Abstract
Efficient and controlled gene delivery from biodegradable materials can be employed to stimulate cellular processes that lead to tissue regeneration. In this report, a substrate-mediated approach was developed to deliver DNA from hyaluronic acid-collagen hydrogels. The hydrogels were formed by crosslinking HA with poly(ethylene glycol) diglycidyl ether. Poly(ethylene imine)(PEI)/DNA complexes were immobilized to the substrate using either biotin/neutravidin or non-specific adsorption. Complexes were formed in the presence or absence of salt to regulate complex size, and resulted in complexes with z-average diameters of 1221.7±152.3 and 139.4±1.3 nm, respectively. During 48-h incubation in PBS or hyaluronidase, DNA was released slowly from the hydrogel substrate (<30% of immobilized DNA), which was enhanced by incubation with conditioned media (≈50% of immobilized DNA). Transgene expression mediated by immobilized, large diameter complexes was 3 to 7-fold greater than for small diameter complexes. However, the percentage of cells expressing the transgene was greater for small diameter complexes (48.7%) than for large diameter complexes (22.3%). Spatially controlled gene transfer was achieved by topographically patterning the hydrogel to pattern cell adhesion. Biomaterial-based gene delivery can be applicable to numerous tissue engineering applications, or as a tool to examine tissue formation.
Keywords: Gene delivery, Polyethyleneimine, Neutravidin, Biotin, Reverse transfection, Solid plase
1. Introduction
Biomaterials for controlled delivery of plasmid DNA can provide a fundamental tool to promote localized transgene expression, which can be employed to direct cellular processes for numerous tissue engineering applications. Strategies to enhance non-viral gene delivery typically involve complexation of plasmids with cationic polymers or lipids. Cationic polymers or lipids can self-assemble with DNA to form particles that are capable of being endocytosed by cells [1]. These complexes are often delivered as a bolus, such as addition to culture wells in vitro or injection into a tissue in vivo. Mass transport or deactivation processes, such as degradation, aggregation, or clearance from the tissue, can limit bolus delivery of these complexes. Biomaterial systems for controlled delivery can overcome these barriers to promote localized gene transfer. In tissue engineering, these biomaterials can fulfill standard functions (e.g., maintaining space, supporting cell adhesion and migration) while the localized gene transfer can increase expression of tissue inductive factors to direct progenitor cell responses.
Controlled delivery of DNA complexes from biomaterials can enhance gene transfer by maintaining an elevated concentration of DNA within the cellular microenvironment via sustained release or substrate-immobilization [2]. Sustained release systems are designed to maintain elevated concentrations locally by supplying DNA to balance the loss by degradation and clearance. Alternatively, DNA can be immobilized within or to a biomaterial scaffold, a strategy that has been used by viruses that associate with extracellular matrix molecules (e.g., fibronectin) [3,4]. Synthetic systems are being developed that specifically bind viruses [5,6] or non-viral DNA complexes [7,8] to a polymeric substrate. Immobilization of DNA to the ARTICLE IN biomaterial to which cells adhere maintains the DNA in the cell microenvironment for subsequent cellular internalization.
Delivery strategies based on the immobilization of non-viral DNA complexes to biomaterials require that the DNA be maintained locally, yet allow for cellular internalization. In this report, we have developed procedures to immobilize non-viral DNA complexes to hydrogels and examined substrate binding, release profiles, and transgene expression. Plasmid DNA was complexed with poly(ethylene imine) (PEI) and immobilized to a hyaluronic acid-collagen hydrogel using non-covalent interactions (biotin/avidin and non-specific adsorption). Complex size was examined as a variable mediating substrate binding and transgene expression. The mechanism of gene transfer from the substrate was characterized by examining DNA release from the substrate and the incorporation of RGD peptides into the complex. Furthermore, topographically patterned hydrogels were used to orient cell growth and spatially regulate transgene expression on the hydrogel.
2. Materials and methods
2.1. Materials
Sodium hyaluronan (HA) was a gift from Genzyme Corporation (1330 kDa, Cambridge, MA). The cross-linker poly(ethylene glycol) diglycidyl ether (CH2OCH-(CH2CH2O)n-CHOCH2, PEGDG, n=200) was purchased from Polysciences (Warrington, PA). Adipic acid dihydrazide (AAD), N-(3-dimethylpropyl)-N-ethylcar-bodiimide hydrochloride (EDC) and N-hydroxysulfo-succinimide sodium salt (sulfo-NHS) were purchased from Sigma (St Louis, MO). Collagen was isolated from rat tails according to the procedure described by Choe et al. [9]. Plasmid DNA encoding for luciferase or green fluorescent protein was purified from bacteria culture using Qiagen (Santa Clara, CA) reagents and stored in Tris-EDTA buffer solution (pH=7.4). Linear PEI, biotinylated linear PEI (PEI-Biotin), and arginine–glycine–aspartic acid modified linear PEI (RGD-PEI) were purchased from PolyTransfection (7mm amine content, Strasbourg, France). Neutravidin (NA) and biotin reagents for HA modification were purchased from Pierce (Rockford, IL). All other reagents were obtained from Fisher Scientific (Fairlawn, NJ) unless otherwise noted.
2.2. Synthesis of biotinylated-hyaluronic acid
HA was biotinylated by first modifying its backbone with adipic acid dihydrazide (AAD) and then reacting the pendant hydrazide groups with sulfo-NHS-LC-biotin (NHS-Biotin) [10]. Briefly, EDC (0.5 equivalents) and sulfo-NHS (0.5 equivalents) were added as solids to the HA solution (10 mg/ml), followed by AAD (30 equivalents) addition. The reaction proceeded at room temperature for 16 h with mixing. HA-AAD was precipitated and washed in cold ethanol. The precipitate was stored at 4°C until use. Biotin was introduced to HA-AAD (HA-Biotin) using NHS-Biotin. NHS-Biotin (100 mg) was added as a solid to HA-AAD (300 mg, 3 mg/ml in PBS, pH 7.4). The reaction proceeded for 16 h at room temperature with stirring, and the product was purified by ethanol precipitation.
2.3. Hyaluronic acid/collagen hydrogel synthesis
HA-collagen hydrogels were formed using the reaction of the alcohols of biotinylated HA with the PEGDG crosslinker as previously described [10]. The hydrogels were formed with a final collagen concentration of 0.19 mg/ml. A topographical pattern was introduced into the hydrogel using a pattern transfer method. NA was bound by hydration of the hydrogel in a 1 µm NA solution in PBS for 1 h, followed by PBS wash.
2.4. Size and zeta potential measurements
Size and zeta potential of DNA/PEI complexes were measured using a Zetasizer Nano ZS instrument (Malvern, Worcestershire, UK). For complexation, DNA (50 µl, 40 µg/ml) was mixed with PEI at nitrogen to phosphate (N/P) ratios ranging from 2.5 to 20. Complexes were formed in either water or 150 mm NaCl in a volume of 100 µl. After a 10-min incubation, complexes were diluted 10 times with either water or 150 mm NaCl. To examine complex aggregation, complexes were formed in water and diluted 10 times with 150 mm NaCl, with size monitored over time.
2.5. DNA/PEI complex immobilization
DNA complexes were immobilized to HA-collagen hydrogels by incubation of complexes with the NA bound HA-collagen hydrogel. DNA (50 µl of 40 µg/ml in 150 mm NaCl or water) was complexed with PEI (50 µl of 150 mm NaCl or water) at a N/P ratio of 5. Mixing biotinylated and non-biotinylated PEI prior to mixing with DNA varied the degree of biotinylation for the complex. The complexes were incubated for 10 min, transferred to NA bound HA-collagen surfaces, and incubated for 2 h at room temperature. The unbound complexes were removed and the surfaces were washed extensively with PBS buffer.
Surface binding of the DNA/PEI complexes was examined using labeled DNA. Plasmid DNA was radiolabeled with α-32P dATP using a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NY) [8]. 32P-DNA/PEI complexes were formed and immobilized to the surface using the described procedures. Following immobilization, the hydrogels were placed in scintillation cocktail (5 ml, ScintiVerse II) and radioactivity measured with a scintillation counter. The measured activity was correlated to DNA mass with a standard curve. For visualization of immobilized DNA/PEI complexes, DNA was labeled with tetramethyl rhodamine using a commercially available kit (label IT TM-Rhodamine, Mirus, Madison, WI). Images of the complexes on HA after extensive washing with PBS buffer were captured using a fluorescence microscope (Leica, Bannockburn, IL) equipped with a digital camera.
2.6. DNA/cationic polymer release
The percentage of DNA/PEI complexes released from the hydrogel was determined using 32P-DNA complexes immobilized to NA bound HA-collagen hydrogels. After washing the hydrogels, PBS (pH 7.4), conditioned media (DMEM with 10% FBS from 2-day NIH/3T3 culture) or hyaluronidase (HAase, 100 U/ml) was added (200 µl) and the surfaces were incubated at 37°C in a humid chamber. The HAase concentration was chosen to obtain significant hydrogel degradation during the 8-day study. At predetermined time-points, solution (100 µl) was removed with replacement, to minimize hydrogel disruption. The activity of the collected sample was measured in a scintillation counter, with the hydrogel also measured at the final time point. The percentage of DNA released was calculated by dividing the amount released at a given time-point by the initial amount immobilized.
2.7. Cell culture and transfection
All transfection studies were performed using a N/P ratio of 5 with complexes formed in either 150 mm NaCl or water. NIH/3T3 cells, a fibroblast cell line, were used to simulate in vivo delivery to fibroblasts [11]. Cells were plated (10,000 cells/well) on the modified hydrogel substrates and cultured for 2 days at 37°C and 5% CO2 in DMEM (Invitrogen, Gaithersburg, MD) supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin. The ability of complexes released from the hydrogel to promote gene transfer was examined by placing the hydrogels in culture above NIH/3T3 cells (15,000 cells/well) in 48-well plates. The DNA-modified hydrogels were not in direct contact with the adhered cells. Additionally, cellular interactions with the immobilized DNA complex was investigated through incorporation of an RGD peptide into the complex, which increase cellular interactions with the complex through integrin receptor binding to RGD [12]. Complexes were formed with a constant amount of PEI-Biotin (25%) and varying amounts of RGD-PEI (10–75%).
The extent of transgene expression elicited by the DNA-modified surfaces was examined using the reporter gene luciferase. For measurements of luciferase activity, the cells were lysed and assayed for luciferase enzymaticac tivity (Promega, Madison, WI). The luminometer was set for a 3-s delay and an integration of the signal for 10 s. Transgene expression of luciferase is reported as RLU per number of cells seeded.
The number and distribution of transfected cells was determined with green fluorescent protein (GFP) expression and fluorescence microscopy. Complexes with 25% biotinylated PEI were immobilized onto the hydrogel surface. NIH/3T3 cells (7000 cells/well) were plated on topographically patterned-DNA modified hydrogels and cultured for 48 h. The cytoskeleton and nucleus of cells were stained with rhodamine phalloidin (1 U/200 µl) and Hoechst (1 µg/ml) stains, respectively (Molecular Probes, Eugene, OR). Five pictures per triplicate were taken with a fluorescence microscope and analyzed. The percentage of transfected cells was calculated as the ratio of GFP-positive cells to total number of nuclei (Hoechst).
3. Results
3.1. DNA/PEI complex characterization
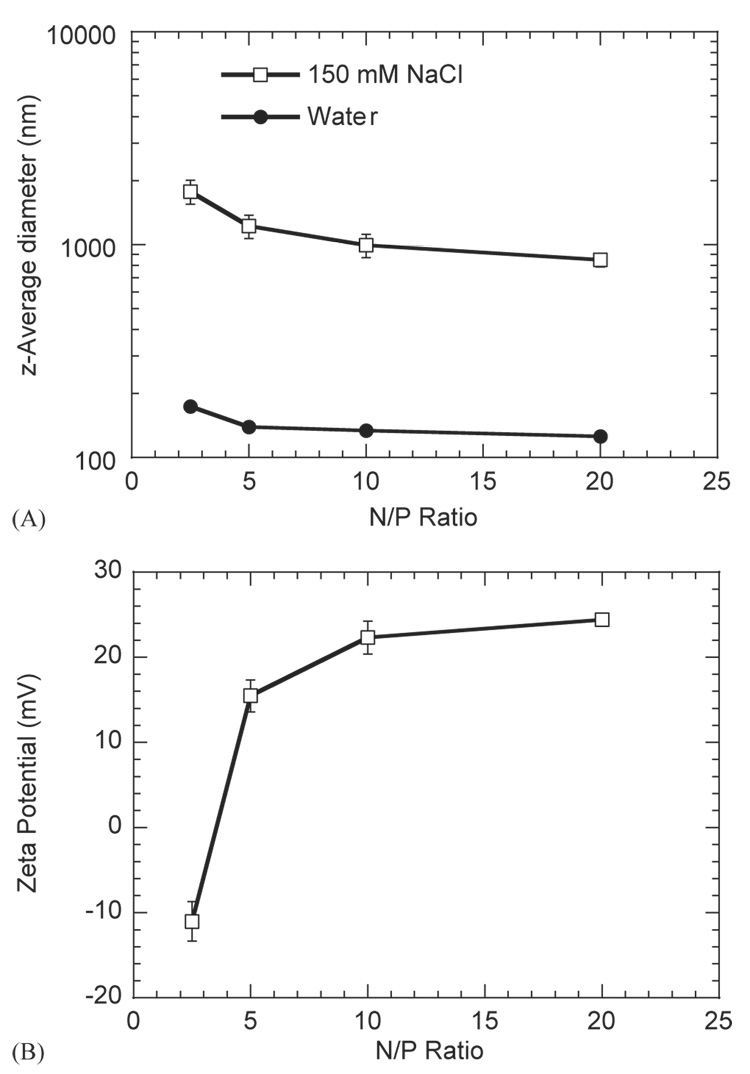
The dependence of the z-average diameter (dz) for the DNA complexes was determined at two salt concentrations (150 mm NaCl, pure water) and varying N/P ratio. dz for PEI/DNA complexes was an order of magnitude larger in the presence of salt (Fig. 1A). At an N/P ratio of 5, dz was equal to 139.4±1.3 nm in water, and 1221.7±152.3 nm in 150 mm NaCl. Additionally, complex diameter decreased with increasing N/P ratio in both 150 mm NaCl and water (Fig. 1A). For 150 mm NaCl, dz ranged from 848.9±63.6 nm (N/P=20) to 1778±227.7 nm (N/P=2.5). In water, dz ranged from 125.6±2.3 nm (N/P=20) to 173.9±2.2 nm (N/P=2.5). The zeta potential of complexes in 150 mm NaCl also increased with increasing N/P ratio with values ranging from −11.0±2.3 mV to 24.4±0.6 mV for complexes formed with 2.5 and 20 N/P ratios, respectively (Fig. 1B).
Fig. 1.
(A) z-average diameter of DNA/PEI complexes at N/P ratios of 2.5, 5, 10 and 20 for complexes formed in 150 mm NaCl or water. (B) Zeta potential of DNA/PEI complexes at N/P ratios of 2.5, 5, 10 and 20 for complexes formed in 150 mm NaCl. Plotted data is an average of triplicate conditions ± SD.
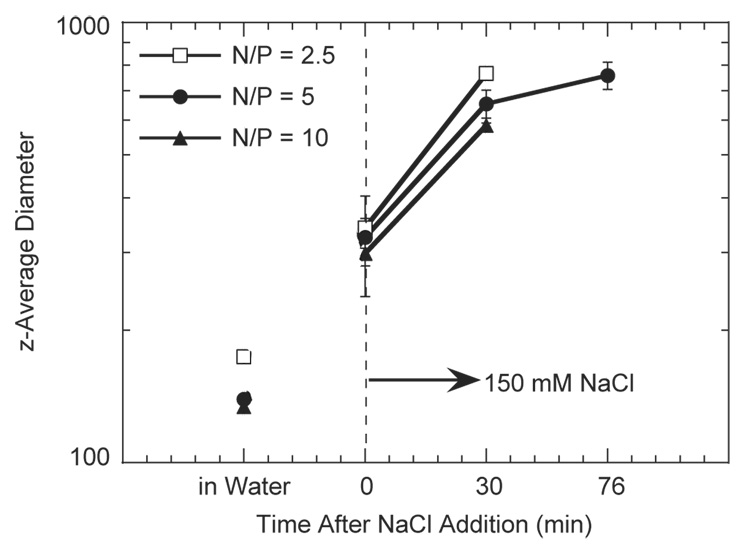
Complexes formed in the absence of salt aggregated upon addition of salt. The dz was monitored before and after salt addition for a total time of 76 min (Fig. 2). The complexes increased in size upon salt addition, with dz increasing from 139.7 ± 1.3 nm to 342.7 ± 12.8 nm immediately after salt addition for an N/P ratio of 5. PEI/DNA complex dz increased further with increased incubation time, reaching 758.7 ± 53.8 nm after 76 min of incubation. This trend of increasing complex diameter with salt and time was observed for all N/P ratios (Fig. 2).
Fig. 2.
DNA/PEI complex aggregation. Complexes were formed in water at different N/P ratios and complex size was monitored following addition of salt to the complexes. Plotted data is an average of triplicate conditions ± SD.
3.2. Surface immobilization
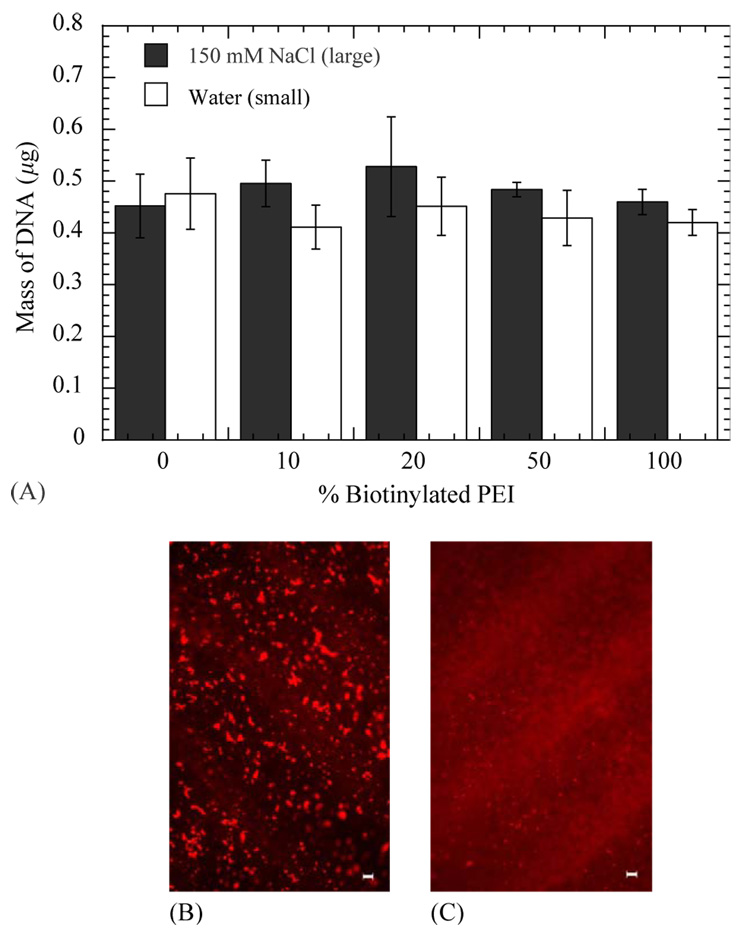
DNA immobilization to the hydrogel was independent of salt content during complex formation and the extent of biotinylation. Quantities immobilized to the hydrogels ranged from 0.411 ± 0.042 to 0.484 ± 0.014 µg DNA, with no significant difference between conditions (p > 0.05; Fig. 3A). Immobilization preserved the complex size that was observed in solution. Complexes formed in salt and deposited on the surface were visualized as discrete particles on the surface (Fig. 3B). Conversely, complexes formed in water resulted in more diffuse fluorescence without large aggregates, consistent with small particle deposition (Fig. 3C).
Fig. 3.
Surface densities of DNA/PEI complexes immobilized to HA-collagen- NA hydrogels for complexes formed in 15 0mm NaCl (B) or water (C). Plotted data is an average of triplicate conditions±SD. Fluorescence photomicrographs of immobilized biotinylated DNA/PEI complexes (N/P=5) to HA-collagen-NA hydrogels. Biotinylated complexes formed in water (B) or 150mm NaCl (C) were incubated on the substrate for 120 min. Images were captured after washing the substrate with PBS buffer.
3.3. Cellular transfection
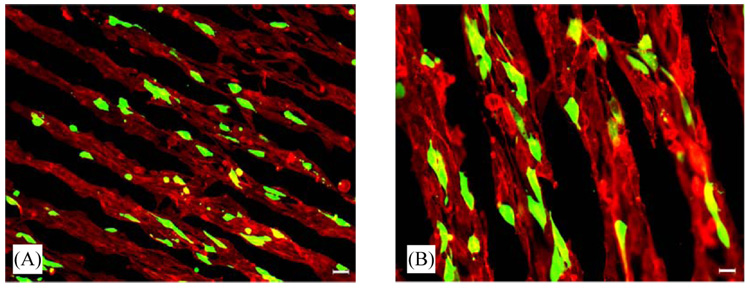
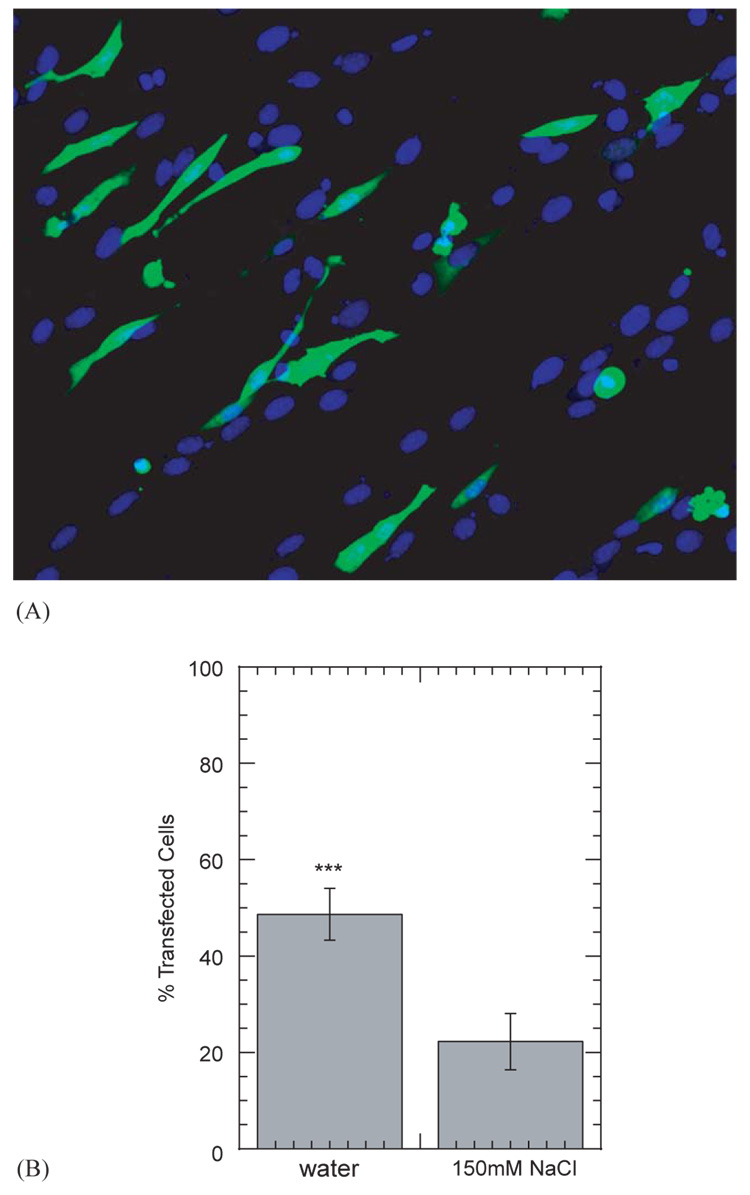
Complexes with plasmid encoding for GFP were immobilized to the topographically patterned hydrogel substrates in order to visualize the distribution of transfected cells. NIH/3T3 cells plated and cultured on HA-collagen hydrogels oriented consistently with the pattern on the hydrogel surface. Visualization of the cytoskeleton (phalloidin) by fluorescence microscopy demonstrated that the cells were aligned with the pattern. Additionally, NIH/3T3 cells cultured on the DNA-modified hydrogel substrates were found to express the transgene in this oriented conformation (Fig. 4).
Fig. 4.
Spatially controlled gene transfer of NIH/3T3 cells plated on topographically patterned HA-collagen-NA hydrogels with immobilized DNA/PEI complexes. Overlay of GFP positive cells and rhodamine phalloidin staining are shown. Magnifications correspond to 100 × (A) and 200 × (B).
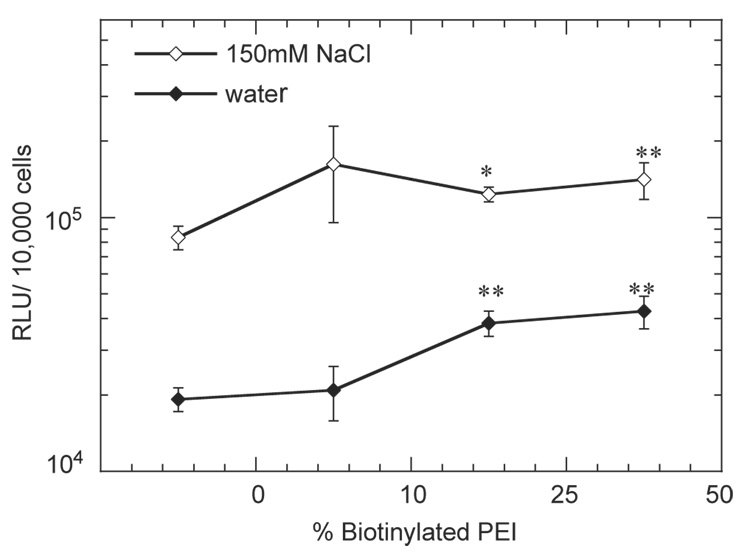
The role of complex size, which was regulated by salt content during complex formation, in gene transfer for substrate-mediated delivery was examined by measuring the extent of transgene expression and percentage of transfected cells. Complex size significantly affected transgene expression, with large diameter complexes (formed in NaCl) resulting in increased luciferase expression relative to small diameter complexes (formed in water, Fig. 5). Maximal transgene expression levels were 1.62 ± 0.67 × 105 and 4.28 ± 0.62 × 104 (RLU/10,000 cells) for the large and small-diameter complexes, respectively (Fig. 5). Furthermore, complexes formed with biotinylated PEI resulted in statistically higher luciferase expression than non-biotinylated complexes (p<0.05; Fig. 5). No difference in expression was observed for complexes formed with different extents of biotinylation (p > 0.05). The percentage of transfected cells by the different complexes was subsequently examined to determine if increased luciferase production correlated with a higher percentage of transfected cells (Fig. 6A). However, small diameter complexes resulted in transfection of 48.7 ± 5.4% of adhered cells, whereas large diameter complexes resulted in a transfection percentage of 22.3 ± 5.8% (Fig. 6B).
Fig. 5.
Transgene expression in NIH/3T3 cells mediated by immobilized complexes to HA-collagen-NA substrates. % biotinylated PEI indicate the percent biotinylation of the complexes. The symbols * and ** represents a statistically significant level of p<0.05 and <0.01; respectively, for single comparisons between biotinylated complexes and non-biotinylated complexes. Complexes were formed at an N/P ratio of 5. Plotted data is an average of triplicate conditions ± SD.
Fig. 6.
GFP transgene expression in NIH/3T3 cells mediated by immobilized complexes to HA-collagen-NA substrates. The cell nuclei are stained with HOESCHT dye for visualization (A). The percentage of transfected cells was calculated by dividing the number of GFP positive cells by the total number of nuclei (B). The symbol *** represents a statistically significant level of p<0.0001 for a single comparison between large and small complexes. Complexes were formed at an N/P ratio of 5. Plotted data is an average of triplicate conditions ± SD.
3.4. Mechanism of DNA internalization
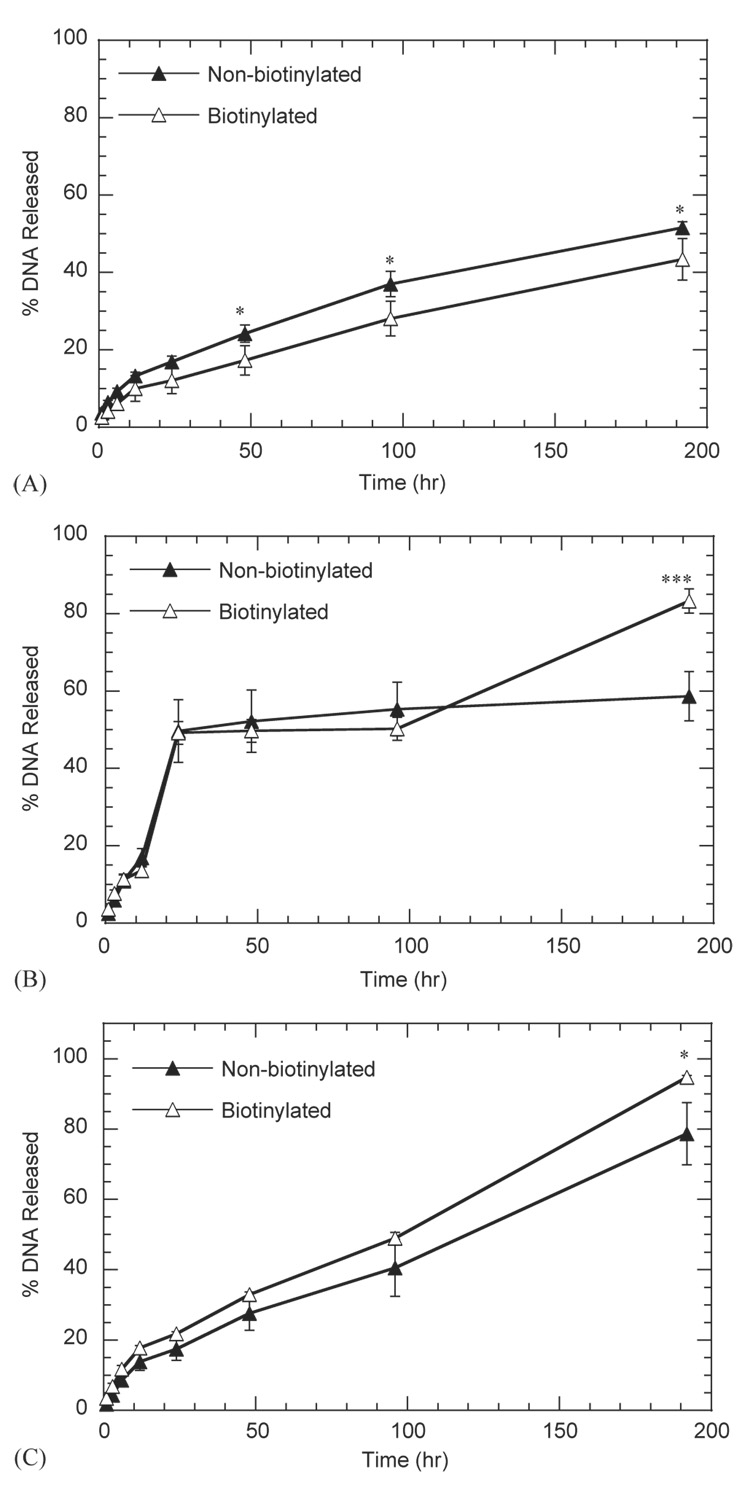
The mechanism of DNA internalization from the substrate was examined by determining release from the substrate, the ability of released complexes to transfect cells, and the role of specific cellular interactions with the complexes. Incubation of hydrogels with immobilized DNA complexes in PBS (Fig. 7A), conditioned media (Fig. 7B), and hyaluronidase (Fig. 7C) indicated that complexes were released from the substrate, with conditioned media producing the maximal release. After 24-h incubation, conditioned media induced release for approximately 50% of the immobilized DNA, which is substantially greater than release in either PBS or hyaluronidase. Release of biotinylated or non-biotinylated complexes was significantly different (p<0.05) primarily at the 8-day time point, with the greatest difference observed for release in conditioned media. After 8-day incubation in PBS, conditioned media, and hyaluronidase, the percentage of non-biotinylated complexes released was 51.6 ± 1.5, 58.7 ± 6.4, and 78.6 ± 8.8%, respectively. For biotinylated complexes, the percentage of released complexes was 43.4 ± 5.3, 83.3 ± 3.1, and 94.7 ± 0.5% for substrates incubated in PBS, conditioned media, and hyaluronidase, respectively.
Fig. 7.
Release kinetics of biotinylated and non-biotinylated complexes immobilized to HA-collagen-NA hydrogels against PBS (A), conditioned media (B) or hyaluronidase (C). Complexes containing 0% and 25% biotinylated PEI were immobilized to the hydrogel substrates. The symbols * and *** represents a statistically significant level of p<0.05 and p<0.001; respectively, for single comparisons between biotinylated complexes and non-biotinylated complexes. Plotted data is an average of triplicate conditions ± SD.
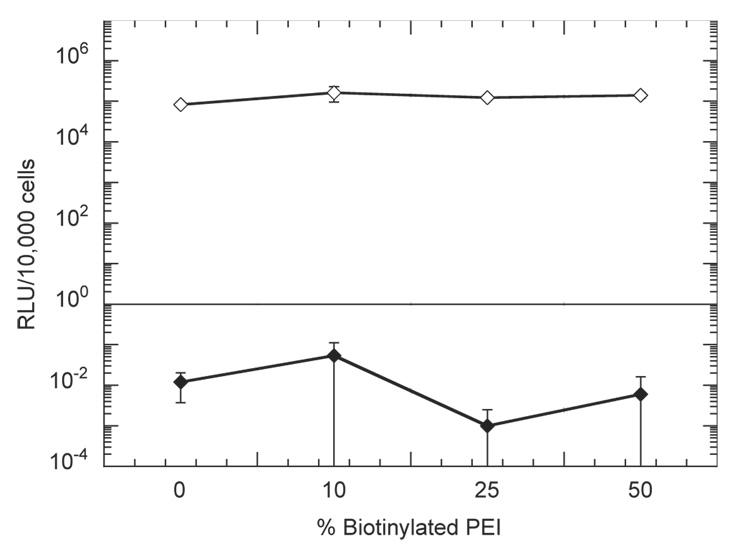
Incubation of DNA-immobilized hydrogels with NIH/3T3 cells cultured on tissue culture polystyrene was performed to assess the role of cellular adhesion to the substrate in gene transfer. Complexes released from the hydrogel did not efficiently transfect cells that were not cultured on the substrate (Fig. 8). Luciferase expression was <1 RLU/10,000 cells by incubation of hydrogels with cells cultured on polystyrene, which is significantly less than expression levels observed when cells were cultured on the hydrogel (> 105RLU/10,000 cells).
Fig. 8.
Luciferase transgene expression in NIH/3T3 cells mediated by immobilized complexes to HA-collagen-NA substrates for cells directly attached to the hydrogel (open diamonds) or not in contact with the hydrogel (closed diamonds). % biotinylated PEI indicate the percent biotinylation of the complexes. All complexes were formed at an N/P ratio of 5. Plotted data are an average of triplicate conditions ± SD.
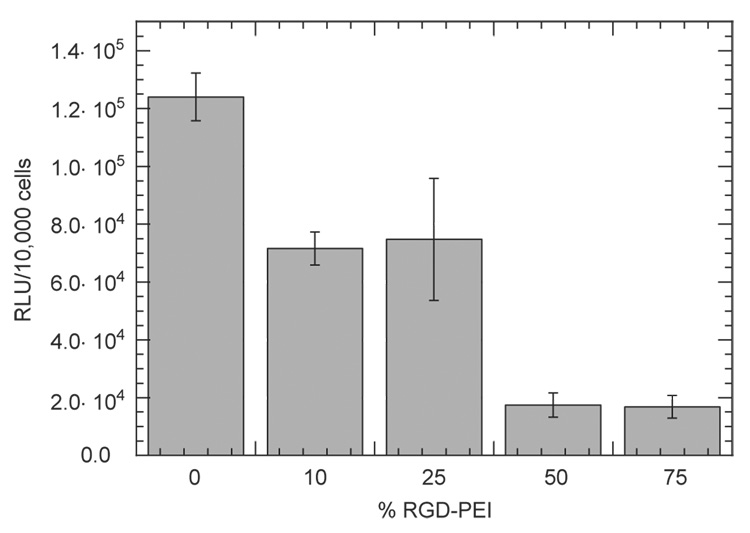
Incorporation of RGD peptides into the DNA complexes, which was hypothesized to increase cellular interactions with the complexes through RGD binding to integrin receptors, resulted in lower transgene expression than unmodified complexes (Fig. 9). For all the conditions tested, the presence of RGD in the complex decreased transgene expression relative to the absence of RGD peptides. In the absence of RGD peptides, transgene expression was 1.24 ± 0.08 × 105 RLU/10,000 cells. The presence of RGD at levels of 10% and 25% resulted in mean expression levels of approximately 7 × 104 RLU/10,000 cells. Further increases in PEI-RGD content to 50% and 75% reduced mean transgene expression to less than 2 × 104 RLU/10,000 cells.
Fig. 9.
Luciferase transgene expression in NIH/3T3 cells mediated by immobilized complexes to HA-collagen-NA substrates. % RGD-PEI indicates the percent RGD modification of the complexes. DNA/PEI complexes were formed (N/P=5) with mixtures of three types of PEI: biotinylated, RGD-modified and unmodified. The percentage of biotinylated PEI was kept constant at 25% and the percentage of RGD-PEI was varied from 0% to 75% to obtain different quantities of RGD in the complex. Plotted data is an average of triplicate conditions ± SD.
4. Discussion
Efficient and controlled gene delivery from biomaterials has the potential to enhance the applicability of gene transfer in tissue engineering. In this report, a substrate-mediated strategy was used to deliver DNA complexes from hyaluronic acid-collagen hydrogels. Hydrogels were formed by crosslinking HA with poly(ethylene glycol) diglycidyl ether, and a topography was introduced using pattern transfer. DNA/PEI complexes of varied sizes were immobilized to the hydrogel substrate using biotin/neutravidin binding and non-specific adsorption. Complexes formed in the presence of salt had a z-average diameter equal to 1221.7 ± 152.3 nm, while formation in water produced complexes with z-average diameter of 139.4±1.3 nm. Complexes immobilized to the hydrogel had sizes consistent with their size in solution, and did not aggregate. The quantity of complexes immobilized was independent of the complex size and the extent of biotinylation. DNA release from the hydrogel substrate was enhanced by incubation with conditioned media, with release at 48 h equal to 49.7 ± 8.1% and 49.2 ± 2.9% for non-biotinylated and biotinylated complexes, respectively. Transfection studies showed that transgene expression mediated by large complexes was 3- to 7-fold greater than with small complexes. However, the number of cells expressing the transgene was increased with small complexes (48.7%), relative to large complexes (22.3%). Finally, the hydrogel was patterned topographically, which guided and oriented cell attachment to the substrate and resulted in spatial patterns of transfected cells on the hydrogel.
Manipulating complex properties for substrate-mediated delivery can be employed to regulate the transfection profile (number of transfected cells, transgene expression). Complex size was modulated by forming complexes in the absence or presence of salt. Large complexes increased the extent of transgene expression relative to small complexes, whereas small complexes increased the number of transfected cells relative to large complexes. The aggregation of complexes in solution has limited the ability to correlate transfection to complex size at a specific N/P ratio [13–15]. Cationic polymer/DNA (e.g., DNA/PEI) complexes aggregate in polyelectrolyte solutions (150 mm NaCl) [16–18] and/or in solutions containing serum proteins such as albumin, fibronectin, and fibrinogen [19–22]. Strategies to stabilize complex size in solution involve steric stabilization [15,16,19,22–24], caging [18,25], and anionic polymer/DNA particles [26,27]. The increase in complex size with time in the presence of salts and/or serum proteins has been hypothesized to result from increased inter-complex interactions, which create bridges between complexes [13]. Nevertheless, transfection results obtained here are consistent with a previous report showing large complexes (600 nm) achieved up to 500 fold greater luciferase expression than small complexes (60 nm); however, the difference in the percentage of transfected cells was less substantial between large complexes and small complexes [14]. The dependence of the transfection profile on complex size has been attributed to mass transport to the cell surface, and altered endosomal release [14]. For substrate-immobilization, mass transport limitations are likely insignificant and thus the transfection profile would be affected by endosomal release. Enhanced transgene expression for the large complexes may result from an increased content of DNA and PEI per complex, which can facilitate endosomal escape and increase the probability of DNA transport to the nucleus. Furthermore, these results suggest that small complexes are more effectively internalized resulting in larger numbers of transfected cells, but are not as effectively transported through the cell and thus lower transgene expression.
In this report, immobilization of DNA complexes to the substrate occurred through non-specific adsorption and biotin–neutravidin binding. Non-specific adsorption of proteins to surfaces occurs through a variety of interactions, such as hydrophobic, electrostatic, and van der Waals [28]. The adsorption of DNA complexes likely occurs through similar mechanisms, and is hypothesized to depend on the molecular composition of the vector (e.g., cationic polymer) and substrate. Cationic polymers such as polylysine and PEI have been used to coat tissue culture plastic for in vitro studies, and PEI/DNA complexes interact with serum proteins [19–22]. Previous reports demonstrated that the presence of biotin groups in the complex resulted in substantially increased binding to the NA-coated polystyrene substrate and transgene expression compared with nonbiotinylated complexes [7,8]. In this report, biotin residues have no significant effect on binding to the substrate, while DNA release and transgene expression are only moderately enhanced. For release in conditioned media and hyaluronidase, biotinylated complexes had an increased release (≈15%) after 8 days with no significant effect at earlier times. Transgene expression, which is analyzed at two days, is enhanced by a factor of 2 or less with biotinylated complexes. These observations regarding in vitro binding, release, and transfection suggests that immobilization primarily occurs by non-specific interactions between the DNA/PEI complexes and the substrate.
Release studies of the immobilized complexes in different release mediums indicate that complexes are released during incubation, and that released complexes transfect only cells that are adhered to the substrate. Immobilization of DNA complexes occurred through complexes interacting with the substrate, which contained NA (biotinylated complexes only), HA, and collagen. Hyaluronidase treatment would release DNA by degrading the HA backbone, the primary component of the substrate. Alternatively, conditioned media contains serum proteins and other cellular products that could induce release enzymatically or through competitive protein adsorption. Although approximately 50% of DNA/PEI complexes are released in conditioned media after 48 h, gene transfer at 48 h was only observed in cells that were directly attached to the hydrogel substrate (Fig. 8). The transfection of cells cultured on the hydrogel, but not incubated with the hydrogel, likely results from an elevated concentration in the cellular microenvironment. For cells cultured with the hydrogel, released complexes may be deactivated (e.g., aggregation) prior to cellular internalization, or mass transport limitations may minimize cellular uptake [29].
The presence of RGD peptides in the DNA complex, which was hypothesized to increase cellular binding to the complexes through integrin receptors, resulted in lower transgene expression than unmodified complexes. The modification of cationic polymers with receptor ligands such as transferrin [30], RGD [12,31], and galactose [32,33], enhance binding and internalization for complexes delivered as a bolus. Reduced transgene expression with ligand-modified complexes has been seen previously for complexes with a net positive charge [12,30–33]. For substrate-mediated delivery, transgene expression decreased by an order of magnitude for complexes formed with 0% and 75% RGD-PEI, respectively. To our knowledge, this reduction in transgene expression as RGD content is increased at a fixed N/P has not been previously reported. A reduction in transgene expression with the presence of RGD peptides suggests that cellular interactions with the complex are not a factor limiting transgene expression. Reduction in transgene expression with increased RGD content may result from the RGD peptide affecting intracellular transport or reduced complex internalization as a result of multiple cellular interactions occurring per complex, which could limit internalization.
Biomaterial scaffolds that support cell adhesion and are capable of efficient gene delivery can provide a fundamental tool for localized transgene expression, which can stimulate and direct cellular processes that lead to tissue formation. The scaffold is typically biodegradable, functions to create and maintain a space for tissue formation, provides a support for cell adhesion and migration, and allows integration of the regenerating tissue with the host [34]. DNA delivery from the scaffold frequently targets infiltrating fibroblasts, which can serve as bioreactors for the localized production of tissue inductive factors [11]. Several strategies employ biomaterials to provide a sustained release of DNA or DNA complexes [35]; however, DNA complexes can also be immobilized to concentrate the DNA at the biomaterial surface and prevent distribution to non-target tissues. This report demonstrates a strategy to immobilize DNA to hydrogels for delivery to adherent cells, with the properties of the immobilized complexes regulating the transfection profile (transgene expression, number of transfected cells). The appropriate expression profile will likely depend upon the physiological system and gene of interest. For example, transplanting engineered cells that secrete low levels of vascular endothelial growth factor (VEGF) produced a mature vascular network, which was not observed by transplantation of cells secreting high levels of VEGF [36]. For genes that act intracellularly, such as tumor suppressor genes or transcription factors, the delivery system must transfect large numbers of cells [37,38].
This report also demonstrates that immobilization enables spatial patterning of gene transfer by transfecting cells that attach and orient along a topographical pattern. The engineering of tissue structures, such as the intricate networks of the vascular and nervous systems, will require methods for controlling the physical placement of molecular signals [39]. The ability to spatially regulate gene delivery on the scaffold can be employed to create spatial gradients in the expression of various tissue inductive factors (e.g., growth factors, matrix molecules), which are characteristic of many developing tissues. Opportunities for spatially controlled gene delivery include directing neurite outgrowth, or orienting cell growth for tissues such as muscle [39]. This spatial control of cell attachment and transfection can be a powerful tool with broad applicability to tissue engineering.
5. Conclusions
Efficient and controlled DNA delivery from biomaterial substrates has the potential to enhance the applicability of gene delivery to tissue engineering. We have fabricated hydrogels with the ability to transfect adherent cells using immobilized DNA complexes, which can be manipulated to alter the number of transfected cells and the extent of transgene expression. Additionally, these hydrogels can be patterned to regulate cell attachment and orient cell growth, which results in spatial patterns of transfected cells.
Acknowledgements
Support for this research was provided in part by grants from the NSF (BES-0092701, LDS), NIH (RO1 GM066830), and the Henderson Dissertation Fellowship (TS). LDS is a member of the Robert H. Lurie Comprehensive Cancer Center.
References
- 1.Kabanov AV, Kabanov VA. DNA complexes with polycations for the delivery of genetic material into cells. Bioconjug Chem. 1995;6(1):7–20. doi: 10.1021/bc00031a002. [DOI] [PubMed] [Google Scholar]
- 2.Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17(6):551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 3.Williams DA. Retroviral-bronectin interactions in transduction of mammalian cells. Ann NY Acad Sci. 1999;872:109–113. doi: 10.1111/j.1749-6632.1999.tb08457.x. [DOI] [PubMed] [Google Scholar]
- 4.Julkunen I, Vartio T, Keski-Oja J. Localization of viral-envelope-glycoprotein-binding sites in fibronectin. Biochem J. 1984;219(2):425–428. doi: 10.1042/bj2190425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy RJ, Song C, Tallapragada S, DeFelice S, Hinson JT, Vyavahare N, et al. Localized adenovirus gene delivery using antiviral IgG complexation. Gene Ther. 2001;8(9):659–667. doi: 10.1038/sj.gt.3301452. [DOI] [PubMed] [Google Scholar]
- 6.Honma K, Ochiya T, Nagahara S, Sano A, Yamamoto H, Hirai K, et al. Atelocollagen-based gene transfer in cells allows highthroughput screening of gene functions. Biochem Biophys Res Commun. 2001;289(5):1075–1081. doi: 10.1006/bbrc.2001.6133. [DOI] [PubMed] [Google Scholar]
- 7.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13(3):621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 8.Segura T, Volk MJ, Shea LD. Substrate-mediated DNA delivery: role of the cationic polymer structure and extent of modification. J Control Release. 2003;93(1):69–84. doi: 10.1016/j.jconrel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Choe MM, Sporn PH, Swartz MA. An in vitro airway wall model of remodeling. Am J Physiol Lung Cell Mol Physiol. 2003;285(2):L427. doi: 10.1152/ajplung.00005.2003. [DOI] [PubMed] [Google Scholar]
- 10.Segura T, Anderson BC, Chung PH, Webber RE, Shull KR, Shea LD. Crosslinked hyaluronicacid hydrogels: a strategy to functionalize and pattern. Biomaterials. 2004 doi: 10.1016/j.biomaterials.2004.02.067. in press. [DOI] [PubMed] [Google Scholar]
- 11.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5(7):753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 12.Kunath K, Merdan T, Hegener O, Haberlein H, Kissel T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J Gene Med. 2003;5(7):588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 13.Hagstrom JE. Self-assembling complexes for in vivo gene delivery. Curr Opin Mol Ther. 2000;2(2):143–149. [PubMed] [Google Scholar]
- 14.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5(10):1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 15.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3(3):E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goula D, Remy JS, Erbacher P, Wasowicz M, Levi G, Abdallah B, et al. Size, diffusibility and transfection performance of linear PEI/DNA complexes in the mouse central nervous system. Gene Ther. 1998;5(5):712–717. doi: 10.1038/sj.gt.3300635. [DOI] [PubMed] [Google Scholar]
- 17.Plank C, Tang MX, Wolfe AR, Szoka FC., Jr Branched cationic peptides for gene delivery: role of type and number of cationic residues in formation and in vitro activity of DNA polyplexes. Hum Gene Ther. 1999;10(2):319–332. doi: 10.1089/10430349950019101. [DOI] [PubMed] [Google Scholar]
- 18.Trubetskoy VS, Loomis A, Slattum PM, Hagstrom JE, Budker VG, Wolff JA. Caged DNA does not aggregate in high ionic strength solutions. Bioconjug Chem. 1999;10(4):624–628. doi: 10.1021/bc9801530. [DOI] [PubMed] [Google Scholar]
- 19.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 20.Lucas P, Milroy DA, Thomas BJ, Moss SH, Pouton CW. Pharmaceutical and biological properties of poly(amino acid)/DNA polyplexes. J Drug Target. 1999;7(2):143–156. doi: 10.3109/10611869909085498. [DOI] [PubMed] [Google Scholar]
- 21.Oupicky D, Howard KA, Konak C, Dash PR, Ulbrich K, Seymour LW. Steric stabilization of poly L-Lysine/DNA complexes by the covalent attachment of semitelechelic poly[N-(2-hydroxypropyl)methacrylamide] Bioconjug Chem. 2000;11(4):492–501. doi: 10.1021/bc990143e. [DOI] [PubMed] [Google Scholar]
- 22.Xu B, Wiehle S, Roth JA, Cristiano RJ. The contribution of poly L-lysine, epidermal growth factor and streptavidin to EGF/PLL/DNA polyplex formation. Gene Ther. 1998;5(9):1235–1243. doi: 10.1038/sj.gt.3300719. [DOI] [PubMed] [Google Scholar]
- 23.Wolfert MA, Dash PR, Nazarova O, Oupicky D, Seymour LW, Smart S, et al. Polyelectrolyte vectors for gene delivery: influence of cationic polymer on biophysical properties of complexes formed with DNA. Bioconjug Chem. 1999;10(6):993–1004. doi: 10.1021/bc990025r. [DOI] [PubMed] [Google Scholar]
- 24.Toncheva V, Wolfert MA, Dash PR, Oupicky D, Ulbrich K, Seymour LW, et al. Novel vectors for gene delivery formed by self-assembly of DNA with poly(L-lysine) grafted with hydrophilic polymers. Biochim Biophys Acta. 1998;1380(3):354–368. doi: 10.1016/s0304-4165(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 25.Adami RC, Rice KG. Metabolic stability of glutaraldehyde cross-linked peptide DNA condensates. J Pharm Sci. 1999;88(8):739–746. doi: 10.1021/js990042p. [DOI] [PubMed] [Google Scholar]
- 26.Trubetskoy VS, Budker VG, Hanson LJ, Slattum PM, Wolff JA, Hagstrom JE. Self-assembly of DNA-polymer complexes using template polymerization. Nucleic Acids Res. 1998;26(18):4178–4185. doi: 10.1093/nar/26.18.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trubetskoy VS, Loomis A, Hagstrom JE, Budker VG, Wolff JA. Layer-by-layer deposition of oppositely charged polyelectrolytes on the surface of condensed DNA particles. Nucleic Acids Res. 1999;27(15):3090–3095. doi: 10.1093/nar/27.15.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norde W, Lyklema J. Why proteins prefer interfaces. J Biomater Sci Polym Ed. 1991;2(3):183–202. doi: 10.1080/09205063.1991.9756659. [DOI] [PubMed] [Google Scholar]
- 29.Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18(8):893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 30.Kircheis R, Kichler A, Wallner G, Kursa M, Ogris M, Felzmann T, et al. Coupling of cell-binding ligands to polyethylenimine for targeted gene delivery. Gene Ther. 1997;4(5):409–418. doi: 10.1038/sj.gt.3300418. [DOI] [PubMed] [Google Scholar]
- 31.Harbottle RP, Cooper RG, Hart SL, Ladhoff A, McKay T, Knight AM, et al. An RGD-oligolysine peptide: a prototype construct for integrin-mediated gene delivery. Hum Gene Ther. 1998;9(7):1037–1047. doi: 10.1089/hum.1998.9.7-1037. [DOI] [PubMed] [Google Scholar]
- 32.Plank C, Zatloukal K, Cotten M, Mechtler K, Wagner E. Gene transfer into hepatocytes using asialoglycoprotein receptor mediated endocytosis of DNA complexed with an artificial tetra-antennary galactose ligand. Bioconjug Chem. 1992;3(6):533–539. doi: 10.1021/bc00018a012. [DOI] [PubMed] [Google Scholar]
- 33.Kunath K, von Harpe A, Fischer D, Kissel T. Galactose-PEI-DNA complexes for targeted gene delivery: degree of substitution affects complex size and transfection efficiency. J Control Release. 2003;88(1):159–172. doi: 10.1016/s0168-3659(02)00458-3. [DOI] [PubMed] [Google Scholar]
- 34.Murphy WL, Mooney DJ. Controlled delivery of inductive proteins, plasmid DNA and cells from tissue engineering matrices. J Periodontal Res. 1999;34(7):413–419. doi: 10.1111/j.1600-0765.1999.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 35.Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol Ther. doi: 10.1016/j.ymthe.2004.03.020. in press. [DOI] [PubMed] [Google Scholar]
- 36.Ozawa CR, Banfi A, Glazer NL, Thurston G, Springer ML, Kraft PE, et al. Microenvironmental VEGF concentration, not total dose, determines a threshold between normal and aberrant angiogenesis. J Clin Invest. 2004;113(4):516–527. doi: 10.1172/JCI18420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braddock M. The transcription factor Egr-1: a potential drug in wound healing and tissue repair. Ann Med. 2001;33(5):313–318. doi: 10.3109/07853890109002083. [DOI] [PubMed] [Google Scholar]
- 38.Scholl SM, Michaelis S, McDermott R. Gene therapy applications to cancer treatment. J Biomed Biotechnol. 2003;2003(1):35–47. doi: 10.1155/S1110724303209037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saltzman WM, Olbricht WL. Building drug delivery into tissue engineering. Nat Rev Drug Discov. 2002;1(3):177–186. doi: 10.1038/nrd744. [DOI] [PubMed] [Google Scholar]