Abstract
Gene transfer has many potential applications in basic and applied sciences. In vitro, DNA delivery can be enhanced by increasing the concentration of DNA in the cellular microenvironment through immobilization of DNA to a substrate that supports cell adhesion. Substrate-mediated delivery describes the immobilization of DNA, complexed with cationic lipids or polymers, to a biomaterial or substrate. As surface properties are critical to the efficiency of the surface delivery approach, self-assembled monolayers (SAMs) of alkanethiols on gold were used to correlate surface chemistry of the substrate to binding, release, and transfection of non-specifically immobilized complexes. Surface hydrophobicity and ionization were found to mediate both DNA complex immobilization and transfection, but had no effect on complex release. Additionally, SAMs were used in conjunction with soft lithographic techniques to imprint substrates with specific patterns, resulting in patterned DNA complex deposition and transfection, with transfection efficiencies in the patterns nearing 40%. Controlling the interactions between complexes and substrates, with the potential for patterned delivery, can be used to locally enhance or regulate gene transfer, with applications to tissue engineering scaffolds and transfected cell arrays.
Keywords: Self-assembled monolayers, Gene delivery, Reverse transfection, Solid-phase delivery, Substrate mediated
1. Introduction
Gene transfer has many potential applications in basic and applied sciences, including functional genomics, gene therapy, and tissue engineering. Although plasmid DNA provides transfection in vivo, complexing DNA with non-viral vectors, cationic lipids or polymers, can facilitate internalization and transfection in vitro and in vivo [1–3]. Complexation can facilitate uptake by enhancing interactions between positively charged DNA complexes and the negatively charged cellular membrane, in addition to providing stability against degradation [4]. Complexation agents can also facilitate intracellular trafficking, which includes endosomal escape, cytoplasmic transport, and nuclear entry, while also dissociating from the DNA to allow expression [1,5]. Non-viral vectors are safer and easier to prepare than viral vectors, but typically have lower efficiency and shorter duration of gene expression.
Controlled delivery systems, including polymeric release in which the DNA is released from the polymer, or substrate-mediated delivery, in which DNA is retained at the surface, have the potential to overcome extracellular barriers that limit gene transfer, as well as enhance gene delivery relative to more traditional delivery methods [6]. In substrate-mediated delivery, also termed reverse transfection or solid-phase delivery, plasmid DNA or DNA complexes are immobilized to a surface or biomaterial that supports cell adhesion. Placing the DNA directly in the cellular microenvironment increases its local concentration, which has been shown to enhance gene delivery [7]. Cells cultured on the substrate can internalize the DNA either directly from the surface, or after release of the DNA from the surface.
DNA complexes can be immobilized on the substrate through specific or non-specific interactions for delivery from the surface. Specific interactions can be introduced through complementary functional groups on the vector and surface, such as antigen–antibody or biotin–avidin [8,9]. The effective affinity of the vector for the substrate is determined by the strength of the specific interactions, which may also be influenced by environmental conditions (e.g., ionic strength, pH), binding-induced conformational changes, or vector unpacking. Poly(L-lysine) (PLL) and polyethylenimine (PEI), modified with biotin residues, were complexed with DNA and bound to a neutravidin substrate [9,10], resulting in 100-fold increased transgene expression from the immobilized complexes relative to bolus delivery of complexes [9]. To control vector binding to the substrate, the number of biotin groups and their distribution among the cationic polymer (PLL) were varied. Increasing the number of biotin groups per complex led to increased binding [9]. However, in vitro transfection was maximal when complexes contained biotin residues attached to a small fraction of the cationic polymers [10]. Additionally, transfection was observed only in the location to which complexes were bound, suggesting the possibility of spatially regulating DNA delivery.
Plasmid DNA or DNA complexed with cationic polymers or lipids can also interact with substrates through non-specific, non-covalent mechanisms [11–15], including hydrophobic, electrostatic, and van der Waals interactions. These interactions have been well-characterized for adsorption and release of proteins from polymeric systems [16,17]. Polyplexes and lipoplexes non-specifically immobilized to substrates have been shown to enhance the extent of transgene expression in both cell lines and primary human-derived cells, along with an increased cellular viability [14]. This enhancement was dependent on both the properties of the complex (e.g., complexation agent, N/P ratio), and substrate. However, the properties of the substrate that mediate gene transfer remain poorly understood.
In this report, self-assembled monolayers (SAMs) of alkanethiols on gold were used to investigate substrate-mediated transfection by non-specifically immobilized complexes. SAMs provide a flexible system to regulate the terminal functional group chemistry to examine the complex–substrate interactions [18–20]. Hydrophilic substrates with varying densities of ionic functional groups, as well as hydrophobic substrates, were examined for their ability to bind and release complexes, and to subsequently support transfection. Furthermore, the flexibility in surface chemistry of SAMs permits preparation of patterned surfaces with varying degrees of surface charge and wettability. Microcontact printing (μCP), a soft lithographic technique, was used to imprint gold substrates with specific patterns of SAMs using an elastomeric stamp [21–24], creating regions of different surface properties. Here, we demonstrate the ability to use micropatterning to pattern DNA complex immobilization and transfection.
2. Material and methods
2.1. Gold slide preparation and monolayer self assembly
Gold-coated glass slides, composed of a titanium adhesion layer and 100–500 Å of gold, were prepared using e-beam evaporation (Edwards Electron Beam Evaporator, Wilmington, MA). The gold-coated slides were then cut into smaller pieces with a diamond-tipped glass cutter, so that pieces fit into standard 48-well tissue culture plates. These gold pieces were prepared for SAM formation by treatment with oxygen plasma (Harrick Scientific, Ossining, NY), followed by sonication in ethanol, or alternatively cleaned in acetone and ethanol, with subsequent drying under a stream of nitrogen.
SAMs were formed by immersion of the clean gold substrates into 2 mM ethanolic solutions of alkanethiols for 30 min to overnight in the dark, depending on monolayer characteristics. Monolayers were formed with three different alkanethiols and combinations thereof, including 1-decanethiol (DT10), 11-mercapto-1-undecanol (MUOH), and 11-mercaptoundecanoic acid (MUA) (Aldrich, St. Louis, MO). Alkanethiol solutions were (freshly) prepared in filtered, degassed ethanol. After monolayer formation, SAM samples were rinsed in pure ethanol and dried with nitrogen before further use.
2.2. Verification of surface chemistry
X-ray photoelectron spectroscopy (XPS) was used to analyze the surface functional groups of the SAMs to verify successful modification. XPS spectra were recorded using an Omicron ESCA Probe system with an Al/Mg anode X-ray source. A single survey scan spectrum (range 20–1000 eV) and 3–5 narrow scans for C1s (275–295 eV) and O1s (525–550 eV) were recorded for each sample. Analysis of the data was performed using Multipak software (Physical Electronics, Inc., Eden Prairie, MN). Chemical shifts were referenced to C1s at 285 eV. Surface modification by SAM formation was also analyzed by the measurement of contact angles of water in air on the various SAMs at room temperature with a goniometer (Ramé-Hart, Mountain Lakes, NJ).
2.3. Quantification of DNA complex immobilization and release
Plasmid DNA encoding for luciferase (LUC) and enhanced green fluorescent protein (EGFP) with a CMV promoter was purified from bacteria culture using Qiagen (Santa Clara, CA) reagents and stored in Tris–EDTA buffer (10 mM Tris, 1 mM EDTA, pH = 7.4) for binding, release and transfection experiments. For DNA complex formation, Lipofectamine 2000 (Life Technologies, Gaithersburg, MD) in serum-free cell growth media (DMEM, Life Technologies) was added drop wise to DNA in serum-free cell growth media, mixed by gentle pipeting, and incubated for 20 min.
The binding and release of DNA complexes from various SAMs was monitored using plasmid DNA radiolabeled with α-32P dATP. Briefly, a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NY) was used following the manufacturer’s protocol with minor modifications [10]. The labeled DNA was diluted with unlabeled DNA to a final concentration of 1% and this mixture was then used to form DNA complexes. After SAM preparation, digital photographs of each sample were taken before complex immobilization, and analyzed with ImageJ (NIH) to determine the area of each surface. Complexes (1 μg of DNA at a DNA (μg):lipid (μL) ratio of 1:2) were immobilized by incubation on SAMs for 2 h, followed by two wash steps with serum-free cell growth media. The quantity of DNA immobilized was determined by immersing individual SAM samples in scintillation cocktail (5 mL, ScintiVerse II) for measurement with a scintillation counter. The counts were correlated to DNA mass using a standard curve. The density of DNA immobilized to each SAM sample was determined by normalizing the amount bound to area and reported as the mean ± sem of three replicates.
The release profiles from SAMs with immobilized DNA complexes were determined by incubation with serum-containing cell growth media at 37 °C in a humid chamber. At predetermined time points, half of the media was removed and replaced with fresh media. The activity of the collected sample was measured in a scintillation counter. At the final time point, the counts remaining on the SAM samples were also determined. The percentage of DNA released was calculated as the ratio of the cumulative counts released through a given time divided by the total counts initially on the substrate, thus, the release curves represent the percentage of DNA released relative to the initial amount bound to each surface.
2.4. Transfection on SAMs
Transfection studies were performed with NIH/3T3 (ATCC; Manassas, VA) cells cultured at 37 °C and 5% CO2 in DMEM supplemented with 1% sodium pyruvate, 1% penicillin–streptomycin, 1.5 g/L NaHCO3 and 10% fetal bovine serum. After complex formation and immobilization as described above, SAMs were immediately seeded with 15,000 cells in 48-well plates. Transfection was analyzed following a 48 h culture, characterized through the extent of transgene expression, which was quantified by measuring the luciferase activity using the Luciferase Assay System (Promega, Madison, WI). Cells were lysed and assayed for enzymatic activity after 48 h. The luminometer (Turner Designs, Sunnyvale, CA) was set for a 3 s delay and an integration of the signal for 10 s. Luciferase activity was normalized to the total protein amount determined with the BCA protein assay (Pierce, Rockford, IL). The effect of SAMs on cell morphology and adhesion was determined by cell seeding on SAMs, as described above, in the absence of DNA complex immobilization.
2.5. Patterned SAMs and complex deposition
Microcontact printing (μCP) with a polydimethylsiloxane (PDMS) stamp was used to imprint gold surfaces with specific patterns of hydrophilic alkanethiols, which could be used to regulate the location of DNA deposition and subsequent delivery from the surface. For stamp fabrication, PDMS was prepared in a 10:1 (v:v) ratio of Silicone Elastomer-184 and Silicone Elastomer Curing Agent-184 (Sylgard 184, Dow Corning, Midland, MI) by mixing the base and curing agent at least 50 times using a syringe mixing system. After allowing all air bubbles to escape, the PDMS was poured directly into a metal mesh master mold, situated between two blocks of polyethylene with neoprene spacers, and cured at room temperature for approximately 3 days. The stamp was then removed from the mold, and washed with ethanol and dried under nitrogen before each use. This master produced stamps with approximately 1 mm features. Alternatively, PDMS stamp molds were also fabricated by spin coating SU-8 negative photoresist (MicroChem, Newton, MA) onto a silicon wafer, exposing photoresist to UV through a mask for 10 s, and then developing the photoresist following the manufacturer’s instructions. The mask was prepared using a computer drawing program to create a pattern that was printed onto a transparency [24,25], also with 1 mm features. PDMS (5.5 g) was poured onto the photolithographic mold, cured at 60 °C for 1–3 h, and then carefully peeled away from mold. The stamp was cleaned in an oxygen plasma cleaner, rinsed in acetone and methanol, and dried under nitrogen before each use.
For μCP of alkanethiols, the PDMS stamp was inked with a 2 mM solution of 50% MUA/50% MUOH thiols and then dried under nitrogen. The stamp was then placed in contact with a gold substrate for 1–5 min. The stamped substrate was then carefully peeled from the stamp and immersed into a 2 mM solution of DT10 for 15–60 min. After derivatization with the secondary thiol, the stamped surface was washed twice in ethanol and dried under nitrogen.
Secondary ion mass spectrometry (SIMS) was used to verify the spatial distribution of SAMs on the stamped surface by imaging the two-dimensional distribution of chemical species on the submicron scale. SIMS spectra and images for bulk and patterned surfaces were recorded using PHI TRIFT III ToF-SIMS system (Physical Electronics, Inc., Eden Prairie, MN). The two-dimensional distribution was obtained using stage raster imaging techniques, scanning specifically for OH, S, C2H2O2, Au, CHO2, C2H3O2, and C2H chemical species in negative mode. Furthermore, water droplets termed condensation figures (CFs), which form from regions with different wettabilities [26], were used as an additional method to verify and image SAM patterns on the surface, by simply pipeting MilliQ water onto the stamped surfaces.
Plasmid DNA (pEGFP-LUC) was labeled with tetramethyl rhodamine (Label IT Nucleic Acid Labeling Kit, Mirus, Madison, WI). This DNA was used to form complexes with Lipofectamine 2000, as described above. Complexes were then deposited onto patterned SAM surfaces in CFs formed on the hydrophilic areas. Complexes were allowed to deposit for a period of 1 h, in humid conditions, and then visualized with fluorescence microscopy. Control deposition studies were performed with rhodamine-labeled DNA complexes immobilized on uniform 50% MUA/50% MUOH and DT10 SAMs.
2.6. Patterned transfection
On a patterned SAM surface, 0.5 μg of plasmid DNA (pEGFP-LUC) complexed with lipid (1:2.5, DNA to lipid ratio) in a total volume of 50 μL, was deposited in CFs that formed on the hydrophilic regions over the entire surface for 1 h, in humid conditions. The droplets were then removed with a stream of nitrogen and deposition was repeated for up to five times, with freshly prepared complexes using the same conditions as above. The patterned surface was then washed with media and NIH/3T3 cells were seeded as described above. Transfection was analyzed following a 48 h culture and characterized through the number of transfected cells, using GFP expression. Transfected cells were visualized and manually counted using an epifluorescence microscope (Leica; Bannockburn, IL) with a FITC filter and equipped with a digital camera. The percentage of transfected cells was calculated as the ratio of the number of transfected cells divided by total cell number, which was determined by manual counting of phase images. Control transfection studies were performed on uniform 50% MUA/50% MUOH and DT10 SAMs.
2.7. Statistics
Statistical analysis was performed using JMP software (SAS Institute, Inc., Cary, NC). Comparative analyses were completed using one-way ANOVA with Tukey post-tests, at a 95% confidence level. Mean values with standard error of the mean are reported and all experiments were performed in triplicate.
3. Results and discussion
3.1. Surface characterization
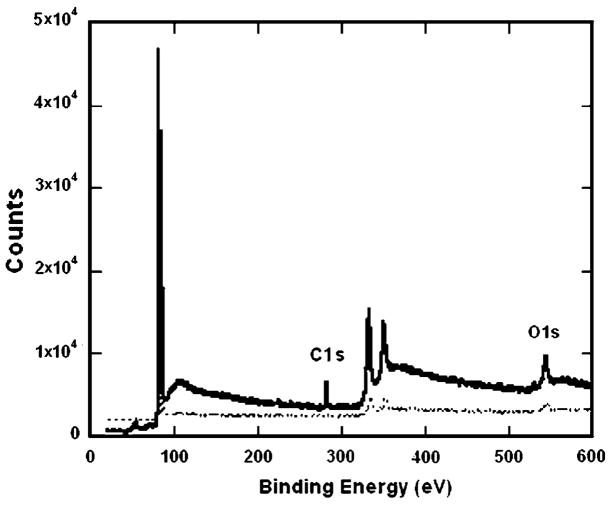
SAM formation was confirmed with XPS and contact angle measurements. The XPS spectra (Fig. 1) indicated that while the gold substrate showed minimal carbon, oxygen, or sulfur peaks, the MUA SAM sample containing carboxylic acid groups had a peak at binding energy ~289 eV, characteristic of the carbon in the COOH groups [27]. Additionally, the O1s peak intensity further verified the presence of carboxylic acid groups in the MUA sample. The low sulfur intensity was similar to XPS spectra previously reported [27]. Spectra for other SAMs also indicated successful modification (data not shown). Substrate modification by SAM formation was further confirmed by the measurement of contact angles of water droplets in air on the various SAMs. The wettability of the surfaces indicated that the DT10 SAMs were hydrophobic (angles greater than 110°), while surfaces with SAMs of MUOH and MUA were hydrophilic (angles less than 30°), as expected [19,27,28]. Additionally, contact angles for all SAMs were different than that of gold, indicating surface modification.
Fig. 1.
XPS verification of SAM formation. XPS spectrum of a MUA surface (—) relative to a gold surface (- - -) indicated increases in carboxylic acid groups at binding energy ~289 eV, characteristic of the carbon in the COOH groups. The O1s peak intensity further verified the presences of the carboxylic acid groups in the MUA sample.
3.2. Quantification of complex immobilization
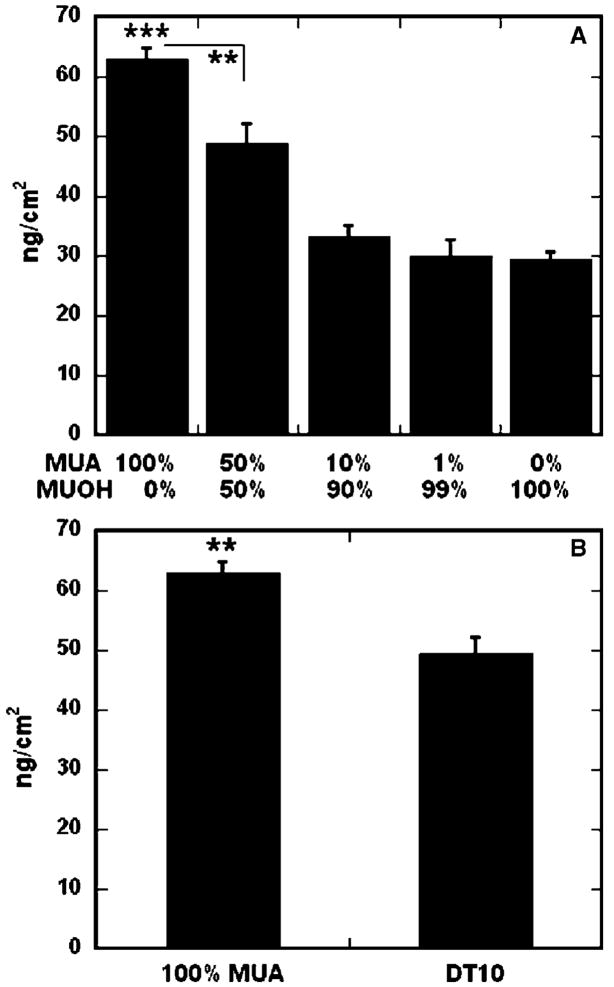
SAMs of alkanethiols on gold were used to investigate the relationship between surface properties and non-specific immobilization of DNA complexes. Both hydrophilic and hydrophobic surfaces were investigated, including hydrophilic substrates with varying densities of charged, carboxylic acid groups. The amount of DNA immobilized to the surfaces ranged from 63 ng/cm2 to 29.5 ng/cm2, as the percentage of carboxylic acid functional groups decreased from 100% (100% MUA) to 0% (0% MUA, 100% MUOH) (Fig. 2A). Binding of DNA complexes on 100% MUA surfaces was significantly greater than 50% MUA SAMs (p < 0.01), as well as 10% MUA, 1% MUA, and 0% MUA SAMs (p < 0.001). Surfaces containing 50% carboxylic acid groups resulted in statistically greater binding than substrates with 10% (p < 0.01) or 1% (p < 0.001) charged groups.
Fig. 2.
DNA-complex binding on SAMs. The amount of immobilized radiolabeled DNA was determined for varying densities of carboxyl groups (A) and surface hydrophobicity (B). Percentage of carboxyl groups (A) refer to a background of MUOH SAMs. All values are reported as the mean ± sem (**p < 0.01,***p < 0.001).
In addition to the percentage of carboxylic acid groups, the hydrophobicity of the SAMs also affected the amount of immobilized DNA complexes (Fig. 2B). While complex binding was statistically greater on a hydrophilic SAM containing 100% carboxylic acid functional groups as compared to a hydrophobic DT10 SAM (49.4 ng/cm2, p < 0.01), the opposite was true for hydrophilic surfaces with few or no charged functional groups. DNA complex adsorption was statistically greater on a hydrophobic substrate (Fig. 2B) than hydrophilic surfaces containing 10% (p < 0.01) or fewer (p < 0.001) carboxylic acid groups (Fig. 2A). For any SAM substrate, the amount bound corresponded to less than 4% of DNA initially added to the surface, which is less than previously reported for similar deposition schemes [14].
Surface ionization and hydrophobicity were both found to mediate complex immobilization. Increasing the density of charged functional groups on the surface increased complex immobilization, suggesting that electrostatic interactions play a major role in binding. However, statistically greater adsorption to a hydrophobic substrate (DT10) as compared to a non-ionic, hydrophilic surface (100% MUOH) suggests that immobilization can also be mediated by hydrophobic interactions. Substrate hydrophobicity has previously been shown to play a significant role in relative local DNA plasmid concentration within deposited spots of an array [19,20,29,30], with higher surface immobilization on hydrophobic polystyrene substrates as compared to more hydrophilic, treated surfaces. DNA polyplex adsorption was also found to be higher on polystyrene substrates, as compared to more hydrophilic, serum-modified surfaces [14].
SAMs present an opportunity to dissect the contributions of specific surface functional groups on complex immobilization and substrate-mediated transfection. Our findings reveal that non-specific DNA complex adsorption is mediated by at least two mechanisms: adsorption by charge–charge interactions and adsorption by hydrophobic interactions, two mechanisms which have been shown to be involved in non-specific protein adsorption [19,20,29]. However, like protein adsorption [19,31], DNA complex immobilization is presumably also affected by properties of the complexes themselves [10,14]. In contrast to our findings, a study by Yamauchi et al. [32] reported that immobilization of DNA complexes on SAMs was independent of surface chemistry, yet indicated that electrostatic interactions were most important for DNA complex immobilization. In addition, hydrophobic regions were shown to have tight interactions [32], which is similar to our finding that surface hydrophobicity and ionization affect DNA complex immobilization.
3.3. Quantification of complex release
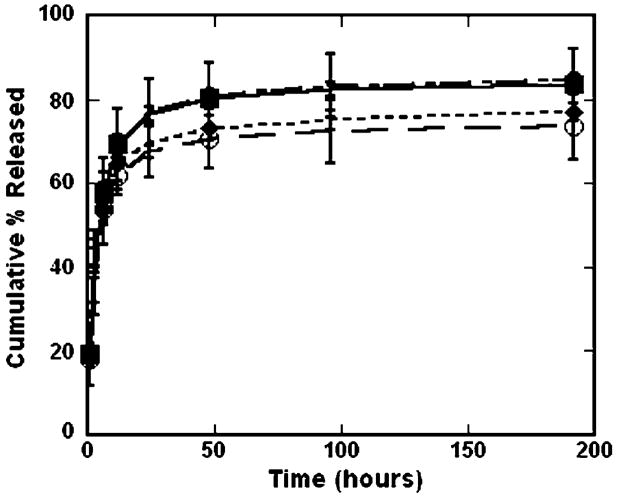
The stability of the interaction between the complexes and surface was investigated through release studies (Fig. 3). Release rates and total amount of DNA complexes released from SAMs was independent of surface chemistry (Fig. 3). Most release occurred by 24 h and after 8 days 70–85% of complexes were released. The release of DNA complexes increased from 73% for hydrophilic, ionic surfaces (100% MUA), 77% for hydrophobic surfaces (DT10), to 84% and 85% for 0% (100% MUOH) and 50% MUA, respectively. Similarly, Yamauchi et al. [32] found at high surface densities that surface chemistry did not affect the release rates of DNA complexes from SAMs. In contrast, they found that lower densities of immobilized complexes resulted in highest release on hydrophilic substrates, containing either carboxylic acid or hydroxyl terminal groups, whereas release was limited on hydrophobic surfaces due to tight interactions [32]. However, their release profiles were determined in PBS, which presumably only disrupted electrostatic interactions [32]. In this report, release profiles were performed in serum-containing growth media. The presence of serum has been shown to significantly enhance the release of non-specifically immobilized complexes relative to incubation with PBS [14]. This enhanced release from the substrate is presumably mediated by competitive binding of serum that displaces the complexes and components. Additionally, serum-containing media represents the most stringent release conditions, as it also contains salts that could disrupt electrostatic interactions, and also best emulates cell culture conditions. Our finding that release is independent of surface chemistry suggests that complex release from the substrate is mediated by competitive binding of serum that displaces the complexes, and components within the serum can bind through both electrostatic and hydrophobic interactions to the underlying SAMs.
Fig. 3.
Surface chemistry and release. Radiolabeled DNA was used to quantify the amount of DNA released from each type of SAM (● 50% MUA, ○ 100% MUA; ■ 100% MUOH, ◆ DT10) into serum-containing media. All values are reported as cumulative percentage released, reported as the mean ± sem at each time point.
3.4. Transfection on bulk SAMs on gold
Substrate-mediated delivery is based on the immobilization of DNA complexes to the culture substrate [9,10,14], resulting in elevated DNA concentrations in the cellular microenvironment, which has been shown to enhance gene delivery [7,33]. While immobilizing DNA to a substrate may seem counterintuitive as cellular internalization is necessary for expression, viral vectors have been shown to associate with the extracellular matrix to facilitate cellular binding and internalization [34,35]. The properties of the substrate are critical determinants of the interaction strength between the complexes and the surface, which subsequently affects the extent of transfection. SAMs were explored to examine the relationship between surface chemistry and substrate-mediated transfection. SAMs of alkanethiols on gold can be used in cell culture for periods of days [25] and have been used in many cell culture studies, particularly to understand molecular surface determinants required for adhesion dependent cell growth and proliferation [36–38].
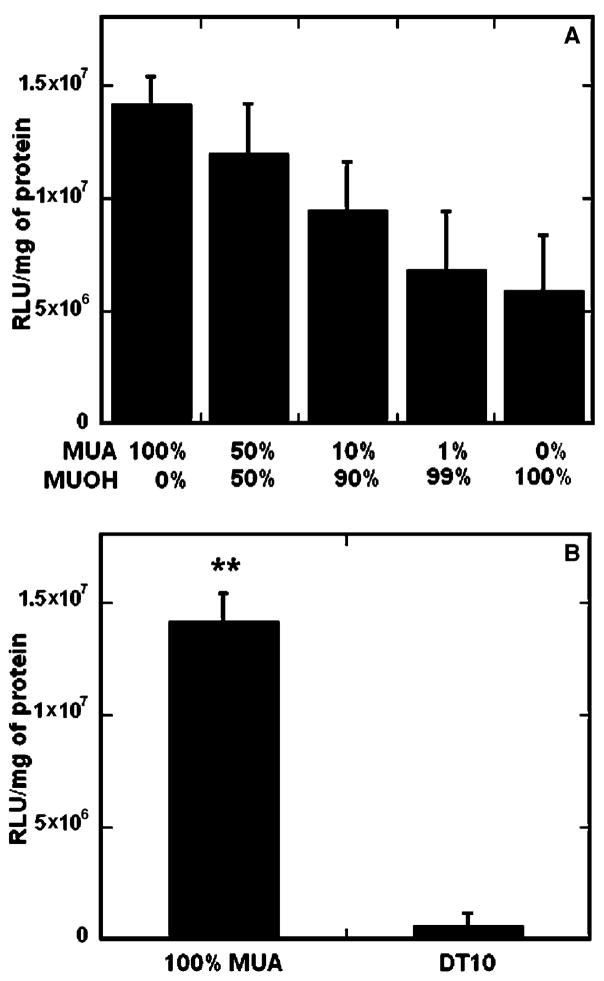
Transfection, like immobilization, was affected by both surface hydrophilicity and ionization (Fig. 4), with the greatest transfection on surfaces containing 100% carboxylic acid functional groups. Increasing the percentage of ionic functional groups presented at the surface increased the extent of transfection, with 2.4- fold greater transfection on 100% MUA surface as compared to a non-ionic hydrophilic substrate (100% MUOH) (Fig. 4A), similar to previous reports [32]. The trend for increased transfection mirrored that of DNA immobilization (Fig. 2A), as over 2-fold more DNA complexes were bound to the 100%MUA surfaces as compared to 100% MUOH. Delehanty et al. [30] also found that an increase in the amount of deposited DNA resulted in a corresponding increase in transfection efficiency.
Fig. 4.
Surface chemistry and substrate-mediated transfection. SAMs were formed with an increasing density of carboxyl groups (A) and varying hydrophobicity (B). All values are reported as the mean ± sem (**p < 0.01).
Complexes immobilized on hydrophilic, ionic surfaces (100% MUA) resulted in statistically greater transfection (p < 0.01) than hydrophobic substrates (Fig. 4B). While DNA complex immobilization was less than 1.5-fold greater on 100% MUA than hydrophobic surfaces (Fig. 2B), transfection was over 2.5-fold lower on the hydrophobic substrate (DT10). Additionally, transfection was greater on all hydrophilic surfaces, including 0–10% MUA, compared to the DT10 surface, in contrast to statistically greater immobilization of DNA complexes on the hydrophobic surface (Fig. 2B). Low transfection on the DT10 substrates could be due to a combination of factors, including insufficient cell adhesion [37] or complex conformation changes induced upon binding, which have been shown to contribute to irreversible binding of proteins to hydrophobic surfaces [39]. Total protein amount on the hydrophobic surfaces, as determined by the BCA assay, was similar or greater than all other surfaces, indicating that low transfection on these substrates was not due to insufficient cell numbers (data not shown). Additionally cell morphology and adhesion were similar on hydrophilic and hydrophobic surfaces (Fig. 5). Yamauchi et al. have proposed that tight interactions of the DNA complexes with hydrophobic substrates can lead to lower transfection efficiencies [32]. For DNA polyplexes or lipoplexes adsorbed to serum-modified, hydrophilic surfaces, the amount bound was similar as to those immobilized on hydrophobic, polystyrene surfaces, however the extent of transfection was greater on the hydrophilic substrates [14], consistent with results reported here.
Fig. 5.
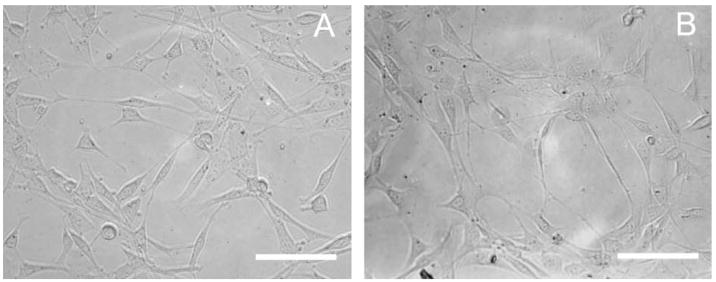
Cell adhesion on SAMs. The morphology and adhesion of cells was observed 24 h after seeding cells on SAMs of (A) 50% MUA and (B) DT10, both lacking immobilized DNA complexes. Scale bars correspond to 200 μm.
Charged, hydrophilic substrates, which may provide reversible interactions between the substrate and complex, provides the most efficient gene delivery, requiring orders of magnitude less DNA on the surface as compared to previous reports [14]. Transfection was highest on the hydrophilic substrates, and transgene expression was significantly increased on hydrophilic substrates with similar or less amounts of DNA relative to hydrophobic substrates. The conformation of the DNA complexes may be altered upon binding to hydrophobic surfaces and explain low transfection levels. Thus, variability in the type of interactions between the complexes and substrate, mediated by changes in hydrophobicity and ionization, result in the different transfection levels observed on SAMs of varying surface chemistries.
3.5. Patterned SAMs and complex deposition
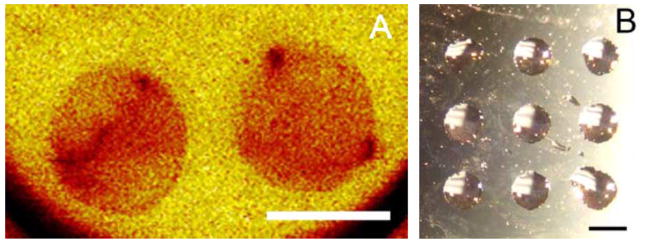
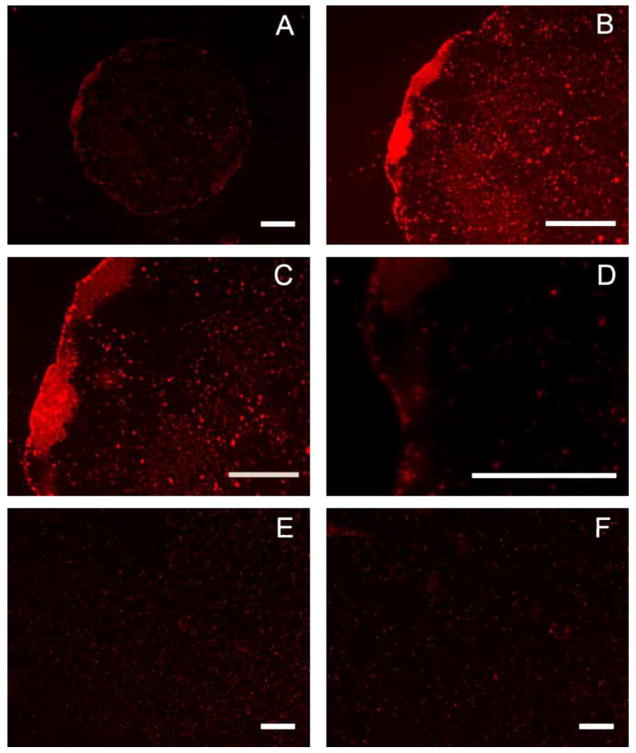
SAMs can be used in conjugation with μCP, a soft lithographic technique, to imprint substrates with specific patterns of SAMs [21,22,24]. A PDMS stamp was used to imprint gold surfaces with specific patterns of hydrophilic alkanethiols (50% MUA) in a background of methyl-terminated alkanethiols (DT10). Secondary ion mass spectrometry (SIMS) verified the spatial carboxylic acid-terminated alkanethiolates. Additionally, the formation of CFs, created by simply pipeting water onto the stamped surface, was used as a method to verify and image SAM patterns on the surface (Fig. 6B). These CFs were also utilized to confine droplets containing DNA complexes to specific regions of the surface, resulting in patterned DNA immobilization on a surface. Rhodamine-labeled DNA complexes were deposited on patterned surfaces in CFs formed on the hydrophilic areas (Fig. 7). Complexes were seen to be confined to the circular patterns created by stamping, with relatively even distributions over the entire circular hydrophilic region (Fig. 7A–D), in contrast to unpatterned surfaces (Fig. 7E and F).
Fig. 6.
Microcontact printing on SAMS. (A) Secondary Ion Mass Spectrometry (SIMS) of stamped surface imaged the relative concentration of the mapped molecules, with red referring to higher concentrations of the hydrophilic alkanethiols. (B) Condensation figures created by pipeting water onto the stamped surface, which quickly collected in the hydrophilic regions. Gold-coated glass slides were used to prepare SAMs (Platypus Technologies, Madison, WI). Scale bars correspond to 1 mm.
Fig. 7.
Patterned complex deposition. Condensation figures were utilized to confine droplets containing rhodamine-labeled DNA complexes to hydrophilic regions of a patterned surface, resulting in patterned DNA immobilization (A–D). Complexes were allowed to deposit for 1 h, in humid conditions, and then visualized with fluorescence microscopy. Control complex deposition was performed on unpatterned 50% MUA (E) and DT10 (F) SAMs. Scale bars correspond to 200 μm (A, B, E, F) and 100 μm (C, D).
3.6. Patterned transfection
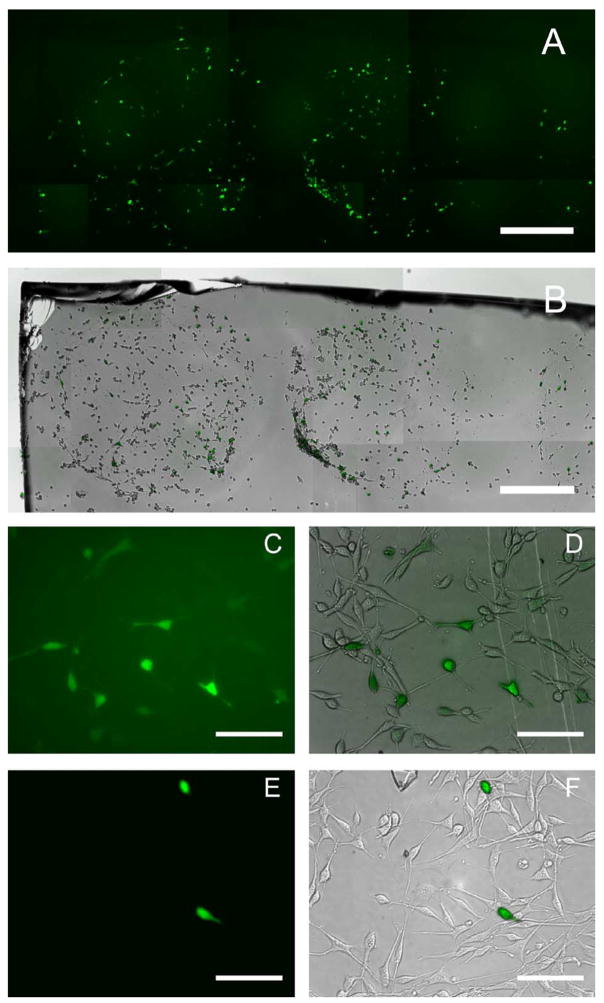
SAM surfaces patterned in regular arrays of hydrophobic and hydrophilic regions result in water condensation preferentially on the hydrophilic regions [26,40]. These hydrophilic/hydrophobic arrays can be used to anchor droplets for concentrating a sample during evaporation, to prepare protein samples for MALDI-MS [41], or to pin aqueous solutions of DNA at specific array locations [42–44]. Additionally, DNA complexes can be deposited in CFs on the hydrophilic regions to spatially regulate immobilization and upon seeding of cells, to pattern transfection. In our study, expression was examined by quantifying the percentage of transfected cells within the patterns (Fig. 8). GFP expression was assayed at 24 and 48 h (data not shown) using fluorescence microscopy. Transfection was confined to the patterns at both time points and cells in these patterns exhibited transfection efficiencies near 40% (Fig. 8A), similar to unpatterned substrates (Fig. 8C–F). Cellular adhesion was also patterned, as adhesion was greater on carboxyl-terminated SAMs (Fig. 8B). This finding supports previous studies reporting the inability of cells to adhere to methyl-terminated SAMs in the presence of serum [36–38,45]. Selective cellular adhesion suggests that hydrophilic/hydrophobic patterning strategies not only aid in the placement of complexes [32,42,43] but also the attachment of cells [36–38], resulting in patterns of transfection. In a similar study, micropatterned SAMs greatly facilitated regionally defined loading of DNAs and their expression in mammalian cells [32], however cellular adhesion was not patterned, presumably due to choice of cell type. We have found that on our patterned substrates, the hydrophilic regions support cell adhesion and high transfection, while the hydrophobic regions limit cellular attachment, properties, which could be important to the fabrication of a transfected cell array.
Fig. 8.
Patterned gene expression. Fluorescence and phase images were acquired and assembled to represent the entire patterned region (A, B). Transfection was confined to patterns and transfection efficiency within patterns was over 30%. Control transfections were performed on uniform 50% MUA (C, D) and DT10 (E, F) SAMs. Gold-coated glass slides were used for SAM preparation (Platypus Technologies). Scale bar corresponds to 500 μm (A, B) and 200 μm (C–F).
Patterned gene delivery from a surface can be translated to a transfected cell array, which represents a high throughput approach to correlate gene expression with functional cell responses [46]. Transfected cell arrays have been formed using a substrate-mediated approach in which plasmids or adenoviruses were mixed with collagen and spotted onto glass slides or into wells [46–48]. Plated cells were transfected and could be analyzed for cellular responses using a variety of imaging or biochemical techniques. These systems have also been used for transfection of siRNAs, which were printed in cationic lipid/Matrigel mixtures [49]. Alternatively, Chang et al. [50] developed a technology they refer to as “surfection”, in which a cationic polymer (PEI) and collagen are coated onto a surface. Plasmid DNA is then mixed with cells that are plated onto this surface, which is divided up into wells by a silicone rubber sheet, to pattern transfection. Also, DNA complexes have been mixed with fibronectin and spotted onto glass slides for arrayed transfection of human mesenchymal stem cells [51]. While transfected cell arrays hold great potential [46–51], further development of a substrate-mediated delivery system that efficiently transfects a wide variety of primary cells and cell lines, while allowing for spatially-controlled delivery within the different domains is required [46,52,53]. Using SAMs on gold to form transfected cell arrays can allow precise control over surface chemistries, interactions between the substrate and DNA complexes, transfection efficiencies, and pattern sizes to create a well-characterized and efficient array delivery system.
4. Conclusions
In substrate-mediated delivery, DNA is immobilized to a substrate for delivery to cells that adhere to the substrate [8,9]. Efficient delivery of DNA complexes from a surface is dependent on balancing the interactions between the substrate and the complexes. SAMs provide a versatile and flexible system to correlate surface chemistry to DNA complex binding, release and transfection. Surface hydrophobicity and ionization were found to mediate both complex immobilization and transfection, while release was independent of substrate properties. Increasing the density of carboxylic acid groups on the surface increased complex immobilization and transfection, suggesting that electrostatic interactions contribute to efficient gene delivery from a substrate. Additionally, μCP was used to imprint substrates with specific patterns of SAMs, creating hydrophilic regions within a hydrophobic background. Condensation figures containing DNA complexes formed preferentially on the hydrophilic regions, resulting in patterned complex immobilization and transfection. The ability to control the interactions between complexes and substrates, combined with patterning strategies, has multiple applications, such as scaffolds for tissue engineering and functional genomic screens in cell-based assays.
Acknowledgments
Support for this research was provided in part by grants from NIH (RO1 GM066830 to L.D.S., CMBD Training Grant to A.K.P.) and NSF (Graduate Research Fellowship to A.K.P.). Gold evaporation was performed at the Materials Processing and Crystal Growth core facility and the XPS measurements were performed at Keck-II/Nuance core facility (Dr. Nick Wu), both at Northwestern University (Evanston, IL). We would like to thank Kinjal Shah, Peter Chung, Zain Bengali, Tiffany Houchin, Dr. Tatiana Segura, Christopher Campbell (Northwestern University), and Dr. Victor Luna-Perez (IIT) for technical assistance.
References
- 1.Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647–52. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 2.Herweijer H, Wol JA. Progress and prospects: naked DNA gene transfer and therapy. Gene Ther. 2003;10:453–8. doi: 10.1038/sj.gt.3301983. [DOI] [PubMed] [Google Scholar]
- 3.Segura T, Shea LD. In: Ann Rev Mater Sci Annual Reviews. Stupp S, editor. 2001. pp. 25–36. [Google Scholar]
- 4.Ledley FD. Pharmaceutical approach to somatic gene therapy. Pharm Res. 1996;13:1595–614. doi: 10.1023/a:1016420102549. [DOI] [PubMed] [Google Scholar]
- 5.Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4:438–46. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- 6.Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol Ther. 2004;10:19–26. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18:893–5. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 8.Levy RJ, et al. Localized adenovirus gene delivery using antiviral IgG complexation. Gene Ther. 2001;8:659–67. doi: 10.1038/sj.gt.3301452. [DOI] [PubMed] [Google Scholar]
- 9.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13:621–9. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 10.Segura T, Volk MJ, Shea LD. Substrate-mediated DNA delivery: role of the cationic polymer structure and extent of modification. J Control Release. 2003;93:69–84. doi: 10.1016/j.jconrel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Bielinska AU, et al. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21:877–87. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- 12.Kneuer C, Sameti M, Bakowsky U, Schiestel T, Schirra H, Schmidt H, et al. A nonviral DNA delivery system based on surface modified silica-nanoparticles can efficiently transfect cells in vitro. Bioconjug Chem. 2000;11:926–32. doi: 10.1021/bc0000637. [DOI] [PubMed] [Google Scholar]
- 13.Manuel WS, Zheng JI, Hornsby PJ. Transfection by polyethyleneimine-coated microspheres. J Drug Target. 2001;9:15–22. doi: 10.3109/10611860108995629. [DOI] [PubMed] [Google Scholar]
- 14.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, et al. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotech Bioeng. 2005;90:290–302. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JT, Chua LS, Lynn DM. Multilayered thin films that sustain the release of functional DNA under physiological conditions. Langmuir. 2004;20:8015–21. doi: 10.1021/la048888i. [DOI] [PubMed] [Google Scholar]
- 16.Putney SD, Burke PA. Improving protein therapeutics with sustained-release formulations. Nat Biotechnol. 1998;16:153–7. doi: 10.1038/nbt0298-153. [DOI] [PubMed] [Google Scholar]
- 17.Norde W, Lyklema J. Why proteins prefer interfaces. J Biomater Sci Polym Ed. 1991;2:183–202. doi: 10.1080/09205063.1991.9756659. [DOI] [PubMed] [Google Scholar]
- 18.Ulman A. Formation and structure of self-assembled monolayers. Chem Rev. 1996;96:1533–54. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 19.Sigal GB, Mrksich M, Whitesides GM. Effect of surface wettability on the adsorption of proteins and detergents. J Am Chem Soc. 1998;120:3464–73. [Google Scholar]
- 20.van der Veen M, Norde W, Stuart MC. Electrostatic interactions in protein adsorption probed by comparing lysozyme and succinylated lysozyme. Colloids Surf B-Biointerf. 2004;35:33–40. doi: 10.1016/j.colsurfb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Whitesides GM. Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol “ink” followed by chemical etching. Appl Phys Lett. 1993;63:2002–4. [Google Scholar]
- 22.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, et al. Engineering cell shape and function. Science. 1994;264:696–8. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 23.Kumar A, Biebuyck HA, Whitesides GM. Patterning self-assembled monolayers: applications in material science. Langmuir. 1994;10:1498–511. [Google Scholar]
- 24.Xia Y, Whitesides GM. Soft lithography. Angew Chem, Int Ed. 1998;37:550–75. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–76. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 26.Lopez GP, Biebuyck HA, Frisbie CD, Whitesides GM. Imaging of features on surfaces by condensation figures. Science. 1993;260:647–9. doi: 10.1126/science.8480175. [DOI] [PubMed] [Google Scholar]
- 27.Troughton EB, Bain CD, Whitesides GM. Monolayer films prepared by the spontaneous self-assembly of symmetrical and unsymmetrical dialkyl sulfides from solution onto gold substrates: structure, properties, and reactivity of constituent functional groups. Langmuir. 1998;4:365–85. [Google Scholar]
- 28.Tsai MY, Lin JC. Surface characterization and platelet adhesion studies of self-assembled monolayer with phosphonate ester and phosphonic acid functionalities. J Biomed Mater Res. 2001;55:554–65. doi: 10.1002/1097-4636(20010615)55:4<554::aid-jbm1049>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 29.Barrias CC, Martins MA, Miranda MA, Barbosa MA. Adsorption of a therapeutic enzyme to self-assembled monolayers: effect of surface chemistry and solution pH on the amount and activity of adsorbed enzyme. Biomaterials. 2005;26:2695–704. doi: 10.1016/j.biomaterials.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 30.Delehanty JB, Shaffer KM, Lin B. A comparison of microscope slide substrates for use in transfected cell microarrays. Biosens Bioelectron. 2004;20:773–9. doi: 10.1016/j.bios.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Wadu-Mesthrige K, Amro NA, Liu GY. Immobilization of proteins on self-assembled monolayers. Scanning. 2000;22:380–8. doi: 10.1002/sca.4950220607. [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi F, Kato K, Iwata H. Micropatterned, self-assembled monolayers for fabrication of transfected cell microarrays. Biochim Biophys Acta. 2004;1672:138–47. doi: 10.1016/j.bbagen.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Luo D, Han E, Belcheva N, Saltzman WM. A self-assembled, modular DNA delivery system mediated by silica nanoparticles. J Control Release. 2004;95:333–41. doi: 10.1016/j.jconrel.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 34.Proctor RA. Fibronectin: a brief overview of its structure, function, and physiology. Rev Infect Dis. 1987;9(Suppl 4):S317–21. doi: 10.1093/clinids/9.supplement_4.s317. [DOI] [PubMed] [Google Scholar]
- 35.Bajaj B, Lei P, Andreadis ST. High efficiencies of gene transfer with immobilized recombinant retrovirus: kinetics and optimization. Biotechnol Prog. 2001;17:587–96. doi: 10.1021/bp010039n. [DOI] [PubMed] [Google Scholar]
- 36.Haddow DB, France RM, Short RD, MacNeil S, Dawson RA, Leggett GJ, et al. Comparison of proliferation and growth of human keratinocytes on plasma copolymers of acrylic acid/1,7-octadiene and self-assembled monolayers. J Biomed Mater Res. 1999;47:379–87. doi: 10.1002/(sici)1097-4636(19991205)47:3<379::aid-jbm13>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 37.Cooper E, Wiggs R, Hutt DA, Parker L, Leggett GJ, Parker TL. Rates of attachment of fibroblasts to self-assembled monolayers formed by the adsorption of alkylthiols onto gold surfaces. J Mater Chem. 1997;7:435–41. [Google Scholar]
- 38.McClary KB, Ugarova T, Grainger DW. Modulating fibroblast adhesion, spreading, and proliferation using self-assembled monolayer films of alkylthiolates on gold. J Biomed Mater Res. 2000;50:428–39. doi: 10.1002/(sici)1097-4636(20000605)50:3<428::aid-jbm18>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 39.Stuart MAC, Fleer GJ, Lyklema J, Norde W, Scheutjens JMHM. Adsorption of ions, polyelectrolytes and proteins. Adv Colloid Interf Sci. 1991;34:477–535. doi: 10.1016/0001-8686(91)80056-p. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Whitesides GM. Patterned condensation figures as optical diffraction gratings. Science. 1994;263:60–2. doi: 10.1126/science.263.5143.60. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y, Watson JT, Bruening ML. Patterned monolayer/polymer films for analysis of dilute or salt-contaminated protein samples by MALDI-MS. Anal Chem. 2003;75:185–90. doi: 10.1021/ac025907p. [DOI] [PubMed] [Google Scholar]
- 42.Brockman JM, Frutos AG, Corn RM. A multistep chemical modification procedure to create DNA arrays on gold surfaces for the study of protein–DNA interactions with surface plasmon resonance imaging. J Am Chem Soc. 1999;121:8044–51. [Google Scholar]
- 43.Gillmore SD, Thiel AJ, Strother TC, Smith LM, Lagally MG. Hydrophilic/hydrophobic patterned surfaces as templates for DNA arrays. Langmuir. 2000;16:7223–8. [Google Scholar]
- 44.Zhang G, Yan X, Hou X, Lu G, Yang B, Wu L, et al. Binary DNA arrays on heterogeneous patterned surfaces. Langmuir. 2003;19:9850–4. [Google Scholar]
- 45.Lopez GP, Albers MW, Schreiber SL, Carroll R, Peralta E, Whitesides GM. Convenient methods for patterning the adhesion of mammalian cells to surfaces using self-assembled monolayers of alkanethiolates on gold. J Am Chem Soc. 1993;115:5877–8. [Google Scholar]
- 46.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–10. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 47.Honma K, et al. Atelocollagen-based gene transfer in cells allows high-throughput screening of gene functions. Biochem Biophys Res Commun. 2001;289:1075–81. doi: 10.1006/bbrc.2001.6133. [DOI] [PubMed] [Google Scholar]
- 48.Michiels F, et al. Arrayed adenoviral expression libraries for functional screening. Nat Biotechnol. 2002;20:1154–7. doi: 10.1038/nbt746. [DOI] [PubMed] [Google Scholar]
- 49.Mousses S, et al. RNAi microarray analysis in cultured mammalian cells. Genome Res. 2003;13:2341–7. doi: 10.1101/gr.1478703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang FH, Lee CH, Chen MT, Kuo CC, Chiang YL, Hang CY, et al. Surfection: a new platform for transfected cell arrays. Nucleic Acids Res. 2004;32:e33. doi: 10.1093/nar/gnh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshikawa T, Uchimura E, Kishi M, Funeriu DP, Miyake M, Miyake J. Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J Control Release. 2004;96:227–32. doi: 10.1016/j.jconrel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Wu RZ, Bailey SN, Sabatini DM. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 2002;12:485–8. doi: 10.1016/s0962-8924(02)02354-1. [DOI] [PubMed] [Google Scholar]
- 53.Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discov Today. 2002;7:S113–8. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]