higher expression rates in vitro. Thus, complete precondensation by peptides prior to the addition of lipid is not necessary for enhanced transfection, as shown in this report, and may in fact be detrimental to the efficient release of DNA within the cell.
Both the scrambled and intact NLS sequences enhanced lipofection to a similar extent and resulted in peptide-lipoplexes with comparable physiochemical properties. NLSs, such as the minimal SV40 T-ag NLS, have previously been used to transfect cells in vitro,40 with several studies indicating a specific enhancement due to peptide sequence, while others report that peptide sequence has minimal effect. The results reported herein indicate that physiochemical properties of the complexes at high peptide/DNA ratios, rather than sequence specificity, is the main contributing factor of enhanced transfection with peptide-lipoplexes. Our results do not preclude an effect due to the sequence. The peptide influences the physiochemical properties of the peptide-modified lipoplexes to increase cellular internalization, which is reportedly the rate-limiting step for lipoplexes. By overcoming cellular internalization, another step in the transfection process (e.g., endosomal escape, vector unpacking) is now rate-limiting. If the rate-limiting step is not nuclear localization, then an effect due to the peptide's nuclear localization ability would not be observed.
CONCLUSION
This report describes enhancement of lipofection by the incorporation of peptides and indicates that the mechanism of lipofection enhancement is an increased cellular association of the lipoplexes. Cell association is a rate-limiting step for lipofection, and this increased cell association correlated with physical properties favorable for cell uptake (e.g., size, zeta potential) and an increased density of surface primary amines. High peptide/DNA ratios are necessary to achieve peptide presentation on the surface of the lipoplex. Peptide/DNA ratios exceeding 108 enhance lipofection despite the inability of the peptide to completely condense DNA. These results suggest a robust mechanism through which lipoplexes can be modified, and illustrates the potential of incorporating modular components into lipoplexes for enhanced gene delivery.
ACKNOWLEDGMENTS
Support for this research was provided in part by the NIH [R01 EB003806-01 (AEB, LDS) and RO1 GM066830 (LDS)], the Institute for BioNanotechnology in Medicine (IBNAM) at Northwestern University, and a Ford Foundation Predoctoral Fellowship (JCR). MALDI-TOF and ESI mass spectrometry were performed in the Analytical Services Laboratory at Northwestern University. We would like to thank Zain Bengali, Angela Pannier, Tiffany Houchin-Ray, Jaclyn Shepard, Nate Brown, Jill Millstone, Savka Stoeva, and Jae-Seung Lee for technical assistance.
REFERENCES
- 1.Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection—A highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasic DD, Templeton NS. Liposomes in gene therapy. Adv Drug Deliver Rev. 1996;20:221–266. [Google Scholar]
- 3.Woodle MC, Scaria P. Cationic liposomes and nucleic acids. Curr Opin Colloid In. 2001;6:78–84. [Google Scholar]
- 4.Templeton NS. Cationic liposome-mediated gene delivery in vivo. Biosci Rep. 2002;22:283–295. doi: 10.1023/a:1020142823595. [DOI] [PubMed] [Google Scholar]
- 5.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J Control Release. 2006;116:255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene-transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 7.Carriere M, Tranchant I, Niore PA, Byk G, Mignet N, Escriou V, Scherman D, Herscovici J. Optimization of cationic lipid mediated gene transfer: Structure-function, physico-chemical, and cellular studies. J Liposome Res. 2002;12:95–106. doi: 10.1081/lpr-120004781. [DOI] [PubMed] [Google Scholar]
- 8.Tranchant I, Thompson B, Nicolazzi C, Mignet N, Scherman D. Physicochemical optimisation of plasmid delivery by cationic lipids. J Gene Med. 2004;6:S24–S35. doi: 10.1002/jgm.509. [DOI] [PubMed] [Google Scholar]
- 9.Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R, Ramsey P, Martin M, Felgner PL. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]
- 10.Mack KD, Walzem RL, Lehmann-Bruinsma K, Powell JS, Zeldis JB. Polylysine enhances cationic liposome-mediated transfection of the hepatoblastoma cell line Hep G2. Biotechnol Appl Biochem. 1996;23:217–220. [PubMed] [Google Scholar]
- 11.Sorgi FL, Bhattacharya S, Huang L. Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther. 1997;4:961–968. doi: 10.1038/sj.gt.3300484. [DOI] [PubMed] [Google Scholar]
- 12.Cheung CY, Murthy N, Stayton PS, Hoffman AS. A pH-sensitive polymer that enhances cationic lipid-mediated gene transfer. Bioconjug Chem. 2001;12:906–910. doi: 10.1021/bc0100408. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, MacDonald RC. New strategy for transfection: Mixtures of medium-chain and long-chain cationic lipids synergistically enhance transfection. Gene Ther. 2004;11:1358–1362. doi: 10.1038/sj.gt.3302297. [DOI] [PubMed] [Google Scholar]
- 14.Koynova R, Wang L, Tarahovsky Y, MacDonald RC. Lipid phase control of DNA delivery. Bioconjug Chem. 2005;16:1335–1339. doi: 10.1021/bc050226x. [DOI] [PubMed] [Google Scholar]
- 15.Narang AS, Thoma L, Miller DD, Mahato RL. Cationic lipids with increased DNA binding affinity for nonviral gene transfer in dividing and nondividing cells. Bioconjug Chem. 2005;16:156–168. doi: 10.1021/bc049818q. [DOI] [PubMed] [Google Scholar]
- 16.De Laporte L, Cruz Rea J, Shea LD. Design of modular non-viral gene therapy vectors. Biomaterials. 2006;27:947–954. doi: 10.1016/j.biomaterials.2005.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma H, Zhu J, Maronski M, Kotzbauer PT, Lee VMY, Dichter MA, Diamond SL. Non-classical nuclear localization signal peptides for high efficiency lipofection of primary neurons and neuronal cell lines. Neuroscience. 2002;112:1–5. doi: 10.1016/s0306-4522(02)00044-1. [DOI] [PubMed] [Google Scholar]
- 18.Simoes S, Slepushkin V, Gaspar R, de Lima MCP, Duzgunes N. Gene delivery by negatively charged ternary complexes of DNA, cationic liposomes and transferrin or fusigenic peptides. Gene Ther. 1998;5:955–964. doi: 10.1038/sj.gt.3300674. [DOI] [PubMed] [Google Scholar]
- 19.Ou Z, Geiger T, Ou JS, Ackerman AW, Oldham KT, Pritchard KA. AP-4F, antennapedia peptide linked to an amphipathic alpha helical peptide, increases the efficiency of lipofectamine-mediated gene transfection in endothelial cells. Biochem Biophys Res Commun. 2003;305:605–610. doi: 10.1016/s0006-291x(03)00803-9. [DOI] [PubMed] [Google Scholar]
- 20.White RE, Wade-Martins R, Hart SL, Frampton J, Huey B, Desai-Mehta A, Cerosaletti KM, Concannon P, James MR. Functional delivery of large genomic DNA to human cells with a peptide-lipid vector. J Gene Med. 2003;5:883–892. doi: 10.1002/jgm.420. [DOI] [PubMed] [Google Scholar]
- 21.Renigunta A, Krasteva G, Konig P, Rose F, Klepetko W, Grimminger F, Seeger W, Hanze J. DNA transfer into human lung cells is improved with Tat-RGD peptide by caveoli-mediated endocytosis. Bioconjug Chem. 2006;17:327–334. doi: 10.1021/bc050263o. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Ranganathan P, Diamond SL. Nuclear targeting peptide scaffolds for lipofection of nondividing mammalian cells. Nat Biotechnol. 1999;17:873–877. doi: 10.1038/12860. [DOI] [PubMed] [Google Scholar]
- 23.Aronsohn AI, Hughes JA. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J Drug Target. 1997;5:163–169. doi: 10.3109/10611869808995871. [DOI] [PubMed] [Google Scholar]
- 24.Ritter W, Plank C, Lausier J, Rudolph C, Zink D, Reinhardt D, Rosenecker J. A novel transfecting peptide comprising a tetrameric nuclear localization sequence. J Mol Med. 2003;81:708–717. doi: 10.1007/s00109-003-0483-2. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz B, Ivanov MA, Pitard B, Escriou V, Rangara R, Byk G, Wils P, Crouzet J, Scherman D. Synthetic DNA-compacting peptides derived from human sequence enhance cationic lipid-mediated gene transfer in vitro and in vivo. Gene Ther. 1999;6:282–292. doi: 10.1038/sj.gt.3300795. [DOI] [PubMed] [Google Scholar]
- 26.Murray KD, Etheridge CJ, Shah SI, Matthews DA, Russell W, Gurling HMD, Miller AD. Enhanced cationic liposome-mediated transfection using the DNA-binding peptide mu (mu) from the adenovirus core. Gene Ther. 2001;8:453–460. doi: 10.1038/sj.gt.3301401. [DOI] [PubMed] [Google Scholar]
- 27.Bremner KH, Seymour LW, Logan A, Read ML. Factors influencing the ability of nuclear localization sequence peptides to enhance nonviral gene delivery. Bioconjug Chem. 2004;15:152–161. doi: 10.1021/bc034140k. [DOI] [PubMed] [Google Scholar]
- 28.Tagawa T, Manvell M, Brown N, Keller M, Perouzel E, Murray KD, Harbottle RP, Tecle M, Booy F, Brahimi-Horn MC, Coutelle C, Lemoine NR, Alton EWFW, Miller AD. Characterisation of LMD virus-like nanoparticles self-assembled from cationic liposomes, adenovirus core peptide mu (mu) and plasmid DNA. Gene Ther. 2002;9:564–576. doi: 10.1038/sj.gt.3301686. [DOI] [PubMed] [Google Scholar]
- 29.Gao X, Huang L. Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry. 1996;35:1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]
- 30.Plank C, Tang MX, Wolfe AR, Szoka FC. Branched cationic peptides for gene delivery: Role of type and number of cationic residues in formation and in vitro activity of DNA polyplexes. Hum Gene Ther. 1999;10:319–332. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 31.Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M, Kishida H, Kudoh M, Mishina Y, Kataoka K, Yamada M, Harashima H. Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery. Gene Ther. 2007;14:682–689. doi: 10.1038/sj.gt.3302910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart JCM. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem. 1980;104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- 33.Read ML, Etrych T, Ulbrich K, Seymour LW. Characterisation of the binding interaction between poly(L-lysine) and DNA using the fluorescamine assay in the preparation of non-viral gene delivery vectors. FEBS Lett. 1999;461:96–100. doi: 10.1016/s0014-5793(99)01435-0. [DOI] [PubMed] [Google Scholar]
- 34.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotechnol Bioeng. 2005;90:290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaysse L, Arveiler B. Transfection using synthetic peptides: Comparison of three DNA-compacting peptides and effect of centrifugation. Biochim Biophys Acta. 2000;1474:244–250. doi: 10.1016/s0304-4165(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 36.Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 2005;12:1023–1032. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- 37.Khalil IA, Kogure K, Futaki S, Harashima H. High density of octaarginine stimulates macropinocytosis leading to efficient intracellular trafficking for gene expression. J Biol Chem. 2006;281:3544–3551. doi: 10.1074/jbc.M503202200. [DOI] [PubMed] [Google Scholar]
- 38.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 39.Akita H, Tanimoto M, Masuda T, Kogure K, Hama S, Ninomiya K, Futaki S, Harashima H. Evaluation of the nuclear delivery and intra-nuclear transcription of plasmid DNA condensed with micro (mu) and NLS-micro by cytoplasmic and nuclear microinjection: A comparative study with poly-L-lysine. J Gene Med. 2006;8:198–206. doi: 10.1002/jgm.839. [DOI] [PubMed] [Google Scholar]
- 40.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Ther. 2005;12:881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]


